Abstract
Primary cultures of human hepatocytes were used to investigate whether the dietary isothiocyanates, sulforaphane (SFN), and phenethyl isothiocyanate (PEITC) can reduce DNA adduct formation of the hepatocarcinogen aflatoxin B1 (AFB). Following 48 h of pretreatment, 10 and 50μM SFN greatly decreased AFB-DNA adduct levels, whereas 25μM PEITC decreased AFB-DNA adducts in some but not all hepatocyte preparations. Microarray and quantitative reverse transcriptase (RT)-PCR analyses of gene expression in SFN and PEITC-treated hepatocytes demonstrated that SFN greatly decreased cytochrome P450 (CYP) 3A4 mRNA but did not induce the expression of either glutathione S-transferase (GST) M1 or GSTT1. The protective effects of SFN required pretreatment; cotreatment of hepatocytes with SFN and AFB in the absence of pretreatment had no effect on AFB-DNA adduct formation. When AFB-DNA adduct formation was evaluated by GST genotype, the presence of one or two functional alleles of GSTM1 was associated with a 75% reduction in AFB-DNA adducts, compared with GSTM1 null. In conclusion, these results demonstrate that the inhibition of AFB-DNA adduct formation by SFN is dependent on changes in gene expression rather than direct inhibition of catalytic activity. Transcriptional repression of genes involved in AFB bioactivation (CYP3A4 and CYP1A2), but not transcriptional activation of GSTs, may be responsible for the protective effects of SFN. Although GSTM1 expression was not induced by SFN, the presence of a functional GSTM1 allele can afford substantial protection against AFB-DNA damage in human liver. The downregulation of CYP3A4 by SFN may have important implications for drug interactions.
Keywords: phenethyl isothiocyanate, sulforaphane, aflatoxin B1, glutathione S-transferase, CYP3A4
Numerous studies in laboratory animals and limited human epidemiological data suggest that a variety of plant-derived compounds (phytochemicals) can lower the risk for certain types of cancer (Pan and Ho, 2008) although the precise mechanisms for such putative chemoprotective effects in humans remain uncertain.
Cruciferous vegetables such as broccoli, cabbage, cauliflower, Brussels sprouts, turnip, and watercress contain the naturally occurring glucosinolate compounds, gluconasturtiin and glucoraphanin (reviewed by Stoewsand [1995]). Once the vegetables are mechanically damaged, for example, during chewing, the biologically active compounds, phenethyl isothiocyanate (PEITC; released from gluconasturtiin) and sulforaphane (SFN; released from glucoraphanin), are released from the glucosinolates by the plant-derived enzyme myrosinase, and further by intestinal microbial myrosinases, albeit to a lower extent (Rungapamestry et al., 2007).
SFN and PEITC protect animals against experimentally induced tumors from a variety of chemical carcinogens (Morse et al., 1991; Zhang et al., 1994). Furthermore, numerous in vitro studies using various mammalian or human models have demonstrated protective effects of SFN and PEITC toward (geno)toxicological end points of several carcinogens/mutagens that require metabolic activation to exert their genotoxic effects (reviewed by Pan and Ho [2008]). Suggested mechanisms for the chemoprotective effects of isothiocyanates include inhibition of bioactivation of procarcinogens and enhanced detoxification of reactive intermediates (Hayes et al., 2008; Juge et al., 2007; Pan and Ho, 2008), modulation of oxidative stress (Kwak et al., 2001), alterations in cell cycle regulation or apoptosis (Gamet-Payrastre et al., 2000), changes in histone acetylation (Myzak et al., 2004), and inhibition of angiogenesis (Bertl et al., 2006).
SFN and/or PEITC may act as modulators of expression and/or catalytic activity of phase I and II biotransformation enzymes that play key roles in the bioactivation of the hepatocarcinogenic mycotoxin aflatoxin B1 (AFB). In particular, cytochromes P450 (CYPs) 1A2 and 3A4/5 are involved in the activation of AFB to the genotoxic aflatoxin B1-8,9-oxide (AFBO) (Eaton and Gallagher, 1994; Gallagher et al., 1996; Guengerich et al., 1998). However, the highly reactive, genotoxic AFBO may be detoxified by glutathione S-transferases (GSTs). Certain alpha-class GSTs in rats (rGSTA5-5) and mice (mGSTA3-3) are highly effective in detoxifying AFBO (Eaton and Gallagher, 1994; Gallagher et al., 1996) and are inducible by diet (Hayes et al., 1998). In contrast, human alpha-class GSTs (hGSTA1, hGSTA2) and other nonhuman primate alpha-class GSTs (Eaton et al., 2001; Wang et al., 2002) lack any measurable activity toward AFBO. However, in the absence of a high-activity alpha-class GST, the low but measurable activity of hGSTM1 may afford some protection against AFBO (Chen et al., 2000; Deng et al., 2005; Guengerich et al., 1998; Long et al., 2005; Kirk et al., 2005; London et al., 1995; Sun et al., 2001). Alternatively, human microsomal epoxide hydrolase (mEH) may also participate in the detoxification of AFBO in the absence of significant GST activity (Dash et al., 2007; Eaton et al., 2001; Kelly et al., 2002; Kirk et al., 2005; London et al., 1995; McGlynn et al., 2003). A large body of evidence indicates that isothiocyanates act as modulators of gene expression and/or catalytic activities of enzymes involved in AFB activation and detoxification. However, few studies have utilized primary human liver cells to evaluate the effects of isothiocyanates on AFB-induced genotoxicity. Using a human-derived test system is important because there are large species differences in susceptibility toward the genotoxic effects of AFB, probably as a result of wide variation in expression, regulation, and substrate specificity of enzymes involved in AFB biotransformation (reviewed by Eaton et al. [2001]). Indeed, a previous study in our laboratory demonstrated that effects of these phytochemicals on gene expression in primary human hepatocytes are quite different from those observed in rodent models or human tumor-derived cell lines (e.g., HepG2 cells) (Gross-Steinmeyer et al., 2004). Thus, the present study utilized human hepatocytes in primary culture to evaluate the putative chemoprotective effects of SFN and PEITC on AFB-DNA adduct formation and to elucidate potential mechanisms underlying the isothiocyanate-mediated modulation of AFB-induced genotoxicity.
MATERIALS AND METHODS
Isothiocyanates and AFB.
PEITC was purchased from Sigma-Aldrich and L-sulforaphane (SFN) was purchased from LKT Laboratories. Both isothiocyanates were of > 99% purity as determined via high pressure liquid chromatography. 3H-AFB was obtained from Moravek Biochemicals. Specific activities of different 3H-AFB product lots ranged from 16 to 28 mCi/mmol.
Preparation, culturing, and treatment of primary human hepatocytes.
Human hepatocytes were isolated from viable human livers that were rejected for transplantation for various reasons. All human subjects’ protocols were reviewed and approved by the University of Washington and the University of Pittsburgh Institutional Review Boards. Hepatocyte isolation was performed as described previously (Strom et al., 1996). Hepatocyte preparations from 16 liver donors were used for this study (liver identification numbers: 970, 985, 987, 990, 1002, 1046, 1072, 1076, 1087, 1095, 1105, 1125, 1165, 1183, 1407, 1410). Hepatocyte cultures were maintained at 37°C under 5% CO2/95% humidified air on a rigid collagen substratum overlaid with Matrigel (Collaborative Biochemicals) in supplemented William's E media as described previously; Matrigel overlay facilitates the maintenance and induction of xenobiotic metabolizing enzymes in primary human hepatocytes (Gross-Steinmeyer et al., 2005). Following a minimum 48-h recovery period, hepatocytes were treated with isothiocyanates (10 or 50μM SFN and 10 or 25μM PEITC) or vehicle, and different end points were evaluated following treatments, as outlined below. During the treatment periods, media containing test compounds was changed after 24 h. Our criteria for selecting the high concentration of each phytochemical were (1) no adverse cytotoxic effects over the exposure period compared with the vehicle-only controls, as measured by lactate dehydrogenase leakage using a commercially available test kit (Promega), (2) maximizing induction effect on CYP1A1 or CYP1A2 as described previously (Gross-Steinmeyer et al., 2004), and (3) maximizing the detection of AFB-DNA adducts. One additional lower concentration was chosen for each compound (Gross-Steinmeyer et al., 2004). Hepatocytes from each individual preparation were genotyped for GSTM1 and GSTT1 gene loci as described previously (Gross-Steinmeyer et al., 2005).
Culture conditions used to distinguish between transcriptional and direct effects of SFN on enzyme activity.
Hepatocytes obtained from three human livers (#1125, 1165, and 1183) were investigated under the following three treatment conditions to discriminate between transcriptional and direct effects of SFN on enzyme activity: (1) following a 48-h pretreatment with 10μM SFN, media was removed, cells were rinsed with PBS, and incubated for 6 h in media containing 3H-AFB but no SFN; (2) following a 48-h pretreatment with 10μM SFN, media was removed, cells were rinsed with PBS, and incubated for an additional 6 h in media containing both 3H-AFB and SFN; (3) following a 48-h treatment with vehicle only (no SFN), media was removed, cells were rinsed with PBS, and incubated for an additional 6 h in media containing both 3H-AFB and 10μM SFN. Condition 1 is designed to detect transcriptional effects, condition 3 detects potential enzyme inhibition effects, and condition 2 detects the overall effect of both. Conditions 1, 2, and 3 were compared with an analogous treatment without SFN application, which served as control (equal to 100%). The control cells were treated for 48 h with vehicle only (no SFN), media was removed, cells were rinsed with PBS, and incubated for an additional 6 h in media containing 3H-AFB only (no SFN). The vehicle (DMSO) concentration in all treatment conditions was identical. AFB-DNA adduct levels were determined as described below.
Measuring AFB-DNA adducts.
Hepatocytes were pretreated for 48 h with SFN (10 or 50μM) or PEITC (10 or 25μM; 50μM PEITC was cytotoxic) followed by a 6-h coincubation of isothiocyanate and 3H-AFB. The final 3H-AFB concentration ranged from 0.2 to 0.4μM, depending on the specific activity of the particular 3H-AFB product lot, to provide sufficient sensitivity for adduct determination. Previous experiments with human hepatocytes that used 0, 0.1, 0.2, 0.3, and 0.4μM 3H-AFB and otherwise the same experimental conditions demonstrated highly linear (R2 = 0.99) correlation between AFB dose and AFB-DNA adduct levels (Gross-Steinmeyer et al., 2009). The AFB concentrations used in our study fell within the aforementioned tested concentration range.
The DMSO (vehicle) concentration was 0.1% in 48-h isothiocyanate pretreatments and 0.2% during subsequent 6-h coincubations with isothiocyanate and 3H-AFB. Harvesting of hepatocytes and DNA isolation were performed using the Qiagen Genomic DNA Purification Kit (Qiagen) according to the manufacturer's recommendation. DNA concentrations were determined as described previously (Gross-Steinmeyer et al., 2009). Liquid scintillation counting was used to quantify covalently bound 3H-AFB. A subset of cells was heated to 95°C for 5 min to inactivate biotransformation enzymes prior to AFB exposure but otherwise treated identically to nonheated samples. The corresponding data were used to correct for noncovalent binding of 3H-AFB. In addition, DNA was isolated in the same manner from hepatocytes not treated with 3H-AFB to correct for background (non-3H-AFB) radioactivity. Final 3H-AFB-DNA adduct levels were calculated in units of (fmol adduct/100 × μmol DNA) which is equivalent to (adducts per 107 nucleotides). A limit of detection was estimated to be 10 adducts per 109 nucleotides, based on cpm measurements that were three times the background levels. In cases where the adduct level was below this limit of detection, a value of five adducts per 109 nucleotides (one-half the limit of detection) was used to calculate mean values.
TaqMan-based reverse transcriptase-PCR analysis.
mRNA expression of human CYPs 1A1, 1A2, 3A4, and 3A5, mEH, GSTM1, and GSTT1 was determined by TaqMan-based reverse transcriptase (RT)-PCR analysis in hepatocytes obtained from eight donors (#970, 985, 987, 990, 1002, 1087, 1095, and 1105). At 0 and 48 h of exposure, the hepatocytes were lysed on the plates, and total RNA was extracted using Trizol reagent (Invitrogen) as recommended by the supplier. The quality of the RNA preparations was assessed electrophoretically via 18S and 28S band intensities. Reverse transcription of 0.1–2 μg total RNA using oligod(T)15 primer and Superscript II RNaseH (GIBCO) was performed as described previously (Gross-Steinmeyer et al., 2009). All gene-specific sequences of primer pairs and probes used in the TaqMan assays have been published previously (Gross-Steinmeyer et al., 2004, 2005, 2009).
Microarray analysis.
Four preparations of primary human hepatocytes (970, 985, 987, 1002) that were of high quality as judged by cell morphology and viability were utilized for array analysis. Hepatocytes were treated for 48 h with SFN (10 or 50μM) or PEITC (10 or 25μM) under the same conditions used during the pretreatment period in the genotoxicity study and TaqMan-based RT-PCR analyses. The commercially available oligonucleotide array platform, CodeLink (GE Healthcare), with approximately 20,000 human genes was used in this study. Oligo arrays assess changes in gene expression of specific members of closely related genes. Given the considerable heterogeneity of the human population, each experiment was performed with hepatocytes from the same liver, that is, each liver served as its own control. Microarray data for only a few selected genes are presented in this article. The microarray data have been deposited with National Center for Biotechnology Information's Gene Expression Omnibus database (accession number GSE20479).
Human CYP1A2 and CYP3A4 inhibition assays.
Methoxyresorufin O-demethylase (MROD) and benzyloxyresorufin O-debenzylase (BROD) assays were conducted utilizing microsomes isolated from a recombinant yeast strain expressing human CYP1A2, commercial Supersomes expressing human CYP3A4 (Gentest), and human liver microsomes. The recombinant yeast strain was derived from the parental Saccharomyces cerevisiae strain yHE2, and yeast microsomes were prepared as previously described (Eugster and Sengstag, 1993). Human liver microsomes derived from viable human livers that were rejected for transplantation were prepared according to a standard protocol. Standard enzyme activity assay protocols using the yeast microsomes and human liver microsomes as well as Supersomes were performed as described previously (Gross-Steinmeyer et al., 2004). A concentration range of 0.15–50μM isothiocyanate was assessed for inhibitory effects and compared with the noninhibited (vehicle) control, as well as to corresponding negative controls. Two independent experiments were performed, and six measurements were taken per condition within each experiment.
Statistical analyses.
Statistical comparisons of treatments versus controls (e.g., modulated DNA adduct levels, modulated transcriptional expression, enzyme inhibition) were determined by one-way ANOVA using Dunnett's posttest. Other comparisons (e.g., genotyped groups) were evaluated by unpaired Student's t-test with equal variances.
Microarray data were normalized using the quantile normalization method (Bolstad et al., 2003) on probes of type Discovery (see Gene Expression Omnibus platform GPL1313 for details of probe types). Probes with normalized intensity values less than 70 were removed from further analysis. To assess the pairwise differences between concentrations, a modified t-test for microarray analysis was performed (Storey and Tibshirani, 2003). Statistical significance was determined by randomly permuting the microarray labels, recomputing the t statistics, calculating p values based on a pooled null distribution, and adjusting the p values for multiple hypotheses testing (Storey and Tibshirani, 2003). Our microarray analysis used the 48-h control as the baseline. Genes with p < 0.05 and a greater than 1.5-fold change (up or down) in expression were selected.
RESULTS
Effects of Isothiocyanates on AFB-DNA Adduct Formation
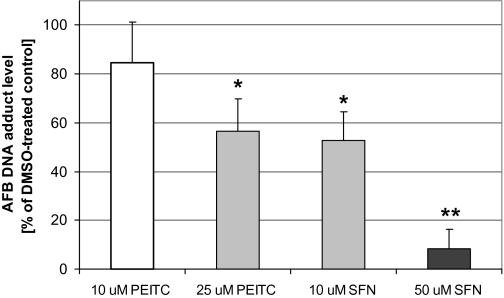
Primary cultures of isolated human hepatocytes were treated with PEITC (10 or 25μM) or SFN (10 or 50μM) for 48 h and subsequently coincubated for 6 h with 3H-AFB and isothiocyanates. No significant cytotoxicity was observed at these concentrations, as measured by lactate dehydrogenase release. Perhaps not surprisingly, human hepatocytes seem to be less sensitive to the cytotoxic effects of SFN than immortalized cell lines such as HepG2 or Hepa1c1c7 cells (Gerhauser et al., 1997), perhaps, because of more efficient glutathione conjugation. The AFB-DNA adduct level in vehicle-treated control cells averaged 5.9 adducts per 107 nucleotides (n = 6 hepatocyte preparations), ranging from 2.2 to 10.7 adducts per 107 nucleotides. PEITC and SFN treatments significantly reduced DNA adduct formation (Fig. 1). SFN treatment was highly effective in reducing AFB-mediated genotoxicity; at 10 and 50μM SFN, AFB-DNA adducts were reduced on average by 47 and 92%, respectively. In five of six hepatocyte preparations pretreated with 50μM SFN, the AFB-DNA levels were below the limit of detection (∼10 adducts per 109 nucleotides); thus, a value of one-half the limit of detection (5 adducts per 109 nucleotides) was used to determine the average. Therefore, the actual extent of reduction in AFB-DNA adducts by 50μM SFN was greater than 92% in most of the hepatocyte preparations. The protective effects of PEITC on AFB-DNA adduct formation showed a large interindividual variation. The average AFB-DNA adduct decrease mediated by 10 and 25μM PEITC was 15 and 44%, respectively; however, in one hepatocyte preparation, AFB-DNA adduct formation was reduced by 93% at 25μM PEITC.
FIG. 1.
Effects of SFN and PEITC on AFB-DNA adduct formation in human hepatocytes. Hepatocytes were treated with two concentrations of each phytochemical for 48 h and subsequently coincubated with 0.4μM 3H-AFB and phytochemical. Control represents nonmodulated AFB-DNA adduct level, where cells were treated with vehicle instead of phytochemical. AFB-DNA adduct levels are expressed as percentages of control and represent means and SEs from six independent experiments (i.e., hepatocytes from six donors). The 100% control value for six hepatocyte preparations was 5.9 adducts per 107 nucleotides (range 2.2–10.7 adducts per 107 nucleotides). “*” Denotes 0.01 < p < 0.05; “**” denotes p < 0.01 comparing vehicle control (100%) versus phytochemical incubations.
Effects of Isothiocyanates on Expression of Genes Involved in AFB Biotransformation and DNA Repair
We carried out microarray analysis of hepatocytes treated with SFN (10 or 50μM) or PEITC (10 or 25μM) for 48 h to assess their effect on global transcription. Because the number of cells from each donor was limited, not all treatment conditions could be performed on the cells from the same donors. Thus, although hepatocytes from a total of four donors (970, 982, 985, 1002) were used, only hepatocytes from three donors were used for each of the SFN (982, 985, 1002) and PEITC (970, 985, 1002) treatments. Microarray results for genes involved in the biotransformation of AFB and DNA repair and genes with a known functional antioxidant response element are shown in Table 1.
TABLE 1.
Effect of SFN and PEITC on Expression of AFB and ARE Relevant Genes by Microarray Analysis
| NCBI_ACCN | Symbol | SFN 10μMa |
SFN 50μMa |
PEITC 10μMa |
PEITC 25μMa |
||||||||
| Fold ch | p Value | SD | Fold ch | p Value | SD | Fold ch | p Value | SD | Fold ch | p Value | SD | ||
| AFB biotransformation DNA repair | |||||||||||||
| NM_003689 | AKR7A2 | 0.8 | 0.1205 | 0.0374 | 0.7 | 0.0477 | 0.0345 | 0.9 | 0.5301 | 0.0599 | 1.0 | 0.6548 | 0.1174 |
| NM_012067 | AKR7A3 | 0.5 | 0.0017 | 0.0316 | 0.4 | 0.0069 | 0.0580 | 0.5 | 0.0009 | 0.0363 | 0.5 | 0.0063 | 0.0753 |
| NM_000499 | CYP1A1 | 1.5 | 0.0102 | 0.0567 | 1.5 | 0.0531 | 0.1207 | 1.5 | 0.0068 | 0.0354 | 1.8 | 0.0031 | 0.0567 |
| NM_000761 | CYP1A2 | 1.3 | 0.0808 | 0.1345 | 1.3 | 0.1098 | 0.0197 | 1.2 | 0.2627 | 0.1551 | 1.3 | 0.1094 | 0.1841 |
| NM_017460 | CYP3A4 | 0.8 | 0.2164 | 0.2840 | 0.1 | 0.0014 | 0.0397 | 1.2 | 0.2391 | 0.1584 | 1.1 | 0.5894 | 0.1556 |
| NM_000777 | CYP3A5 | 1.1 | 0.8679 | 0.2364 | 0.6 | 0.0808 | 0.1375 | 0.9 | 0.2319 | 0.2814 | 1.1 | 0.7334 | 0.5546 |
| NM_001923 | DDB1 | 0.7 | 0.0857 | 0.2069 | 0.9 | 0.4309 | 0.1641 | 0.6 | 0.0213 | 0.1872 | 0.7 | 0.0787 | 0.2607 |
| NM_000107 | DDB2 | 1.3 | 0.1180 | 0.1040 | 0.9 | 0.4908 | 0.1765 | 1.2 | 0.2222 | 0.1042 | 1.2 | 0.1381 | 0.0747 |
| AF253417 | EPHX1 | 0.8 | 0.1500 | 0.0379 | 0.7 | 0.0375 | 0.0121 | 0.9 | 0.2147 | 0.0104 | 0.8 | 0.0517 | 0.0215 |
| NM_001983 | ERCC1 | 1.1 | 0.5120 | 0.1633 | 2.0 | 0.0605 | 0.5057 | 1.0 | 0.8500 | 0.1018 | 1.2 | 0.2611 | 0.1824 |
| NM_000400 | ERCC2 | 1.2 | 0.2510 | 0.0582 | 1.2 | 0.2642 | 0.0375 | 1.1 | 0.2866 | 0.0760 | 1.3 | 0.0988 | 0.0856 |
| NM_000122 | ERCC3 | 1.4 | 0.0312 | 0.0333 | 1.4 | 0.1197 | 0.1935 | 1.3 | 0.1679 | 0.1636 | 1.3 | 0.0907 | 0.0573 |
| L76568 | ERCC4 | 1.5 | 0.0376 | 0.1595 | 1.5 | 0.1805 | 0.2329 | 1.5 | 0.0127 | 0.1458 | 1.5 | 0.0067 | 0.1327 |
| NM_000123 | ERCC5 | 2.0 | 0.0025 | 0.2080 | 1.4 | 0.1654 | 0.2198 | 1.7 | 0.0155 | 0.2357 | 1.8 | 0.0098 | 0.2048 |
| NM_000124 | ERCC6 | 1.0 | 0.8000 | 0.0960 | 1.6 | 0.0543 | 0.1927 | 1.0 | 0.6800 | 0.0915 | 1.1 | 0.68 | 0.1591 |
| NM_145740 | GSTA1 | 1.4 | 0.0687 | 0.1794 | 1.2 | 0.8280 | 0.3438 | 1.2 | 0.1625 | 0.0897 | 1.5 | 0.0893 | 0.3805 |
| NM_000561 | GSTM1b | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NM_000853 | GSTT1b | 0.7 | 0.0193 | 0.0224 | 0.6 | 0.1263 | 0.0489 | 0.7 | 0.7638 | 0.0500 | 0.7 | 1 | 0.0508 |
| NM_001513 | GSTZ1 | 0.8 | 0.1252 | 0.0625 | 0.5 | 0.0299 | 0.0920 | 0.8 | 0.1435 | 0.0721 | 0.9 | 0.4153 | 0.0815 |
| NM_006502 | POLH | 1.2 | 0.1031 | 0.0387 | 2.2 | 0.0109 | 0.2821 | 1.2 | 0.0980 | 0.0325 | 1.3 | 0.0440 | 0.0574 |
| NM_005053 | RAD23A | 0.7 | 0.0216 | 0.0305 | 0.9 | 0.6662 | 0.0587 | 0.6 | 0.0041 | 0.0176 | 0.6 | 0.0171 | 0.0591 |
| NM_000380 | XPA | 0.9 | 0.4798 | 0.1762 | 0.7 | 0.1604 | 0.1394 | 0.9 | 0.5303 | 0.1578 | 1.0 | 0.6250 | 0.2073 |
| NM_004628 | XPC | 1.2 | 0.1715 | 0.0706 | 1.3 | 0.2155 | 0.1263 | 1.2 | 0.1427 | 0.0949 | 1.1 | 0.5737 | 0.1057 |
| ARE regulated genes | |||||||||||||
| NM_001498 | GCLC | 1.7 | 0.0178 | 0.2121 | 3.0 | 0.0019 | 0.1283 | 1.6 | 0.0374 | 0.3127 | 1.8 | 0.0213 | 0.3053 |
| NM_002061 | GCLM | 1.3 | 0.1083 | 0.1278 | 2.3 | 0.0060 | 0.1980 | 0.9 | 0.3982 | 0.0803 | 1.2 | 0.2683 | 0.1185 |
| NM_000852 | GSTP1 | 0.8 | 0.0420 | 0.0276 | 0.6 | 0.0180 | 0.0361 | 0.8 | 0.0383 | 0.0223 | 0.9 | 0.2588 | 0.1129 |
| NM_002133 | HMOX1 | 0.8 | 0.1103 | 0.1918 | 1.0 | 0.6805 | 0.2356 | 0.8 | 0.1946 | 0.2456 | 1.1 | 0.6798 | 0.4070 |
| NM_000903 | NQO1 | 2.8 | 0.0001 | 0.1149 | 2.4 | 0.0288 | 0.5142 | 2.3 | 0.0006 | 0.1931 | 6.6 | 0.0000c | 1.6774 |
| NM_003329 | TXN | 1.7 | 0.0037 | 0.0413 | 1.8 | 0.0321 | 0.2188 | 1.4 | 0.0214 | 0.0217 | 1.8 | 0.0044 | 0.1069 |
| NM_003330 | TXNRD1 | 2.1 | 0.0010 | 0.1635 | 4.4 | 0.0024 | 0.9034 | 1.3 | 0.1361 | 0.1436 | 2.0 | 0.0396 | 0.7375 |
Note. NA, not applicable; fold ch, absolute fold changes relative to vehicle treated controls are given; data in bold indicates genes whose expression changed at least 1.5-fold (up or down) and p < 0.05.
Hepatocytes from three donors were used for each treatment condition and one array was used for each hepatocyte preparation.
Samples were from both GSTM1-null and GSTM1-positive genotypes; therefore, no data are provided; all samples used were GSTT1 positive.
p = 0.0000345.
Table 1 shows the effect of SFN and PEITC on human genes with a known functional ARE. SFN induced expression of cysteine ligase catalytic subunit (GCLC), glutamate cysteine ligase modifier subunit (GCLM), NAD(P)H:quinone oxidoreductase (NQO1), thioredoxin, and thioredoxin reductase 1, but not GSTP1 or heme oxygenase (HO-1), at least at the time point examined in this study. Similarly, PEITC induced expression of GCLC, NQO1, thioredoxin, and thioredoxin reductase 1, but not GCLM, GSTP1, or HO-1. Gene regulation is complex and it is unlikely that any of these “environmental response” genes are regulated solely by the antioxidant response element (ARE)/Keap-1/Nrf-2 mechanism. Other cis and trans acting factors are involved in the regulation of these genes, and this may explain why some were not induced by SFN and PEITC under the conditions tested. It is also possible that induction of some of these genes occurred quickly but had returned to baseline by 48 h following treatment. This could well be an explanation for the lack of apparent increase in HO-1 and other genes with rapid induction responses.
Although SFN and PEITC increased expression of some genes involved in antioxidant response, neither SFN nor PEITC increased expression of genes involved in the detoxication of AFB intermediates (GSTs, mEH, AKR7A2/3; Table 1). In contrast, the microarray results indicated that treatment with 50μM SFN resulted in a significant decrease of CYP3A4 mRNA, whereas PEITC had no significant effect on CYP3A4 expression in the three hepatocyte preparations examined by microarray analysis (Table 1). Neither isothiocyanate modulated CYP1A2 or CYP3A5 mRNA levels in the microarray analysis. Interestingly, SFN (10μM but not 50μM) and PEITC (10 and 25μM) increased mRNA levels of the two nucleotide excision repair genes ERCC4 and ERCC5.
Effects of Isothiocyanates on Expression of Genes Involved in Biotransformation of AFB as Assessed by TaqMan-Based RT-PCR Analysis
In addition to the semi-quantitative but global assessment of transcriptional effects of SFN and PEITC by microarray analysis, we also determined mRNA expression of genes involved in biotransformation of AFB by the highly quantitative TaqMan-based RT-PCR assay. mRNA expression of CYP1A1, CYP1A2, CYP3A4, CYP3A5, GSTM1, GSTT1, and mEH were determined in hepatocyte preparations derived from eight different donors (970, 985, 987, 990, 1002, 1087, 1095, 1105; Table 2). RT-PCR data from GSTM1- and GSTT1-null individuals were omitted from this analysis because no gene expression could occur and including the null genotypes would have skewed the results.
TABLE 2.
Modulation of Transcriptional Gene Expression by Isothiocyanates in Cultured Human Primary Hepatocytes as Determined by TaqMan-Based RT-PCR Analysis
| Treatment | CYP1A1 | CYP1A2 | CYP3A4 | CYP3A5 | mEH | GSTM1a | GSTT1a |
| PEITC 10μM | 3.7 ± 1.2 (8) | 3.6 ± 0.6 (8) | 0.92 ± 0.07 (8) | 0.83 ± 0.10 (8) | 1.8 ± 0.3 (8) | 0.67 ± 0.10 (4) | 1.2 ± 0.3 (6) |
| PEITC 25μM | 21.0 ± 5.9 (8)* | 7.0 ± 2.6 (8)** | 0.56 ± 0.11 (8)** | 0.70 ± 0.07 (8)** | 1.9 ± 0.5 (8) | 0.66 ± 0.22 (4) | 1.3 ± 0.5 (6) |
| SFN 10μM | 0.8 ± 0.1 (8) | 0.86 ± 0.21 (8) | 0.69 ± 0.11 (8)** | 1.1 ± 0.2 (8) | 1.1 ± 0.2 (8) | 0.98 ± 0.35 (4) | 0.70 ± 0.10 (6)* |
| SFN 50μM | 2.2 ± 0.6 (8) | 0.58 ± 0.14 (8)* | 0.13 ± 0.06 (8)*** | 1.1 ± 0.3 (8) | 1.1 ± 0.2 (8) | 0.48 ± 0.22 (4) | 0.46 ± 0.13 (6)* |
Note. Hepatocytes were treated with two concentrations of SFN or PEITC for 48 h. Control represents nonmodulated transcription level and was treated with vehicle for 48 h. Numerical values represent means of fold alterations relative to vehicle control (with the control being equal to 1) and SEs. The number of individual hepatocyte preparations used for each calculation is displayed in parentheses.
Includes only samples that had a positive genotype (e.g., were not homozygous null for the gene deletion).
“*” Denotes 0.01 < p < 0.05, “**” denotes 0.001 < p < 0.01, and “***” denotes p < 0.001 (comparing vehicle control [equal to 1.0] vs. isothiocyanate incubations).
Consistent with a previous study from our laboratory (Gross-Steinmeyer et al., 2005), PEITC significantly induced the expression of both CYP1A1 and CYP1A2, whereas SFN had no effect on CYP1A1 and actually decreased CYP1A2 mRNA by 44% at 50μM. Microarray analysis also identified the increase of CYP1A1 by PEITC (Table 1). However, it did not indicate the increase and decrease of CYP1A2 message by PEITC and SFN as determined by RT-PCR analysis, respectively (Tables 1 and 2). This is likely because of (1) different donors were used for the microarray and the RT-PCR analyses and (2) inherent differences between microarray technology and TaqMan-based RT-PCR analysis. In contrast to microarray analysis, RT-PCR analysis is a highly sensitive and quantitative methodology to measure mRNA levels.
Consistent with our microarray results, SFN treatment produced a dramatic downregulation of CYP3A4 (Table 2). This effect was seen consistently in all eight hepatocyte preparations. Additional incubations with 25μM SFN in five hepatocyte preparations revealed clear concentration-dependent effects (data not shown). The average transcriptional CYP3A4 levels, relative to the solvent control, were 69, 20, and 13% at 10, 25, and 50μM SFN, respectively. CYP3A4 mRNA levels in some SFN-treated hepatocyte samples were as low as 2% of the vehicle control; 25μM PEITC also significantly decreased CYP3A4 mRNA, but to a lesser extent than SFN, and was also not evident by microarray analysis. Although SFN had no effect, PEITC significantly decreased CYP3A5 mRNA by 30%. It should be noted that a common polymorphism in CYP3A5 results in a lack of expression of a functional mRNA in approximately 80% of humans. However, the RT-PCR technique used here did not discriminate between functional and nonfunctional CYP3A5 mRNA. Hepatocyte samples were not genotyped for CYP3A5 polymorphism; therefore, the functional significance, if any, of the modest decrease in CYP3A5 mRNA by PEITC is uncertain.
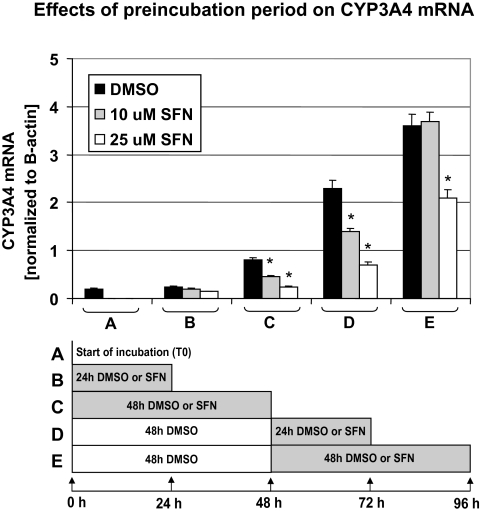
CYP3A4 expression is relatively low for at least the initial 24 h following hepatocytes isolation but appears to continually increase over 96 h of incubation (Fig. 2). Because SFN treatment reduced AFB-DNA adducts more effectively than PEITC, we evaluated the time-dependent effects of SFN on CYP3A4 expression at 0, 24, 48, 72, and 96 h incubation, adding SFN for only the last 24 or 48 h of each incubation period (Fig. 2). Both 10 and 25μM SFN treatment led to a concentration-dependent reduction in CYP3A4 expression. More specifically, cells that were treated for 48 h with 25μM SFN had approximately the same level of CYP3A4 mRNA as measured at time point 0, whereas the vehicle control showed a marked increase (Fig. 2C). Similarly, cells treated for 48 h with the vehicle only, followed by 24 h treatment with 25μM SFN exhibited CYP3A4 expression essentially unchanged from the 48 h vehicle treatment. In contrast, the corresponding time vehicle control showed a significant increase in CY3A4 expression (Fig. 2D). Although a similar trend was observed when cells were preincubated for 48 h with vehicle and then treated for an additional 48 h with SFN (25μM), the effect was less pronounced (Fig. 2E). This may be because of effective elimination (metabolism) of SFN over the 48-h treatment period as it was not replenished during that time.
FIG. 2.
Effects of SFN on CYP3A4 mRNA expression in cultured human primary hepatocytes over 96 h. CYP3A4 mRNA levels of hepatocytes were determined at the following times and treatments: (A) at 0 h (no DMSO or SFN treatment); (B) following 24 h treatment with 10 or 25μM SFN or DMSO; (C) following 48 h of SFN or DMSO treatment; (D) 48 h in culture with DMSO (no SFN) followed by 24 h of SFN or DMSO treatment; (E) 48 h in culture with DMSO (no SFN) followed by 48 h of SFN or DMSO treatment. Bars represent means of normalized CYP3A4 mRNA expression, including corresponding SD from three individual cell culture vessels from a single hepatocyte donor. “*” Denotes p < 0.05 comparing vehicle control versus SFN incubations within each condition.
SFN treatment also caused a significant and concentration-dependent downregulation of GSTT1 transcription (30 and 54% decrease in mRNA at 10 and 50μM SFN in GSTT1-positive samples, respectively) and a nonsignificant decrease of GSTM1 mRNA in four GSTM1-positive subjects (52% decrease at 50μM SFN). PEITC had no significant effect on GSTM1 or GSTT1 expression. Neither PEITC nor SFN had a significant effect on mEH expression (Table 2).
Direct Effects of Isothiocyanates on AFB Biotransformation Enzyme Catalytic Activity
We further explored the ability of these isothiocyanates to inhibit catalytic activities of key enzymes involved in the oxidation of AFB: CYP1A2 (AFB-epoxide formation [activation] and aflatoxin M1 formation [detoxication]) and CYP3A4 (aflatoxin Q1 formation [detoxication] and AFB-epoxide formation [activation]), using recombinant yeast strain expressing human CYP1A2 and commercially available CYP3A4-expressing Supersomes (Gentest). PEITC inhibited both human CYP1A2-mediated MROD and CYP3A4-mediated BROD activities in a concentration-dependent manner; 15 and 50μM PEITC resulted in approximately 20 and 50% reduction of MROD activity in CYP1A2 expression yeast microsomes (p < 0.05) and a 20 and 60% decrease of CYP3A4 activity in CYP3A4 supersomes (p < 0.05), respectively (data not shown). In contrast, SFN had no effect on either CYP enzyme activity (data not shown).
Modulation of AFB-DNA Adduct Formation by SFN under Different Treatment Conditions
The findings described above demonstrated that SFN treatment caused a more extensive reduction in AFB-mediated genotoxicity than PEITC (Fig. 1) and also resulted in a more substantial decrease in expression (mRNA levels) of CYP enzymes involved in AFB activation (Table 2). Therefore, we limited the remaining investigations to SFN.
We further explored to what extent the transcriptional effects (i.e., induction/repression) versus potential enzyme inhibitory effects contributed to the SFN-mediated decrease in AFB-DNA adduct formation in human hepatocytes. We measured adducts under three conditions: Under condition 1, cells were pretreated with SFN for 48 h, after which time the media was removed, cells were rinsed with PBS, and incubated for 6 h in media containing 3H-AFB but no SFN. Condition 2 was as condition 1, with the exception that cells were coexposed for the last 6 h with both 3H-AFB and SFN; this was the same protocol that was used to generate the data displayed in Figure 1. Condition 3 was as condition 2, except cells were not treated with SFN for the first 48 h, but with vehicle only. Condition 1 was designed to detect mainly transcriptional effects resulting in repression of CYP3A4 and CYP1A2, whereas condition 3 was chosen to assess effects that were because of direct interaction (inhibition) of SFN with CYP enzymes. Effects measured under condition 2 detected transcriptional as well as direct enzyme interaction effects.
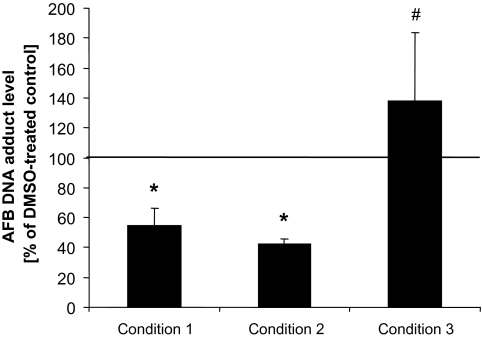
The results shown in Figure 3 demonstrate that the decrease in AFB-mediated genotoxicity by SFN was almost identical between condition 1 (SFN pretreatment and subsequent treatment with 3H-AFB) and condition 2 (SFN pretreatment and subsequent cotreatment with SFN and 3H-AFB). Both values were significantly different from the corresponding control (no pretreatment and subsequent treatment with 3H-AFB). Condition 3 (no pretreatment and subsequent cotreatment with SFN and 3H-AFB) had no significant effect on AFB-DNA adduct levels compared with control, which was not surprising as SFN had no inhibitory effect on CYP1A2 or CYP3A4 enzyme activities involved in AFB bioactivation (data not shown). These findings further support the hypothesis that it is SFN's effect on gene expression, and not catalytic activity, that is responsible for the reduction in AFB-DNA adduct formation.
FIG. 3.
Modulation of AFB-DNA adduct formation by SFN in cultured human primary hepatocytes at different treatment conditions. To assess contribution of transcriptional versus enzyme inhibition effects, we measured AFB-DNA adduct levels under three different conditions: (1) following a 48-h pretreatment with SFN, cells were incubated for 6 h in media containing 3H-AFB but no SFN (to detect transcriptional effects); (2) following a 48-h pretreatment with SFN, cells were incubated for an additional 6 h in media containing 3H-AFB and SFN (to detect both transcriptional effects and enzyme inhibition effects); (3) following a 48-h treatment with vehicle only (no SFN), cells were incubated for an additional 6 h in media containing 3H-AFB and SFN (to detect enzyme inhibition effects). SFN was applied at 10μM. Conditions 1, 2, and 3 were compared with an analogous treatment without SFN application, which served as control (equal to 100%), for example, following a 48-h treatment with vehicle only (no SFN), cells were incubated for an additional 6 h in media containing 3H-AFB only (no SFN). AFB-DNA adduct levels are expressed as percentages of control and represent means and SDs from three independent experiments (e.g., hepatocytes from three individual preparations). Conditions 1 and 2 were not significantly different from each other, but each was different from control (p < 0.05), marked as “*.” Condition 3 was significantly different from conditions 1 and 2 (p < 0.05) but not from the control (#).
Modulation of AFB-DNA Adduct Formation by SFN and GSTM1 Genotype
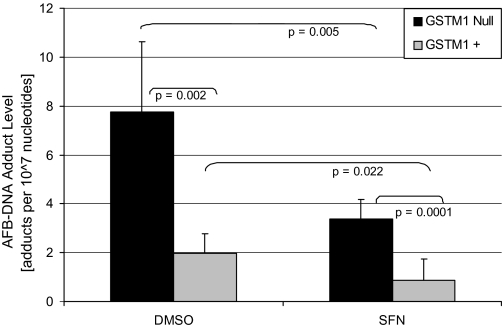
In the initial evaluation of AFB-DNA binding in human hepatocytes, four of the six hepatocytes preparations (#1046, 1072, 1076, 1087, 1095, 1105) evaluated were from individuals homozygous for the GSTM1 gene deletion (GSTM1 null) and the other two were GSTM1 positive (one or two functional alleles; the genotyping method does not distinguish between hemizygotes and homozygotes, although homozygotes are relatively rare). Interestingly, the two individuals that were GSTM1 positive appeared to have substantially lower levels of AFB-DNA adducts than the four that were GSTM1 null. To further investigate the possible role of the GSTM1 genotype/phenotype on AFB-DNA adduct formation, we completed DNA adduct assays in five additional human liver samples (#1125, 1165, 1183, 1407, and 1410), three of which were GSTM1 positive and two of which were GSTM1 null, in the presence and absence of pretreatment/cotreatment with 10μM SFN under conditions identical to those used for the data in Figure 1. The results from these 5 livers were combined with the previous 6 to yield AFB-DNA data from 11 human hepatocyte preparations, 6 that were GSTM1 null and 5 that were GSTM1 positive (Fig. 4). The presence of the GSTM1 gene was associated with 74.5% reduction in AFB-DNA adducts at baseline (DMSO only) relative to hepatocytes lacking the GSTM1 gene. AFB-DNA adducts were decreased to the same extent by pretreatment with 10μM SFN in both GSTM1-null hepatocytes (56.2%) and GSTM1-positive hepatocytes (55.9%).
FIG. 4.
Modulation of AFB-DNA adduct formation in the context of the GSTM1 genotype status. A total of 11 different hepatocyte preparations were examined for AFB-DNA binding in the presence or absence of 10μM SFN pretreatment. Six of the samples were GSTM1 null and five were GSTM1 positive. AFB-DNA adducts per 107 nucleotides were calculated and are shown. Each bar represents the mean and SEM. ANOVA indicated variances that were not statistically different among the groups. Statistical significance was determined by unpaired t-test with equal variances. All groups were significantly different (p < 0.05), with p values for individual comparisons shown.
There was no significant effect of GSTT1 polymorphism on AFB-DNA adduct formation, although only three subjects were GSTT1 null (data not shown). Of the six GSTM1-null samples, five were also GSTT1 positive and one was GSTT1 null. AFB-DNA adduct levels in the one GSTT1-null and GSTM1-null sample was 4.1 adducts per 107 nucleotides, whereas the average AFB-DNA adduct level in the 5 GSTM1-null and GSTT1-positive samples was 8.5 adducts per 107 nucleotides (DMSO-vehicle controls). For hepatocyte samples that were GSTM1 positive, 2 were GSTT1 null, with an average control AFB-DNA adduct level of 2.4 adducts per 107 nucleotides, whereas the 3 GSTT1-positive samples averaged 1.7 adducts per 107 nucleotides.
DISCUSSION
SFN, and to a lesser extent PEITC, reduced AFB-DNA adduct formation in human hepatocytes in a concentration-dependent manner. The two primary chemopreventive mechanisms against AFB-induced carcinogenesis proposed in the literature are induction of GSTs that detoxify AFBO and/or inhibition of CYP-mediated activation of AFB (Kwak et al., 2001; Langouet et al., 1995). SFN's protection against AFB-DNA adduct formation required treatment of hepatocytes prior to AFB exposure (Fig. 3), suggesting that SFN altered expression of enzymes involved in AFB biotransformation, rather than inhibiting catalytic activities.
Certain rodent alpha-class GSTs detoxify AFBO effectively, thereby providing protection against AFB-induced hepatocarcinogenesis (Eaton et al., 2001; Hayes et al., 1998; Kwak et al., 2001). In contrast, hepatic human alpha-class GSTs have no detectable activity toward AFBO (Eaton et al., 2001; Guengerich et al., 1998). However, human mu class GSTM1-1 has low but detectable activity toward AFBO (Guengerich et al., 1998). Some human epidemiological studies indicated that GSTM1-1 may provide protection against AFB-induced hepatocarcinogenesis (Chen et al., 1996; Deng et al., 2005; Kirk et al., 2005; London et al., 1995; Long et al., 2005; Omer et al., 2001), whereas others found little or no protective effect (Chen et al., 2002; Hsieh et al., 1996; McGlynn et al., 2003; Sun et al., 2001; Yu et al., 1995). Thus, induction of GSTM1 may reduce AFB carcinogenicity in GSTM1-positive individuals. However, in the present study, neither SFN nor PEITC induced GSTM1 expression (Table 2). In addition, we demonstrated previously that neither PEITC nor SFN affected expression of GSTA1 in human hepatocytes (Gross-Steinmeyer et al., 2004). Therefore, the protective effect of SFN or PEITC cannot be attributed to the induction of GSTs in this study.
We provide, for the first time, direct laboratory evidence that hGSTM1 is capable of protecting human hepatocytes from AFB-DNA adduct formation. hGSTM1-positive cells had 75% fewer adducts than GSTM1-null cells, independent of SFN treatment (Fig. 4). Pretreatment of hepatocytes with 10μM SFN reduced AFB-DNA adducts by 56% in both GSTM1-positive and GSTM1-null cells. Thus, the relative protective effect of a functional GSTM1 gene was more potent than 10μM SFN treatment and, interestingly, had a nearly identical effect on the level of reduction (74.5 vs. 74.3%) in AFB-DNA adducts in DMSO-treated versus SFN-treated hepatocytes. Taken together, these observations suggest that SFN treatment has no more effect in individuals with a functional GSTM1 gene than individuals that are homozygous null. These data support the epidemiological observations that individuals with the GSTM1-null genotype are more susceptible to AFB-induced hepatocarcinogenesis. Steck and Hebert (2009) recently reviewed the literature pertaining to isothiocyanate-mediated protection against the carcinogenic and mutagenic effects of dietary heterocyclic aromatic amines and commented that “it may be sulforaphane's ability to induce the GSTs rather than its role as a substrate for the GSTs that is most crucial in cancer prevention. This is consistent with findings from some studies in the United States showing that intake of Brassica vegetables (the majority being broccoli) is associated with greater cancer risk reduction in individuals with the active forms of the GST genes as compared with individuals with the inactive forms” (Steck and Hebert, 2009). The results of this study do not directly support a role of induction of GSTM1 in protection against AFB-induced hepatocarcinogenesis, although the presence of a functional allele of GSTM1 does provide substantial protection against AFB-DNA adduct formation, the presumed initiating event in AFB-induced carcinogenesis. However, it should be noted that we did not measure GSTM1 protein or activity following these treatments, and it is possible that SFN could have increased GSTM1 activity (in those subjects with a functional GSTM1 allele) via protein stabilization or potentially through an induction of mRNA that was short lived and had returned to baseline by 48 h.
Only two samples in our study were both GSTM1 positive and GSTT1 positive, and only one sample was both GSTM1 null and GSTT1 null. Therefore, possible “gene-gene” interactions could not be evaluated statistically. Nevertheless, the highest adduct levels were seen in cells that were GSTM1 null and GSTT1 positive, suggesting that GSTT1 may not play a significant role in AFBO detoxication.
Another route of detoxication in humans may be the enzymatic hydrolysis of AFBO to the less toxic AFB-dihydrodiol catalyzed by mEH (Dash et al., 2007; Kelly et al., 2002; Kirk et al., 2005; London et al., 1995; McGlynn et al., 2003). However, SFN did not affect mEH expression. Therefore, increased mEH detoxification is an unlikely mechanism by which SFN afforded protection.
It is also possible that SFN induced DNA repair pathways, thereby enhancing the rate of repair of AFB-DNA lesions. Indeed, microarray analysis indicated an increase in expression of two nucleotide excision repair genes ERCC4 and ERCC5 following SFN (10μM but not 50μM) and PEITC (10 and 25μM; Table 1) treatments. However, given that 50μM SFN greatly reduced AFB-DNA adduct levels (Fig. 1), but did not increase ERCC4 or ERCC5 expression, it is unlikely that these genes are involved in the major mechanism by which SFN reduced AFB-induced genotoxicity.
A major effect of SFN was the reduction of CYP3A4 mRNA levels at both 10 (31% decrease) and 50μM (87% decrease). Although 10μM SFN did not result in a significant decrease in CYP1A2 mRNA, 50μM concentration did (42% decrease; Table 2). CYP1A2 and 3A4 are the two major enzymes activating AFB (Gallagher et al., 1994). Thus, the most likely mechanism by which SFN alleviated AFB-induced genotoxicity is by reducing AFB activation. Although we did not directly measure whether CYP3A4 proteins were decreased proportionately to CYP3A4 mRNA levels in these hepatocytes, baseline CYP3A4 expression and protein levels following the isolation procedure is very low for the first 24–48 h following isolation. The treatment with SFN basically prevented mRNA expression from recovering, and thus, it is very likely that CYP3A4 protein levels remained low. Indeed, in a recent series of experiments using both broccoli extracts and 10 and 25μM SFN, we evaluated CYP3A4 protein levels via Western blot analysis and found a reduction of CYP3A4 protein to 33 ± 5% and 37 ± 8%, respectively, for the 10 and 25μM SFN concentrations (Gross-Steinmeyer, Tracy, and Eaton, in preparation). In the same experiment, 13 ± 5% and 41 ± 9% mRNA reduction was observed at the corresponding concentrations.
Hepatic AFB concentrations encountered by humans through diet are less than 1μM. Previous in vitro kinetic analysis in human microsomes suggested that CYP1A2 is the predominant enzyme activating AFB at concentrations below 1μM (Eaton and Gallagher, 1994; Gallagher et al., 1996). In contrast to SFN, 25μM PEITC not only lead to a 44% reduction of CYP3A4 but also a sevenfold induction of CYP1A2 mRNA levels (Table 2). At the same time, 25μM PEITC lowered AFB-DNA adducts by 44% (Fig. 1), mirroring CYP3A4 but not CYP1A2 mRNA levels. Unlike SFN, 50μM PEITC inhibited catalytic activities of both CYPs 1A2 and 3A4 by ∼50 and 60%, respectively (data not shown). Let us assume that 25μM PEITC inhibited AFBO formation of both CYPs by approximately 50% (which is an over-estimation). If CYP1A2 were the major activator of AFB, the overall effect of 25μM PEITC treatment would most likely result in an increase of AFB-DNA adduct formation, not a decrease. Therefore, in human hepatoctyes in primary culture, CYP3A4 appears to be the predominant activator of AFB, and SFN alleviated genotoxicity mainly by reducing CYP3A4 expression. These results are in contrast to our previously reported kinetic data, predicting CYP1A2 to be the major enzyme activating AFB at the low concentrations used in this study (Gallagher et al., 1996). However, that kinetic investigation utilized microsomal preparations, whereas this study was performed with intact human hepatocytes. Microsomes prepared from human livers reflect in vivo like CY1A2 and CYP3A4 levels, and the latter is expressed more highly than the former in most human livers. In addition, expression of CYPs dramatically decrease following isolation of primary hepatocytes and recovers steadily over time in culture, and recovery of CYP1A2 expression does not occur as rapidly and extensively as that of CYP3A4 (Hewitt et al., 2007). Thus, in the absence of an inducer (endogenous or exogenous), CYP1A2 constitutive expression in hepatocyte cultures is very low and is thus less important in AFB activation than CYP3A4 in primary hepatocyte cultures. We demonstrated that the Ah Receptor ligand 3,3′-diindolylmethane (DIM) is an effective inducer of CYP1A2 (up to 90-fold) but not CYP3A4 expression. In addition, AFB-DNA adducts increased up to sixfold following DIM treatment, indicating that CYP1A2 can indeed contribute to AFBO formation in human hepatocytes (Gross-Steinmeyer et al., 2009).
We recently reported that SFN is an effective antagonist of the pregnane X-receptor (PXR or NR1I2) (Zhou et al., 2007). It inhibits CYP3A4 expression via interaction with PXR, which is the predominant mediator of hepatic CYP3A4 expression. This is consistent with our observation that SFN blocked recovery of CYP3A4 expression, following hepatocyte isolation, over the initial 96 h in culture. A previous study also reported that SFN treatment reduced CYP3A4 mRNA, protein, and activity in human hepatocytes (Maheo et al., 1997), although they did not assess the capacity of SFN to affect AFB-DNA adduct formation.
Whether SFN could reach a concentration in the liver sufficient to inhibit CYP3A4 expression following dietary exposure is uncertain. However, a plasma concentration of 1–2μM SFN was achieved 1 h following a single oral dose of 200 μmoles of SFN (Ye et al., 2002), and it is likely that substantially higher peak concentrations would be seen in the liver because of extensive first pass clearance of SFN by the liver. SFN has been identified by the National Cancer Institute as a candidate for chemoprevention studies; it is one of several compounds selected for study in the National Cancer Institute's “Rapid Access to Preventive Intervention Development” Program and is currently being used in several preclinical and clinical trials. Thus, it is conceivable that concentrations of SFN in the liver exceeding 10–20μM might be possible, especially if used in relatively high doses for chemopreventive purposes. CYP3A4 is involved in biotransformation of over 50% of all therapeutic drugs. Thus, if SFN were to inhibit constitutive expression of CYP3A4 in vivo at doses used in chemoprevention, it could have important implications for adverse reactions of CYP3A4 metabolized drugs and/or other PXR-regulated biotransformation pathways.
In summary, our data suggest that SFN dramatically decreased AFB-induced DNA adduct formation in human hepatocytes by repressing CYP3A4 expression. Our data do not support GST induction by SFN as a protective mechanism. However, it provides evidence that the presence of a functional GSTM1 alleles can afford protection. The apparent downregulation of CYP3A4 by SFN may have important implications for drug–drug interactions.
FUNDING
National Institutes of Health (R01ES05780, R01GM079280-01A1); Cancer Research and Prevention Foundation (fellowship to K.G.-S.); Microarray analysis, genotyping, and RT-PCR was conducted by the Functional Genomics Core of the Center for Ecogenetics and Environmental Health with support from the National Institute of Environmental Health Sciences Center (P30ES007033) and the National Center for Research Resources (UW CTSA grant UL1RR025014). Primary human hepatocytes were provided through the liver tissue and cell distribution system funded by N01 DK 7-0004 (to S.C.S.).
References
- Bertl E, Bartsch H, Gerhauser C. Inhibition of angiogenesis and endothelial cell functions are novel sulforaphane-mediated mechanisms in chemoprevention. Mol. Cancer Ther. 2006;5:575–585. doi: 10.1158/1535-7163.MCT-05-0324. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Yu MW, Liaw YF, Wang LW, Chiamprasert S, Matin F, Hirvonen A, Bell DA, Santella RM. Chronic hepatitis B carriers with null genotypes of glutathione S-transferase M1 and T1 polymorphisms who are exposed to aflatoxin are at increased risk of hepatocellular carcinoma. Am. J. Hum. Genet. 1996;59:128–134. [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Chen CJ, Tsai WY, Ahsan H, Liu TY, Lin JT, Santella RM. Associations of plasma aflatoxin B1-albumin adduct level with plasma selenium level and genetic polymorphisms of glutathione S-transferase M1 and T1. Nutr. Cancer. 2000;38:179–185. doi: 10.1207/S15327914NC382_6. [DOI] [PubMed] [Google Scholar]
- Chen SY, Wang LY, Lunn RM, Tsai WY, Lee PH, Lee CS, Ahsan H, Zhang YJ, Chen CJ, Santella RM. Polycyclic aromatic hydrocarbon-DNA adducts in liver tissues of hepatocellular carcinoma patients and controls. Int. J. Cancer. 2002;99:14–21. doi: 10.1002/ijc.10291. [DOI] [PubMed] [Google Scholar]
- Dash B, Afriyie-Gyawu E, Huebner HJ, Porter W, Wang JS, Jolly PE, Phillips TD. Determinants of the variability of aflatoxin-albumin adduct levels in Ghanaians. J. Toxicol. Environ. Health A. 2007;70:58–66. doi: 10.1080/15287390600748880. [DOI] [PubMed] [Google Scholar]
- Deng ZL, Wei YP, Ma Y. Polymorphism of glutathione S-transferase mu 1 and theta 1 genes and hepatocellular carcinoma in southern Guangxi, China. World J. Gastroenterol. 2005;11:272–274. doi: 10.3748/wjg.v11.i2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DL, Bammler TK, Kelly EJ. Interindividual differences in response to chemoprotection against aflatoxin-induced hepatocarcinogenesis: implications for human biotransformation enzyme polymorphisms. Adv. Exp. Med. Biol. 2001;500:559–576. doi: 10.1007/978-1-4615-0667-6_85. [DOI] [PubMed] [Google Scholar]
- Eaton DL, Gallagher EP. Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994;34:135–172. doi: 10.1146/annurev.pa.34.040194.001031. [DOI] [PubMed] [Google Scholar]
- Eugster HP, Sengstag C. Saccharomyces cerevisiae: an alternative source for human microsomal liver enzymes and its use in drug interaction studies. Toxicology. 1993;82:61–73. doi: 10.1016/0300-483x(93)90060-6. [DOI] [PubMed] [Google Scholar]
- Gallagher EP, Kunze KL, Stapleton PL, Eaton DL. The kinetics of aflatoxin B1 oxidation by human cDNA-expressed and human liver microsomal cytochromes P450 1A2 and 3A4. Toxicol. Appl. Pharmacol. 1996;141:595–606. doi: 10.1006/taap.1996.0326. [DOI] [PubMed] [Google Scholar]
- Gallagher EP, Wienkers LC, Stapleton PL, Kunze KL, Eaton DL. Role of human microsomal and human complementary DNA-expressed cytochromes P4501A2 and P4503A4 in the bioactivation of aflatoxin B1. Cancer Res. 1994;54:101–108. [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li P, Lumeau S, Cassar G, Dupont MA, Chevolleau S, Gasc N, Tulliez J, Terce F. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 2000;60:1426–1433. [PubMed] [Google Scholar]
- Gerhauser C, You M, Liu J, Moriarty RM, Hawthorne M, Mehta RG, Moon RC, Pezzuto JM. Cancer chemopreventive potential of sulforamate, a novel analogue of sulforaphane that induces phase 2 drug-metabolizing enzymes. Cancer Res. 1997;57:272–278. [PubMed] [Google Scholar]
- Gross-Steinmeyer K, Stapleton PL, Liu F, Tracy JH, Bammler TK, Quigley SD, Farin FM, Buhler DR, Safe SH, Strom SC, et al. Phytochemical-induced changes in gene expression of carcinogen-metabolizing enzymes in cultured human primary hepatocytes. Xenobiotica. 2004;34:619–632. doi: 10.1080/00498250412331285481. [DOI] [PubMed] [Google Scholar]
- Gross-Steinmeyer K, Stapleton PL, Tracy JH, Bammler TK, Lehman T, Strom SC, Eaton DL. Influence of Matrigel-overlay on constitutive and inducible expression of nine genes encoding drug-metabolizing enzymes in primary human hepatocytes. Xenobiotica. 2005;35:419–438. doi: 10.1080/00498250500137427. [DOI] [PubMed] [Google Scholar]
- Gross-Steinmeyer K, Stapleton PL, Tracy JH, Bammler TK, Strom SC, Buhler DR, Eaton DL. Modulation of aflatoxin B1-mediated genotoxicity in primary cultures of human hepatocytes by diindolylmethane, curcumin, and xanthohumols. Toxicol. Sci. 2009;112:303–310. doi: 10.1093/toxsci/kfp206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Johnson WW, Shimada T, Ueng YF, Yamazaki H, Langouet S. Activation and detoxication of aflatoxin B1. Mutat. Res. 1998;402:121–128. doi: 10.1016/s0027-5107(97)00289-3. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ, Ellis EM, McLeod R, James RF, Seidegard J, Mosialou E, Jernstrom B, Neal GE. Regulation of rat glutathione S-transferase A5 by cancer chemopreventive agents: mechanisms of inducible resistance to aflatoxin B1. Chem. Biol. Interact. 1998;111–112:51–67. doi: 10.1016/s0009-2797(97)00151-8. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Kelleher MO, Eggleston IM. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 2008;47(Suppl. 2):73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- Hewitt NJ, Lechon MJ, Houston JB, Hallifax D, Brown HS, Maurel P, Kenna JG, Gustavsson L, Lohmann C, Skonberg C, et al. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab. Rev. 2007;39:159–234. doi: 10.1080/03602530601093489. [DOI] [PubMed] [Google Scholar]
- Hsieh LL, Huang RC, Yu MW, Chen CJ, Liaw YF. L-myc, GST M1 genetic polymorphism and hepatocellular carcinoma risk among chronic hepatitis B carriers. Cancer Lett. 1996;103:171–176. doi: 10.1016/0304-3835(96)04209-7. [DOI] [PubMed] [Google Scholar]
- Juge N, Mithen RF, Traka M. Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell Mol. Life Sci. 2007;64:1105–1127. doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EJ, Erickson KE, Sengstag C, Eaton DL. Expression of human microsomal epoxide hydrolase in Saccharomyces cerevisiae reveals a functional role in aflatoxin B1 detoxification. Toxicol. Sci. 2002;65:35–42. doi: 10.1093/toxsci/65.1.35. [DOI] [PubMed] [Google Scholar]
- Kirk GD, Turner PC, Gong Y, Lesi OA, Mendy M, Goedert JJ, Hall AJ, Whittle H, Hainaut P, Montesano R, et al. Hepatocellular carcinoma and polymorphisms in carcinogen-metabolizing and DNA repair enzymes in a population with aflatoxin exposure and hepatitis B virus endemicity. Cancer Epidemiol. Biomarkers Prev. 2005;14:373–379. doi: 10.1158/1055-9965.EPI-04-0161. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Egner PA, Dolan PM, Ramos-Gomez M, Groopman JD, Itoh K, Yamamoto M, Kensler TW. Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat. Res. 2001;480–481:305–315. doi: 10.1016/s0027-5107(01)00190-7. [DOI] [PubMed] [Google Scholar]
- Langouet S, Coles B, Morel F, Becquemont L, Beaune P, Guengerich FP, Ketterer B, Guillouzo A. Inhibition of CYP1A2 and CYP3A4 by oltipraz results in reduction of aflatoxin B1 metabolism in human hepatocytes in primary culture. Cancer Res. 1995;55:5574–5579. [PubMed] [Google Scholar]
- London WT, Evans AA, Buetow K, Litwin S, McGlynn K, Zhou T, Clapper M, Ross E, Wild C, Shen FM, et al. Molecular and genetic epidemiology of hepatocellular carcinoma: studies in China and Senegal. Princess Takamatsu Symp. 1995;25:51–60. [PubMed] [Google Scholar]
- Long XD, Ma Y, Wei YP, Deng ZL. [A study about the association of detoxication gene GSTM1 polymorphism and the susceptibility to aflatoxin B1-related hepatocellular carcinoma] Zhonghua Gan Zang Bing Za Zhi. 2005;13:668–670. [PubMed] [Google Scholar]
- Maheo K, Morel F, Langouet S, Kramer H, Le Ferrec E, Ketterer B, Guillouzo A. Inhibition of cytochromes P-450 and induction of glutathione S-transferases by sulforaphane in primary human and rat hepatocytes. Cancer Res. 1997;57:3649–3652. [PubMed] [Google Scholar]
- McGlynn KA, Hunter K, LeVoyer T, Roush J, Wise P, Michielli RA, Shen FM, Evans AA, London WT, Buetow KH. Susceptibility to aflatoxin B1-related primary hepatocellular carcinoma in mice and humans. Cancer Res. 2003;63:4594–4601. [PubMed] [Google Scholar]
- Morse M, Eklind K, Hecht S, Chung F. Inhibition of tobacco-specific nitrosamine 4-(N-nitrosomethylamino)-1-(3-pyridyl)-1-butanone (NNK) tumorigenesis with aromatic isothiocyanates. IARC Sci. Publ. 1991;105:529–534. [PubMed] [Google Scholar]
- Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- Omer RE, Verhoef L, Van't Veer P, Idris MO, Kadaru AM, Kampman E, Bunschoten A, Kok FJ. Peanut butter intake, GSTM1 genotype and hepatocellular carcinoma: a case-control study in Sudan. Cancer Causes Control. 2001;12:23–32. doi: 10.1023/a:1008943200826. [DOI] [PubMed] [Google Scholar]
- Pan MH, Ho CT. Chemopreventive effects of natural dietary compounds on cancer development. Chem. Soc. Rev. 2008;37:2558–2574. doi: 10.1039/b801558a. [DOI] [PubMed] [Google Scholar]
- Rungapamestry V, Duncan AJ, Fuller Z, Ratcliffe B. Effect of meal composition and cooking duration on the fate of sulforaphane following consumption of broccoli by healthy human subjects. Br. J. Nutr. 2007;97:644–652. doi: 10.1017/S0007114507381403. [DOI] [PubMed] [Google Scholar]
- Steck SE, Hebert JR. GST polymorphism and excretion of heterocyclic aromatic amine and isothiocyanate metabolites after Brassica consumption. Environ. Mol. Mutagen. 2009;50:238–246. doi: 10.1002/em.20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoewsand GS. Bioactive organosulfur phytochemicals in Brassica oleracea vegetables—a review. Food Chem. Toxicol. 1995;33:537–543. doi: 10.1016/0278-6915(95)00017-v. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol. Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- Sun CA, Wang LY, Chen CJ, Lu SN, You SL, Wang LW, Wang Q, Wu DM, Santella RM. Genetic polymorphisms of glutathione S-transferases M1 and T1 associated with susceptibility to aflatoxin-related hepatocarcinogenesis among chronic hepatitis B carriers: a nested case-control study in Taiwan. Carcinogenesis. 2001;22:1289–1294. doi: 10.1093/carcin/22.8.1289. [DOI] [PubMed] [Google Scholar]
- Wang C, Bammler TK, Eaton DL. Complementary DNA cloning, protein expression, and characterization of alpha-class GSTs from Macaca fascicularis liver. Toxicol. Sci. 2002;70:20–26. doi: 10.1093/toxsci/70.1.20. [DOI] [PubMed] [Google Scholar]
- Ye L, Dinkova-Kostova AT, Wade KL, Zhang Y, Shapiro TA, Talalay P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin. Chim. Acta. 2002;316:43–53. doi: 10.1016/s0009-8981(01)00727-6. [DOI] [PubMed] [Google Scholar]
- Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109:1266–1273. doi: 10.1016/0016-5085(95)90587-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Poulton EJ, Grun F, Bammler TK, Blumberg B, Thummel KE, Eaton DL. The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol. Pharmacol. 2007;71:220–229. doi: 10.1124/mol.106.029264. [DOI] [PubMed] [Google Scholar]