Abstract
We employed the suppressive subtractive hybridization to identify 41 up- and downregulated transcripts in Jurkat cells after benzo[a]pyrene (BaP) treatment. Among the 21 downregulated transcripts, we found that BaP suppresses the Keap1 transcript by 7.5-fold. Subsequent analyses revealed that BaP significantly suppresses the Keap1 message and protein levels to about 40 and 60%, respectively, of the vehicle controls in Jurkat cells without reactive oxygen species involvement. In addition, the nuclear Nrf2 (nuclear factor erythroid 2-related factor) protein content is significantly increased by 2.6-fold. The same BaP treatment to Hepa1c1c7 cells also downregulates the Keap1 message and protein levels to a similar extent. When we treated Jurkat cells with 3-(4-morpholinyl)propyl isothiocyanate, which is known to increase the amount of the Nrf2 content, we found that there is no change in the Keap1 message, but the amount of the Keap1 (kelch-like ECH-associated protein 1) protein is reduced to 75% of the vehicle controls. Although both Nrf2 target messages nqo1 and gstp1 are upregulated by BaP in Jurkat cells, only GSTP1 is upregulated at the protein level. Unlike Hepa1c1c7 cells, Jurkat cells have no detectable aryl hydrocarbon receptor and BaP metabolites, minimal CYP1A1 activity, and no quinone oxidoreductase 1 (NQO1) activity. We concluded that BaP, but not its metabolites, increases the amount of the nuclear Nrf2 protein by downregulating the Keap1 message in Jurkat cells.
Keywords: benzo[a]pyrene, gene regulation, suppressive subtractive hybridization, Keap1, Nrf2
Benzo[a]pyrene (BaP) is a polycyclic aromatic hydrocarbon (PAH) that is formed through incomplete combustion of organic materials and is found in air, tobacco smoke, automobile exhaust, and food (Bostrom et al., 2002; Phillips, 1999). Depending on the sources, BaP may enter our body by inhalation, dermal absorption, or ingestion (Faustman and Omenn, 2008). Human exposure to BaP may be an acute event; but generally, individuals are exposed to a low level of BaP over an extended period of time. The food chain is considered the dominant pathway for routine human exposure and accounts for about 97% of the total daily intake of BaP (Hattemer-Frey and Travis, 1991). Inhalation and consumption of contaminated water are considered minor pathways of BaP exposure for the general population, except for tobacco users or workers in the coal industry (Miller and Ramos, 2001).
It is widely accepted that BaP is carcinogenic, mutagenic, cytotoxic, immunotoxic, and teratogenic in various species and tissues (Ellard et al., 1991; Miller and Ramos, 2001). It causes lung carcinogenesis and atherosclerosis in cigarette smokers (Alexandrov et al., 2002). BaP is metabolically activated by CYP1, epoxide hydrolase, and aldo-keto reductases to cause oxidative damage and DNA adduct formation (Conney et al., 1994; Knize et al., 1999; Shimada, 2006). Similar to many PAHs, BaP is an aryl hydrocarbon receptor (AhR) agonist. BaP toxicity is believed to be AhR-dependent because binding of BaP to AhR induces expression of CYP1 enzymes, which are necessary to initiate BaP bioactivation (Baird et al., 2005; Shimada et al., 2003). However, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), which is the best-known AhR ligand, causes apoptosis in T-cell lines in the absence of AhR (Hossain et al., 1998), suggesting that cellular effects of an AhR ligand may not be related to AhR.
Nrf2 (nuclear factor erythroid 2-related factor) has been widely accepted to be responsible for the chemoprotective effect of many phytochemicals (Hu et al., 2009). Accumulation of the Nrf2 protein in the nucleus occurs in response to oxidative stress and the presence of reactive electrophiles in a cell (Itoh et al., 2003). Nrf2 is a transcription factor that activates antioxidant response element (ARE)-dependent transcription of target genes, such as heme oxygenase-1 (HO-1) (Stewart et al., 2003), glutathione S-transferase (GST) (Hayes et al., 2000), quinone oxidoreductase 1 (NQO1) (Venugopal and Jaiswal, 1996), UDP-glucuronosyl transferase (Buckley and Klaassen, 2009), ABC transporters (Adachi et al., 2007), and others (Aleksunes and Manautou, 2007). Many of the induced enzymes degrade the bioactivated molecules and minimize protein and DNA damages. Currently, it is believed that Keap1 (kelch-like ECH-associated protein 1) binds Nrf2 in the cytoplasm and targets it to proteasomal degradation by the Cullin-3-dependent E3 ligase complex (Kaspar et al., 2009; Nguyen et al., 2009). The Keap1 protein contains a number of cysteine residues that are sensitive to reactive oxygen species (ROS) and modification by reactive electrophiles (Wakabayashi et al., 2004). This cysteine-dependent modification may serve as a signal for the Keap1 proteasomal degradation, and subsequently, Nrf2 is accumulated in the nucleus to activate gene transcription. In addition, there is some evidence suggesting that phosphorylation may play a role in the Nrf2 function. For example, protein kinase C (PKC) and phosphatidylinositol 3-kinase have been shown to be involved in the ARE-driven gene transcription (Keum et al., 2008). Phosphorylation of Nrf2 at Ser-40 by PKC has been shown to reduce the Keap1/Nrf2 interaction (Bloom and Jaiswal, 2003), and casein kinase 2 triggers the Nrf2 clearance from the nucleus (Pi et al., 2007).
In this study, we exposed Jurkat cells to BaP and examined whether BaP would cause cellular responses independent of AhR using suppressive subtractive hybridization (SSH) PCR. We discovered a number of novel transcripts that are affected by BaP. In particular, the Keap1 message is downregulated, leading to the increase of the Nrf2 protein in the Jurkat cell nuclei. Here, we have provided evidence to support that BaP, but not its metabolites, suppresses the Keap1 message and in turn increases the amount of the functionally active nuclear Nrf2 protein.
MATERIALS AND METHODS
Reagents.
BaP and cell culture media were purchased from Sigma (St Louis, MO). 3-Morpholinopropyl isothiocyanate (3MP-ITC) was purchased from Alfa Aesar (Ward Hill, MA). Other cell culture reagents were purchased from Invitrogen (Carlsbad, CA). Fetal calf serum was purchased from Tissue Culture Biologicals (Tulare, CA). Jurkat cells were grown in RPMI-1640 medium supplemented with 10% fetal calf serum, 10 U/ml of penicillin, and 10 μg/ml of streptomycin. The cell population was maintained at densities between 1 × 105 and 1 × 106 cells/ml. MCF-7 and Hepa1c1c7 cells were grown in DMEM medium supplemented with 10% fetal calf serum, 10 U/ml of penicillin, and 10 μg/ml of streptomycin. All cells were maintained at 37°C and 5% CO2. Anti-AhR polyclonal rabbit and goat immunoglobulin G (IgG) (H-211 and N-19) and anti-Nrf2 rabbit IgG (H-300) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Keap1 monoclonal mouse IgG MAB3024 was purchased from R&D Systems (Minneapolis, MN). Anti-GSTP1 rabbit polyclonal IgG (4413) was purchased from ProSci (Poway, CA). Anti-NQO1 goat polyclonal IgG (ab2346) was purchased from Abcam (Cambridge, MA). Anti-GAPDH rabbit polyclonal IgG G9545 was purchased from Sigma. Anti-acetyl-histone H4 rabbit polyclonal IgG 06-866 was purchased from Upstate Biotechnology (Upstate, NY). Secondary IgG conjugated with IRDye 800CW or 680 was purchased from LI-COR Bioscience (Lincoln, NE). 3-Hydroxy-BaP, 7-hydroxy-BaP, 9-hydroxy-BaP, BaP-7,8-dihydrodiol, and BaP-7,8-dione were purchased from NCI Chemical Carcinogen Standard Reference Repository (Midwest Research Institute, Kansas City, MO).
BaP exposure to Jurkat and Hepa1c1c7 cells.
Jurkat cells were prepared in a 75-cm2 flask containing 40 ml of complete media at a concentration of 2 × 105 cells/ml. On the following day, when the cell density reached at 4 × 105 cells/ml, BaP in the DMSO vehicle was added to the cells to a final concentration of 2.5μM. Control cells were treated with the vehicle DMSO. After 48 h at 37°C, cells were harvested to initiate SSH PCR. As for Hepa1c1c7 cells, they were seeded at about 30% confluent 1 day before treatment with 2.5μM BaP or DMSO.
SSH and complementary DNA library construction.
Total RNA was extracted from the vehicle control or the BaP-treated cells using the TRI reagent (Applied Biosystems, Foster City, CA), and then, poly(A)RNA was isolated from the total RNA using the Oligotex mRNA kits (Qiagen, Valencia, CA). Synthesis of complementary DNA (cDNA) and SSH PCR were performed using the PCR-Select cDNA subtraction kit (Clontech, Mountain View, CA). Eight micrograms of poly(A)RNA from the control or the BaP-treated cells was used for the first-strand cDNA synthesis. The whole reverse transcriptase reaction was subjected to the second strand synthesis. Ten micrograms of the cDNA library was used to start the SSH PCR method according to the manufacturer's recommendation. For the forward subtraction to identify the upregulated genes, cDNA from the control cells was used as the driver, whereas cDNA from the BaP-treated cells was used as the tester. The opposite would apply for the reverse subtraction to identify the downregulated genes. The PCR products obtained from the forward and reverse subtractions were cloned into the pGEM-TA plasmid (Promega, Madison, WI). The ligated products were transformed into XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA) for blue/white screening.
Secondary screen using DNA dot blot hybridization.
Each of the pGEM-cloned plasmids was extracted from a 3 ml overnight bacterial culture of a single white colony. To an overnight culture, the routine miniprep solutions I–III were used to release the plasmid into the solution. After centrifugation at 16,000 × g for 30 min at 4°C, the supernatant was directly used for dot blot hybridization. Supernatant (2 μg) was spotted onto an 82 mm GE nylon membrane. DNA dot blot hybridization was performed using a BrightStar psoralen-biotin-labeled subtracted cDNA probe (Applied Biosystems) generated from cDNA of either the forward or the reverse subtraction. The blot membranes were prehybridized at 42°C for 30 min in the ultrasensitive hybridization buffer (Applied Biosystems), and then the denatured probe was added to the buffer, followed by hybridization at 42°C overnight. After hybridization, the membranes were washed twice in 2X saline-sodium citrate (2×SSC) with 0.1×SDS at 42°C for 5 min, twice in 0.1×SSC with 0.1×SDS at 42°C for 15 min. After the final wash, the membrane was blocked with 5% bovine serum albumin (BSA) in 25mMTris, pH 7.6, 0.15M NaCl, 0.05% Tween-20 (1×TBST) for 1 h and then incubated for an additional 1 h in 1×TBST containing streptavidin peroxidase (1:10,000; Sigma) and 5% BSA. Afterward, the membrane was washed six times with 1×TBST for 5 min. The signal was visualized using Pierce SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL). The signal intensities were analyzed using the Un-Scan-It software. Clones with a fivefold difference in signal intensity between the forward and the reverse probe hybridization were sequenced and identified using the BLAST database.
Real-time quantitative PCR.
To determine whether the isolated up- and downregulated genes were authentic, we performed real-time qPCR using primers corresponding to the highest and the lowest fold difference in our forward and reverse subtraction products. Primer sequences for each clone were obtained from the Primer Bank (Spandidos et al., 2010). In addition, we performed real-time qPCR to quantify the Keap1 message. The cDNA library of the DMSO- or BaP-treated Jurkat cells were reversed transcribed from 1 μg of the extracted total RNA using random primers (Epicentre, Madison, WI). Real-time qPCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) on a Bio-Rad iCycler thermal cycler. PCR conditions (40 cycles) were as follows: 90°C for 10 s and 60°C (annealing and extension) for 1 min. SYBR Green fluorescence readings were taken at 60°C when the fluorescence intensity corresponded solely to the PCR product of interest. Normalized fold increase of the endogenous transcript was determined by the 2−ΔΔCT method using 18S as the internal standard (Jensen et al., 2006).
EROD assay.
We performed our ethoxyresorufin-O-deethylase (EROD) assay based on a similar concept in a published protocol (Donato et al., 1993). We induced Jurkat, MCF7, and Hepa1c1c7 cells (1 × 106) with 2.5μM BaP or 1μM 3-methylcholanthrene (3MC) in a 1.5-ml microfuge tube containing 1 ml of the complete media. Cells were rotated at 40 cpm at 37°C for 5 h. After induction, cells were precipitated at 600 × g for 5 min and then resuspended in 1 ml of media containing 2.2μM ethoxyresorufin and 10μM dicumarol. After rotating at 40 cpm for 30 min at 37°C, the cells were centrifuged at 600 × g for 5 min and 750 μl of the media was mixed with 2 ml of ethanol. After centrifugation at 1500 × g for 10 min, the solution was subjected for fluorescence analysis using a Shimadzu RF-5301 PC spectrofluorophotometer (with the excitation wavelength at 535 nm and the emission wavelength at 590 nm).
Quantitative Western blot analysis.
Whole-cell extract (WCE) were prepared from BaP- or DMSO-treated Jurkat, Hepa1c1c7, and MCF-7 cells as follows: To a 75-cm2 flask, 300 μl of lysis buffer (25mM HEPES, pH 7.4; 0.4 M KCl; 1mM EDTA; 1mM dithiothreitol [DTT]; 10% glycerol; 1mM PMSF; and 2 μg/ml of leupeptin) was used to harvest the cells. After three cycles of freeze/thaw, lysate was kept on ice for 30 min and then centrifuged at 14,000 × g at 4°C for 10 min. The supernatant was defined as WCE. Nucleus extract was prepared from BaP- or DMSO-treated Jurkat and Hepa1c1c7 cells as follows: Cells from a 75-cm2 flask were resuspended into 200 μl of the resuspension buffer (25mM HEPES, pH 7.4; 5mM KCl; 0.5mM MgCl2; 1mM DTT; 0.5% NP-40; 1mM PMSF; and 2 μg/ml of leupeptin), followed by rotation at 4°C for 15 min. After centrifugation at 3000 × g for 2 min, the pellet was then resuspended into 100 μl of the nuclear extract buffer (25mM HEPES, pH 7.4; 350mM NaCl; 10% sucrose; 0.05% NP-40; 1mM DTT; 1mM PMSF; and 2 μg/ml of leupeptin), followed by rotation at 4°C for 1 h. After centrifugation at 14,000 × g for 10 min at 4°C, the supernatant was considered as the nuclear extract. The transferred nitrocellulose membrane was blocked in Odyssey blocking buffer (LI-COR Bioscience) for 1 h. Blots were then incubated with primary antibody in blocking buffer containing 0.1% Tween-20 overnight. Dilutions for antibodies are as follows: AhR (N-19 and H-211), Keap1, and Nrf2 at 1:100, NQO1 and acetyl-histone H4 at 1:1000, GSTP1 at 1:2,000, and GAPDH at 1:10,000. After (primary and secondary) antibody incubation, membrane was washed with PBS containing 0.1% Tween-20 four times (5 min each). Incubation of the secondary antibody conjugated with IRDye (1:10,000) was performed in Odyssey blocking buffer containing 0.1% Tween-20 for 1 h. Secondary IgG conjugated with IRDye 800CW was used to detect the specific antigen for each Western, whereas anti-rabbit IgG conjugated with IRDye 680 was simultaneously used to detect the loading control GAPDH or acetyl-histone H4. Membrane was scanned and quantified using an Odyssey Infrared Imaging System (LI-COR Bioscience).
Cell survival assay.
In a 24-well plate, cells (2 × 105cells, 1 ml) were seeded 1 day before treatment. At different time points (12–48 h), cells in media were transferred into a 15-ml conical tube and then centrifuged at 600 × g for 5 min. The resulting cell pellet was resuspended in 100 μl of PBS. Cells were stained with trypan blue (0.4%, 10 μl) for 3 min, and then 10 μl of resuspended cells was counted using a hemacytometer. Percent cell survival was determined by dividing the number of unstained cells with the total number of cells.
BaP metabolism in Jurkat and Hepa1c1c7 cells.
The metabolites of BaP (namely 3-, 7-, and 9-OH BaP, BaP-7,8-dihydrodiol, and BaP-7,8-dione) were examined in Jurkat and Hepa1c1c7 cells. Hepa1c1c7 or Jurkat cells (3 × 107) were incubated in 2 ml of phenol red-free DMEM or RPMI, respectively, in the presence of either 5μM BaP or DMSO at 37°C and 90 rpm in a shaking incubator. After 12 h, two volumes (4 ml) of ethyl acetate were added to each tube, followed by vigorous mixing. The content was transferred to a borosilicate glass tube and then centrifuged at 600 × g for 5 min to separate the aqueous from the organic phase. The organic phase was transferred to a new glass tube and was evaporated under argon gas to dryness. Methanol (100 μl) was used to reconstitute the sample. BaP and its metabolites were quantified on an Applied Biosystems 4000 Q Trap mass spectrometer with atmospheric pressure chemical ionization source equipped with a LEAP CTC PAL autosampler (Carrboro, NC) and an Agilent 1200 series binary pump (Santa Clara, CA). The mobile phase consisted of water plus 0.1% formic acid (A) and acetonitrile plus 0.1% formic acid (B). A linear gradient was used as follows: 95% A to 45% A over 0.5 min; 45% A for 1 min; 45% A to 30% A over 1 min; 30% A to 20% A over 20 min; 20% A for 10 min; 20% A to 5% over 20 min; 5% A for 10 min. The identity of BaP metabolites was determined using external standards as follows: m/z 269 → 251 for 3-OH and 7-OH BaP (retention time was 33.7 min) and m/z 287 → 239 for BaP-7,8-dihydrodiol (retention time was 15 min). The flow rate was 0.5 ml/min through a Zorbax-ODS C18, 5 μm, 4.6 × 250 mm column (Wilmington, DE).
Fluorescence microscopy to detect intracellular ROS production.
Hepa1c1c7 cells were grown in four-well chambered cover glass dishes and treated when they were 60–80% confluent. Jurkat cells were grown in 4-cm plates and treated when they reached a concentration of 0.5–1 × 106 cells/ml. Cells were subjected to various treatments. Prior to analysis, media was aspirated, followed by incubation of 5μM 5-(and-6)carboxy-2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes, Carlsbad, CA) in 1×PBS for 30 min in the dark. Cells were then washed in 1×PBS, resuspended in fresh media, and allowed to incubate for 30 min in the dark prior to visualization. Fluorescence was observed under a Leica DMIRE2 inverted fluorescence microscope using a Yokogawa CSU-X1 confocal scanner unit.
Statistical analysis.
We performed unpaired one-tailed t-test to determine the statistical significance using the GraphPad Prism 5 software.
RESULTS
AhR Expression and EROD Activity in BaP- and DMSO-Treated Jurkat Cells
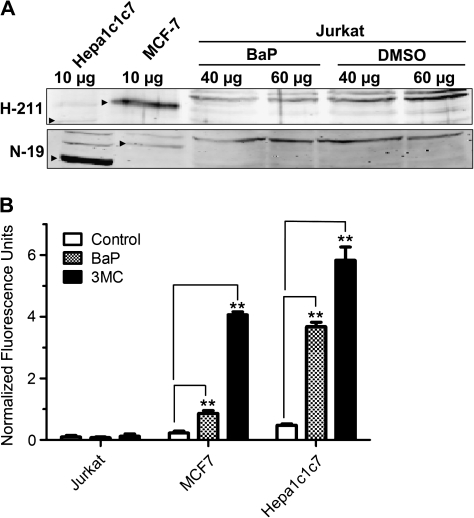
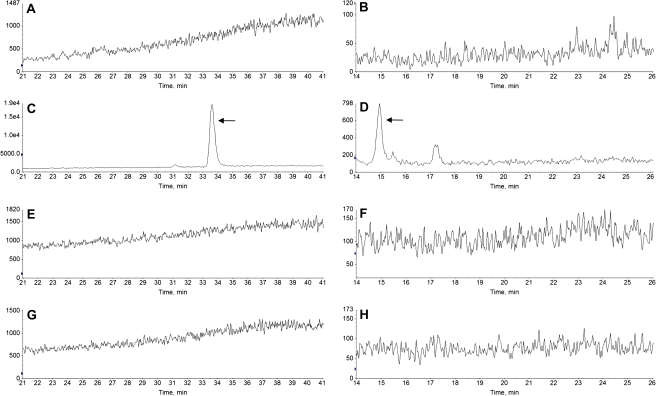
To determine whether our Jurkat cells expressed AhR, WCE from BaP- or DMSO-treated Jurkat cells were prepared for Western blot analysis. WCE from MCF-7 and Hepa1c1c7 cells was used to show the AhR expression. No detectable AhR protein was observed in both BaP- and DMSO-treated Jurkat WCE up to 60 μg of protein, whereas 10 μg of MCF-7 or Hepa1c1c7 WCE showed a strong signal for the AhR protein (Fig. 1A), indicating that there was minimal, if any, AhR in Jurkat cells. We used two different AhR-specific antibodies H-211 and N-19 because H-211 is best to show the human AhR band, whereas N-19 is more sensitive to the mouse AhR. Both antibodies were used to unambiguously show that AhR was expressed in our human breast (MCF-7) and mouse hepatoma (Hepa1c1c7) cells but was clearly absent in Jurkat cells. Next, we examined whether there was any CYP1A1 activity that might bioactivate BaP in Jurkat cells. Results from the EROD assay showed that there was minimal (but measurable) CYP1A1 activity in Jurkat cells in the presence or absence of an inducer: the CYP1A1 activities for vehicle, 2.5μM BaP, and 1μM 3MC-treated Jurkat cells were 0.099 ± 0.088, 0.075 ± 0.042, and 0.120 ± 0.077 fluorescence units/106 cells, respectively (Fig. 1B). In contrast, 2.5μM BaP induced the CYP1A1 activity 3.8- and 7.3-fold, respectively, in the AhR expressing cell lines MCF-7 and Hepa1c1c7. This induction, however, was even higher when a strong inducer 3MC was used at an optimal concentration (18.0- and 12.5-fold induction in MCF-7 and Hepa1c1c7 cells, respectively). To confirm that BaP is not metabolized in Jurkat cells, we performed LC/MS/MS analysis to look for BaP metabolites in Jurkat cells. No hydroxylated BaP and BaP-7,8-dihydrodiol were detected after Jurkat cells were treated with 5μM BaP for up to 12 h (Fig. 2). Same BaP treatment in the control Hepa1c1c7 cells showed the presence of hydroxylated BaP (3-OH and/or 7-OH BaP) and BaP-7,8-dihydrodiol. Our analysis was not able to differentiate 3-OH and 7-OH BaP because the two standards eluted at the same retention time. In addition, no detection of BaP-7,8-dione and 9-OH BaP was observed from both Jurkat and Hepa1c1c7 cells after BaP treatment. Without much CYP1A1 activity and no detection of any oxidized BaP, minimal turnover of BaP to the reactive DNA alkylator BaP diolepoxide or the superoxide generator BaP-7,8-catechol should be expected when Jurkat cells were treated with 5μM BaP or less.
FIG. 1.
AhR expression and CYP1A1 activity in Jurkat cells. (A) Western blot analysis showing that AhR is not detected in BaP- and DMSO-treated Jurkat cells. Jurkat cells were treated with 2.5μM BaP or DMSO vehicle for 48 h. Different amounts of WCE from BaP- or DMSO-treated Jurkat (40 and 60 μg), MCF-7 (10 μg), and Hepa1c1c7 (10 μg) cells were used to detect the AhR protein using AhR-specific antibodies H-211 (upper panel) and N-19 (lower panel). Arrowheads indicate the human (MCF-7) and mouse AhR (Hepa1c1c7). This Western has been repeated at least once. (B) CYP1A1 activity of Jurkat, MCF7, and Hep1c1c7 cells determined by EROD assay. Cells were treated with DMSO vehicle, 2.5μM BaP, or 1μM 3MC for 5 h at 37°C, followed by EROD assay. The error bars represent ±SD of triplicate samples (n = 3, ** p ≤ 0.005). Normalized fluorescence units were calculated by subtracting each fluorescence number with the number obtained from the same treatment without cells.
FIG. 2.
Representative chromatograms showing BaP metabolites. Hepa1c1c7 (A–D) and Jurkat (E–H) cells were incubated for 12 h in the presence of DMSO (A and B, and E and F) or BaP (C and D, and G and H). Hydroxylated BaP was detected in Hepa1c1c7 cells (C, arrow) but not in Jurkat cells (G). BaP-7,8-dihydrodiol was detected in Hepa1c1c7 cells (D, arrow) but not in Jurkat cells (H). An unidentified metabolite was observed in Hepa1c1c7 cells (D, retention time 17.2 min). (A) and (B) are the corresponding controls for (C) and (D), respectively, whereas (E) and (F) are the corresponding controls for (G) and (H).
Determination of ROS Formation in Jurkat and Hep1c1c7 Cells after BaP Exposure
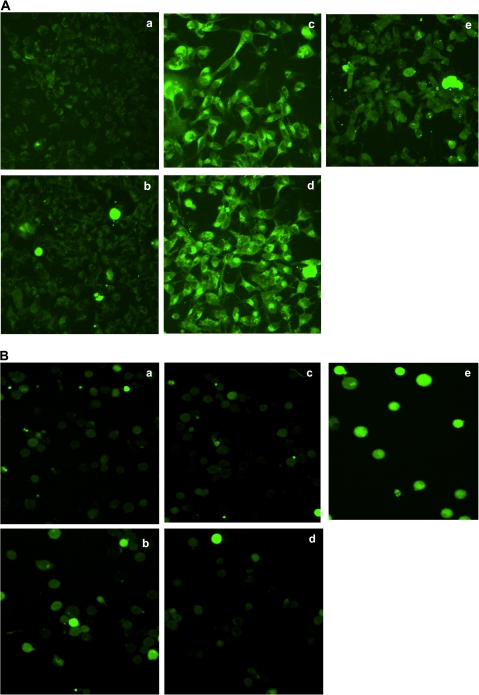
To explore whether ROS is formed after BaP exposure in Jurkat cells, we performed fluorescence staining studies to detect the presence of ROS under various BaP treatments. We observed no ROS staining in Jurkat cells after exposure to 2.5μM BaP for 24 or 48 h, whereas the control Hepa1c1c7 cells showed definitive ROS formation as early as 12 h after BaP exposure (Fig. 3). In addition, we observed no apparent ROS staining in Jurkat cells after they were treated with 2.5μM BaP for 1.5, 3, or 12 h (data not shown). We used chelerythrine as the positive control because we have observed consistent ROS formation when HCT116 cells were treated with chelerythrine (Matkar et al., 2008).
FIG. 3.
ROS detection by fluorescence staining. (A) Fluorescence staining of Hepa1c1c7 cells treated with (a) DMSO for 24 h, (b) 2.5μM BaP for 1.5 h, (c) 2.5μM BaP for 12 h, (d) 2.5μM BaP for 24 h, and (e) 7.5μM chelerythrine for 1.5 h. (B) Fluorescence staining of Jurkat cells treated with (a) DMSO for 24 h, (b) DMSO for 48 h, (c) 2.5μM BaP for 24 h, (d) 2.5μM BaP for 48 h, and (e) 7.5μM chelerythrine for 1.5 h. Untreated cells were identical as the DMSO-treated cells (data not shown).
Determination of the Up- and Downregulated Transcripts in Jurkat Cells after BaP Exposure
The PCR products obtained from the forward and the reverse subtractions were cloned into the pGEM-TA plasmid. Expectedly, we observed less cDNA in the subtracted samples when compared with the corresponding unsubtracted samples (data not shown), indicating that the subtraction process was effective in eliminating the unregulated transcripts. From a total of about 15,000 colonies, we selected 150 and 200 white colonies from the forward and the reverse subtraction, respectively, which corresponded to 150 up- and 200 downregulated genes after BaP treatment. In an effort to minimize false-positive clones, DNA dot blot hybridization was performed using biotinylated probes synthesized from the subtracted cDNA to screen all clones from the forward and the reverse subtractions. Only the clones that had at least a fivefold difference in intensity between the forward and the reverse probes were selected to proceed with further characterization. Forty-one clones ranging from 350 to 1100 base pairs were then selected, identified, and characterized (Tables 1 and 2). Real-time qPCR is generally used to quantify the amount of message of a particular gene in the control and the treated cells to show up- or downregulation of a gene. Because we isolated a number of up- and downregulated transcripts in response to BaP, we wanted to validate our screening method using real-time qPCR. We expected that clones with the highest fold difference in our screening should show a clear induction at the messenger RNA level. In contrast, we might not see a substantial difference at the message level in clones that had a small fold difference in our screening because our PCR approach preferentially amplified rare transcripts that might not be easily detected using conventional methods (Diatchenko et al., 1996). We chose four clones to be analyzed by real-time qPCR: the highest and lowest fold difference of the upregulated (DKC1 and PTP4A2) and downregulated (ERGIC3 and ZAP70) genes were selected to be analyzed using the cDNA library of the control or BaP-treated Jurkat cells as the template. Corresponding primers were generated according to the Primer Bank (Spandidos et al., 2010) (Table 3). Results from our real-time qPCR experiment showed that the highest fold difference clones had a greater difference at the message level as compared with the lowest fold difference clones (Fig. 4). In addition, the up- and downregulated genes of the lowest fold difference from our screening showed a detectable difference at the message level.
TABLE 1.
Upregulated Transcripts by BaP in Jurkat Cells Obtained from Forward Subtraction (tester is BaP-treated cDNA, whereas driver is DMSO-treated cDNA)
| Categories | Regulated genes | Regulation/effect | Fold | Accession number |
| Cell growth | Sigma receptor 1 (SIGMA1) | Up/ND | 26.12 | NM_147157 |
| Small subunit processome component (UTP18) | Up/ND | 10.6 | NM_16001 | |
| GTPase activation protein-binding protein 2 (G3BP2) | Up/↑ growth | 16.94 | NM_203505 | |
| Sialidase 3 (NEU3) | Up/↑ growth | 5.37 | NM_006656 | |
| Protein tyrosine phosphatase 4A2 (PTP4A2) | Up/↑ growth | 5.06 | NM_080391 | |
| DNA repair | General transcription factor IIH2 (GTF2H2) | Up/↑ repair | 8.74 | NM_001515 |
| Ribonuclease H1 (RNASEH1) | Up/↑ repair | 5.98 | NM_002936 | |
| Gene regulation | Dyskeratosis congenita 1 (DKC1) | Up | > 68 | NM_001363 |
| Ribosomal protein L7 (RPL7) | Up | 26.35 | NM_000971 | |
| Ribosomal protein S11 (RPS11) | Up | 19.51 | NM_001015 | |
| Small nuclear ribonucleoprotein D1 (SNRPD1) | Up | 12.75 | NM_006938 | |
| Proteasome activator subunit 1 (PSME1) | Up | 11.27 | NM_006263 | |
| Bromodomain containing 3 (BRD3) | Up | 10.59 | NM_007371 | |
| Zinc finger protein 107 (ZNF107) | Up | 5.15 | NM_001013746 | |
| Metabolism | ELOVL family member 5 (ELOVL5) | Up | 23.1 | NM_021814 |
| Pyrophosphatase 2 (PPA2) | Up | 5.36 | NM_176869 | |
| ND | Chromosome 1 orf 151 (C1orf151) | Up | 9.34 | NM_001032363 |
| Stress response | Farnesyl diphosphate synthase (FDPS) | Up | 7.02 | NM_002004 |
| Hsp70 A8 (HSPA8) | Up | 12.07 | NM_006597 | |
| β-Tubulin 2C (TUBB2C) | Up | 5.78 | NM_006088 |
Note. ND, not determined.
TABLE 2.
Downregulated Transcripts by BaP in Jurkat Cells Obtained from Reverse Subtraction (tester is DMSO-treated cDNA and driver is BaP-treated cDNA)
| Categories | Regulated genes | Regulation/effect | Fold | Accession number |
| Amino acid transport | Solute carrier family 7, member 1 (CAT) | Down/↓ transport | 39.66 | NM_003045 |
| Cell growth | Squamous cell carcinoma antigen (SART1) | Down/↓ growth | > 68 | NM_005146 |
| SNF-related kinase (SNRK) | Down/↓ growth | > 68 | NM_001100594 | |
| Histone cell cycle regulation defective homolog A (HIRA) | Down/↓ growth | 22.5 | NM_003325 | |
| B-cell CLL/lymphoma 2 (BCL2) | Down/↓ growth | 10.05 | NM_000657 | |
| WD repeat domain 6 (WDR6) | Down/↑ growth | 7.42 | NM_018031 | |
| ERGIC and golgi 3 (ERGIC3) | Down/ND | > 68 | NM_015966 | |
| Ring finger protein 146 (RNF146) | Down/ND | 51.35 | NM_030963 | |
| Ring finger protein 130 (RNF130) | Down/ND | 7.43 | NM_018434 | |
| Chromosome 19 ofr 48 (C19orf48) | Down/ND | 5.38 | NM_199250 | |
| Immune response | Transcription factor 7 (TCF7) | Down/↓ response | 26.83 | NM_003202 |
| Dedicator of cytokinesis 2 (DOCK2) | Down/↓ response | 5.55 | NM_004946 | |
| Zeta-chain associated protein kinase 70 (ZAP70) | Down/↓ response | 5.19 | NM_207519 | |
| Gene regulation | XPA binding protein 2 (XAB2) | Down | 21.68 | NM_020196 |
| Eukaryotic translation elongation factor 1 delta (EEF1D) | Down | 67.57 | NM_001960 | |
| Splicing factor arg/ser 4 (SFRS4) | Down | 65.1 | NM_005626 | |
| Kelch-like ECH-associated protein 1 (Keap1) | Down | 7.49 | NM_012289 | |
| ND | Chromosome 10 orf 4 (C10orf4) | Down | 20.91 | NM_145246 |
| Hypothetical protein (LOC100129034) | Down | 9.56 | XM_001726040 | |
| Bardet-Biedl syndrome 9 (BBS9) | Down | 11.13 | NM_001033605 | |
| Protein interaction | Sushi domain containing 4 (SUSD4) | Down | 20.15 | NM_017982 |
Note. ND, not determined.
TABLE 3.
Primer sequences for real-time qPCR. All primer sequences are from Primer Bank except human Keap1 (Ohta et al., 2008), GCLC (Aguirre et al., 2007), GSTP1 (Loignon et al., 2009), and NQO1 (Loignon et al., 2009)
| Gene name | Real-time qPCR primers |
| PTP4A2 | Forward: 5′ TTTCGTGAAGAGCCAGGTTG 3′ |
| Reverse: 5′ CGTAATCGCATCTTAGGTCGG 3′ | |
| DKC1 | Forward: 5′ CCAAAGTTGCTAAGTTGGACACG 3′ |
| Reverse: 5′ TGCAAGAGGTGTATAGTGTGTTG 3′ | |
| ZAP70 | Forward: 5′ TGCCCTTCTTCTACGGCAG 3′ |
| Reverse: 5′ ACGAGCGACAGCACATAGC 3′ | |
| ERGIC3 | Forward: 5′ GGGAGGACTATCCAGGCATTG 3′ |
| Reverse: 5′ AGCTCATAGAGGACGAAGACTC 3′ | |
| Human Keap1 | Forward: 5′ TACGATGTGGAAACAGAGACGTGGACTTTCGTA 3′ |
| Reverse: 5′ TCAACAGGTACAGTTCTGCTGGTCAATCTGCTT-3′ | |
| Mouse Keap1 | Forward: 5′ TACGACGTGGAGACAGAGACCTGGACTTTCGTA 3′ |
| Reverse: 5′ TCAGCAGGTACAGTTTTGTTGATCAATTTGCTT 3′ | |
| GCLC | Forward: 5′ ATGGAGGTGCAATTAACAGAC 3′ |
| Reverse: 5′ ACTGCATTGCCACCTTTGCA 3′ | |
| GSTP1 | Forward 5′ CCCTACACCGTGGTCTATTTCC 3′ |
| Reverse 5′ GAGGCTTTGAGTGAGCCCT 3′ | |
| NQO1 | Forward 5′ TGAAGGACCCTGCGAACTTTC 3′ |
| Reverse 5′ GAACACTCGCTCAAACCAGC 3′ |
FIG. 4.
Real-time qPCR results confirming the up- and downregulated transcripts in Jurkat cells. Relative amount of transcripts with the highest fold difference (DKC1 and ERGIC3) and the lowest fold difference (PTP4A2 and ZAP70) from our screening. DKC1 and PTP4A2 are upregulated transcripts, whereas ERGIC3 and ZAP70 are downregulated transcripts. 18S was used to normalize the message amount. The error bar represents ±SD of three independent experiments [n = 3, †p > 0.05 (not significant) and ** p ≤ 0.005]. This experiment has been repeated at least once.
Determination of the Effect of BaP and 3MP-ITC on Keap1 Expression in Jurkat and Hepa1c1c7 Cells
It is particularly interesting to us that BaP caused downregulation of the Keap1 message in our SSH screening because regulation of the Keap1 function normally occurs at the protein level.
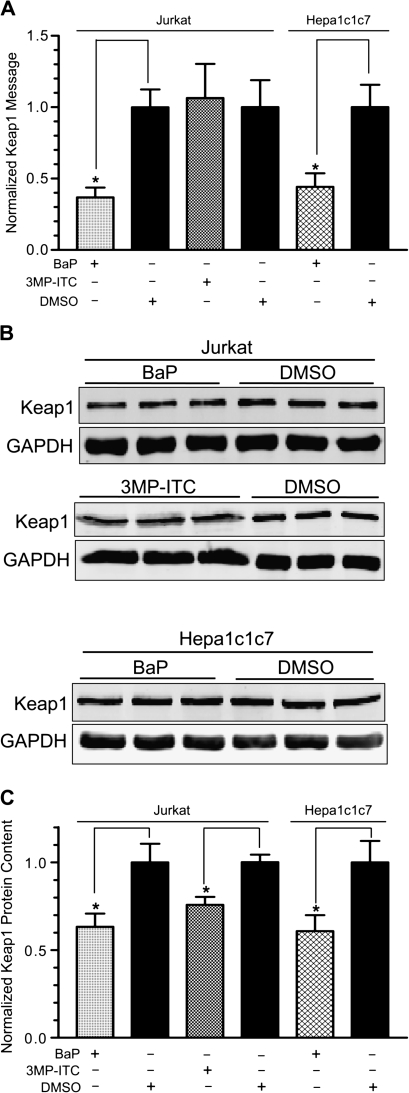
We performed real-time qPCR and quantitative Western analysis to determine whether the message and protein levels of Keap1 are altered by BaP treatment. After cells were treated with 2.5μM BaP or vehicle DMSO for 48 h at 37°C, we found that the Keap1 message was downregulated to about 40% of the controls in both Jurkat and Hepa1c1c7 cells. On the contrary, the Keap1 message was not altered after treatment with 25μM 3MP-ITC for 12 h at 37°C (Fig. 5A). However, the Keap1 protein content in the cytoplasm was significantly reduced by both BaP (60% of control in both Jurkat and Hepa1c1c7 cells) and 3MP-ITC (75% of control in Jurkat cells) (Figs. 5B and 5C), revealing that different mechanisms were involved in regulating the Keap1 protein content. It should be noted that although it may be difficult to observe a visible difference in the images presented in Fig. 5B, these Western data were quantified by the Odyssey imaging system that accurately detects the stoichiometric binding of the near infrared secondary probe to the primary antibody. Therefore, it is more informative to look at the numbers presented in Fig. 5C and to use Fig. 5B as a reference for the triplicate data.
FIG. 5.
Effect of BaP and 3MP-ITC on the Keap1 message and protein levels in Jurkat and Hepa1c1c7 cells. (A) Real-time qPCR results showing downregulation of the Keap1 message in BaP-treated Jurkat and Hepa1c1c7 cells (2.5μM BaP for 48 h), whereas there was no change in the Keap1 massage in 3MP-ITC-treated Jurkat cells (25μM 3MP-ITC for 12 h). 18S was used to normalize the message amount. The Keap1 messages in all three DMSO-treated samples were arbitrarily set as 1. The error bar represents ±SD of three independent experiments (n = 3, *p ≤ 0.05). This experiment has been repeated at least once. (B) Quantitative Western data of three separate Western experiments showing the downregulation of the Keap1 protein by BaP and 3MP-ITC. Twenty micrograms of the BaP/DMSO WCE (top and bottom panels) and 10 μg of the 3MP-ITC/DMSO WCE (middle panel) were loaded in each lane. Triplicate samples are shown with corresponding GAPDH amount. The Western on the BaP treatment in Jurkat cells has been repeated at least once. (C) A graph showing the data in (B). The amounts of the Keap1 protein were normalized with GAPDH. Amounts of the Keap1 protein in DMSO-treated cells were arbitrarily set as 1. The error bar represents ±SD of three independent experiments (n = 3, *p ≤ 0.05).
Determination of the BaP Effect on Nrf2 and Its Target Gene Expression in Jurkat Cells
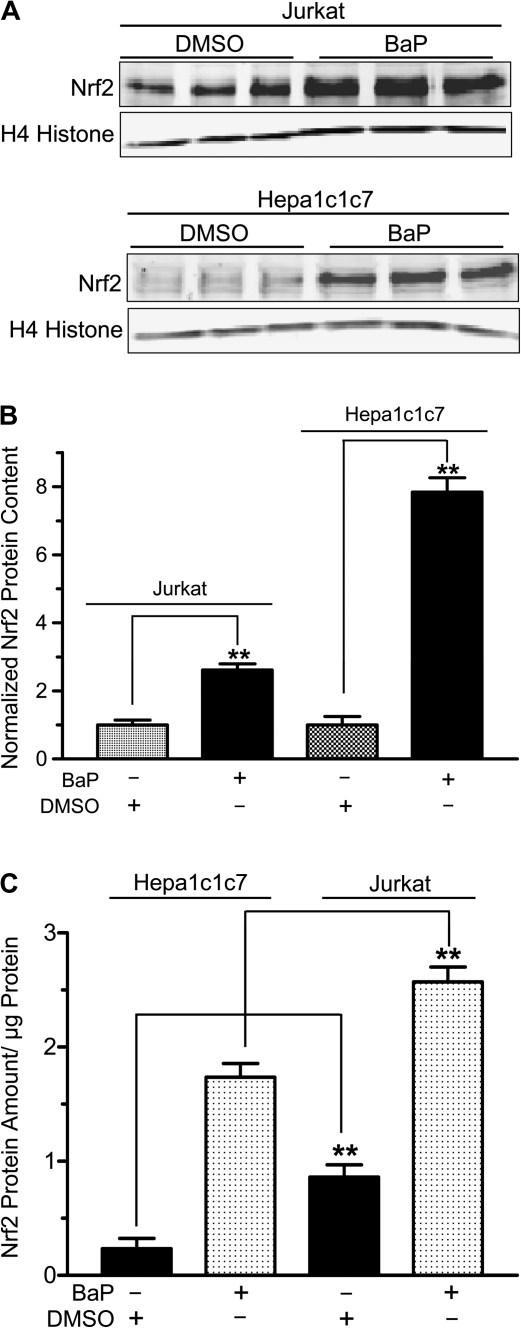
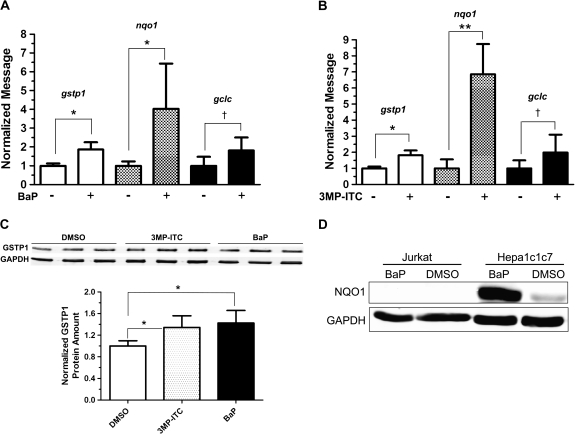
Because Keap1 protein targets Nrf2 for proteasomal degradation, we predicted that BaP should increase Nrf2 protein content in both Jurkat and Hepa1c1c7 cells. After we treated Jurkat and Hepa1c1c7 cells with 2.5μM BaP for 48 h at 37°C, we performed quantitative Western analysis and found that the nuclear Nrf2 content (normalized by the acetyl H4 histone content) increased significantly to 2.6- and 7.8-fold, respectively (Figs. 6A and 6B). In order to compare the relative amount of the Nrf2 protein in Jurkat and Hepa1c1c7 cells, the nuclear extracts were analyzed simultaneously on one gel and normalized by protein content rather than acetyl H4 histone because there might be some differences in the acetyl H4 histone expression in these two cell lines. The resulting quantitative Western data showed that the untreated Jurkat cells had 3.7 times more Nrf2 protein than Hepa1c1c7 cells. After BaP treatment, the Nrf2 protein was still significantly higher (1.5-fold) in Jurkat cells than in Hepa1c1c7 cells (Fig. 6C). Next, we examined whether this increase of the Nrf2 content would translate into target gene expression. We found that two Nrf2 target genes gstp1 and nqo1 were upregulated by two- and fourfold, respectively, at the message level by BaP in Jurkat cells (Fig. 7A). This finding was similar to the results observed in the 3MP-ITC-treated Jurkat cells, which we expected that 3MP-ITC would upregulate the Nrf2 function (Fig. 7B). However, three other Nrf2 target genes glutamate-cysteine ligase catalytic subunit (GCLC), HO-1, and ferritin H were not affected by BaP and 3MP-ITC treatments. The gclc message was detectable by real-time PCR but was not altered significantly by BaP or 3MP-ITC (Figs. 7A and 7B), whereas the HO-1 and ferritin H messages were minimal in Jurkat cells and could not be quantified (data not shown). The GSTP1 protein was consistently induced by BaP in Jurkat cells (Fig. 7C). Similar induction was observed when Jurkat cells were treated with the control 3MP-ITC (Fig. 7C), suggesting that accumulation of the nuclear Nrf2 in Jurkat cells triggers cellular response in protection against electrophiles by upregulating GSTP1 expression. On the contrary, no detectable NQO1 protein was observed in the Jurkat WCE regardless of BaP treatment (Fig. 7D), suggesting that NQO1 is not present and not inducible by BaP in Jurkat cells. To make certain that our NQO1 antibody is functional, we performed Western analysis on Hepa1c1c7 cells with and without BaP treatment and found that the NQO1 protein was pronouncedly induced by 11.7-fold when Hepa1c1c7 cells were exposed to 2.5μM BaP for 48 h at 37°C.
FIG. 6.
Effect of BaP on the Nrf2 protein content in the Jurkat and Hepa1c1c7 cell nuclei. (A) Quantitative Western data of two separate Western experiments (one for Jurkat and the other for Hepa1c1c7) in triplicate showing the amount of the nuclear Nrf2 protein after treatment with 2.5μM BaP or DMSO vehicle for 48 h in Jurkat (upper) and Hepa1c1c7 (lower) cells. Ten micrograms of nuclear extract was loaded per lane, and acetyl H4 histone was used as the loading control. (B) A graph of the data in (A). The amount of the Nrf2 protein was normalized with acetyl H4 histone. The amount of the Nrf2 protein in DMSO-treated Jurkat or Hepa1c1c7 cells was arbitrarily set as 1 to reflect the fold increase in each cell line. (C) A graph showing the relative amount of Nrf2 in Jurkat and Hepa1c1c7 cells. Cells treated with 2.5μM BaP or DMSO vehicle for 48 h and 10 μg of nuclear extract per lane were loaded on the same gel for quantitative Western analysis. For both (B) and (C), the error bar represents ±SD of three independent experiments (n = 3, **p ≤ 0.005). All experiments in this figure have been repeated at least once.
FIG. 7.
Effect of BaP on Nrf2 target genes. Real-time qPCR results showing the effect of BaP (A) or 3MP-ITC (B) on the gstp1, nqo1, and gclc messages in Jurkat cells. Cells were treated with either 2.5μM BaP (48 h), 25μM 3MP-ITC (24 h) or vehicle DMSO. The error bar represents ±SD of three independent experiments (n = 3). BaP experiment has been repeated at least once. (C) Western data showing the upregulation of GSTP1 protein by treatment of either BaP (1.4-fold) or 3MP-ITC (1.3-fold). The images above show the Western images of three independent experiments. The plot below is the quantified data showing the means with the error bars (mean ± SD, n = 3). Ten micrograms of WCE was loaded in each lane, and GAPDH was used as the loading control. (D) Western data showing NQO1 expression in Hepa1c1c7 but not in Jurkat cells. Cells were treated with 2.5μM BaP or DMSO vehicle for 48 h. Twenty micrograms of WCE was loaded in each lane, and GAPDH was used as the loading control. The GSTP1 and NQO1 Western experiments have been repeated at least once. *p ≤ 0.05, **p ≤ 0.005, †p > 0.05 (not significant).
Determination of the BaP Effect on the Viability of Jurkat and Hepa1c1c7 Cells
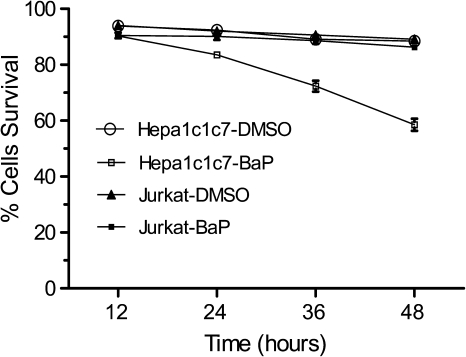
Because we know that unlike Hepa1c1c7 cells, Jurkat cells have no detectable AhR and CYP1A1 activity, we predicted that BaP would be more toxic to Hepa1c1c7 cells than to Jurkat cells. We found that when cells were treated with 2.5μM BaP, there was minimal cell death observed in Jurkat cells. On the contrary, 40% cell death was observed in Hepa1c1c7 cells in the presence of BaP (Fig. 8), suggesting that BaP metabolites and ROS might be responsible for the cell death observed in Hepa1c1c7 cells.
FIG. 8.
Effect of BaP on Jurkat and Hepa1c1c7 cell viability. Percent cell survival was determined at different time points by trypan blue staining of cells after treatment with 2.5μM BaP or DMSO (12–48 h). The error bar represents ±SD of three independent experiments (n = 3).
DISCUSSION
BaP is a well-known AhR ligand that activates the AhR signaling pathway. The BaP-bound receptor modulates the expression of target genes at the transcriptional level, which should explain in part the BaP-mediated toxicities (Nebert et al., 1993). In particular, the upregulation of the AhR-mediated CYP1 protein expression is responsible for the first step of the BaP bioactivation, which leads to DNA damage (Nebert et al., 2004). The AhR is therefore an important component of the BaP-induced gene expression, and the loss of AhR has been shown to inhibit BaP-induced carcinogenicity in mice (Shimizu et al., 2000). However, AhR-independent promotion of apoptosis has been reported for the AhR ligand TCDD in L-MAT cells (Kobayashi et al., 2009). In addition, TCDD-mediated immunotoxicity appears to be AhR independent in Jurkat and L-MAT cells (Hossain et al., 1998). Therefore, we examined the cellular responses to BaP in AhR-deficient Jurkat cells. To confirm that our Jurkat cells do not express AhR, we performed Western blot analysis to show that there is no detectable amount of AhR in them. This finding allowed us to use our Jurkat cells to investigate how AhR-independent gene regulation is affected by BaP. Other investigators had shown that 2.5μM BaP was a noncytotoxic dose for Jurkat cells up to 48-h exposure (Oh et al., 2004). Thus, we used this concentration of BaP for our study because human exposure to the environmental BaP often does not present any acute toxicity. We found that Jurkat cells that were treated with 2.5μM BaP for 48 h appeared normal and showed no sign of cell death. In addition to our finding that there was minimal CYP1A1 activity present in Jurkat cells, we were unable to detect any oxidized BaP metabolite after BaP treatment for 6 and 12 h (Fig. 2 and data not shown). These observations are in agreement with our assumption that there is minimal, if any, BaP metabolism by CYP1A1 occurring in Jurkat cells. However, we cannot conclude definitively that there is no BaP metabolite formation in Jurkat cells because these cells have measurable (but very minimal) EROD activity, and we did not examine all the possible BaP metabolites in our metabolic study.
We investigated genome-wide changes in gene expression in BaP-treated Jurkat cells using SSH PCR. SSH PCR is a powerful and yet simplistic technique to identify genes that are either up- or downregulated (Diatchenko et al., 1996). This PCR-based method selectively suppresses the amplification of unregulated sequences and equalizes for the abundance of cDNAs within a target population by the incorporation of a normalization step. Thus, this approach should enhance the probability of identifying regulation of rare (low abundance) transcripts. Therefore, we expected to identify novel transcripts that had not been reported for BaP treatment using other screening techniques, such as oligonucleotide microarray (Willinger et al., 2006). We screened approximately 7500 clones for each of the up- and downregulated populations by performing blue/white screening and DNA dot blot hybridization to minimize false-positive clones. Arbitrarily, we selected clones with at least a fivefold difference in our dot blot screening to be considered positive. Other groups had used the same cutoff for their SSH screenings (Kang et al., 1998). The 41 identified genes were categorized according to their function—cell growth, DNA repair, gene regulation, immune response, stress response, metabolism, amino acid transport, and protein interaction. It appears that in response to BaP exposure, Jurkat cells alter the machinery for transcription, RNA processing, translation, and protein degradation to suppress cell proliferation and to promote DNA repair. Specifically, 11 regulated transcripts are involved in transcription, RNA splicing, translation, and protein degradation processes, in which 7 transcripts (DKC1, RPL7, RPS11, SNRPD1, PSME1, BRD3, and ZNF107) are upregulated and 4 transcripts (XAB2, EEF1D, SFRS4, and Keap1) are downregulated. Four of them have a direct effect on ribosome assembly and protein synthesis (RPL7, SNRPD1, RPS11, and EEF1D), suggesting that modulation of active protein synthesis is a response to BaP. Genes that are involved in cell growth, DNA repair, and gene regulation contribute to 66% (27 out of 41 transcripts) of the identified transcripts. Among them, there are 14 upregulated genes (SIGMA1, PTP4A2, G3BP2, UTP18, NEU3, GTF2H2, RNASEH1, DKC1, RPL7, RPS11, SNRPD1, PSME1, BRD3, and ZNF107) and 13 downregulated genes (HIRA, SART1, WDR6, BCL2, ERGIC3, RNF130, SNRK, RNF146, C19orf48, XAB2, EEF1D, SFRS4, and Keap1). Although there are similar numbers of transcripts for genes that either promote or suppress growth, the two transcripts SART1 and SNRK that have the highest fold increase (> 68-fold) suppress growth. In addition, two DNA repair–implicated transcripts GTP2H2 (Vonarx et al., 2006) and RNASEH1 (Cerritelli and Crouch, 2009) are upregulated, suggesting that BaP may trigger DNA repair and slow down cell growth in Jurkat cells.
It has been reported that BCL2 and TCF7 are downregulated in response to TCDD (Hossain et al., 1998) and cigarette smoke (Parsanejad et al., 2008), respectively. Results from our screening studies also showed that these two transcripts are downregulated by BaP. Downregulation of the histone regulator HIRA has been shown to suppress cell growth by suppressing the BCL2 gene transcription (Ahmad et al., 2005); this finding is consistent with our observation that both HIRA and BCL2 transcripts are downregulated in our BaP-treated cells. In addition, the TCF7 transcript is downregulated in our screening by more than 26-fold, suggesting that the BaP in the cigarette smoke may cause the downregulation of TCF7.
Downregulation of the Keap1 transcript is surprising because the regulation of the Keap1 function is thought to occur at the protein level. There is much evidence showing that the Keap1 protein is sensitive to modification by ROS and reactive electrophiles and is the key protein to target Nrf2 for proteasomal degradation (Kaspar et al., 2009). Naturally, the amount of the Keap1 protein present in the cell would directly affect the half-life of Nrf2. To our knowledge, there is no evidence in the literature at present suggesting that the amount of the Keap1 protein can be downregulated at the transcriptional level, although it has been reported that arsenic upregulates the Keap1 message and protein (Pi et al., 2003). Here, we clearly showed that the Keap1 message is downregulated by BaP in both Jurkat and Hepa1c1c7 cells, and the amount of the Keap1 protein is subsequently reduced, leading to more Nrf2 present in the nucleus. This Nrf2 regulation by BaP is distinct from the mechanism of Nrf2 upregulation by 3MP-ITC. In the latter case, there is no change of the Keap1 message. The Keap1 message is expected to be unchanged in the case of 3MP-ITC because the isothiocyanate moiety of 3MP-ITC may serve as an electrophile, which possibly allows alkylation of the Keap1 cysteine residues, leading to the release of Nrf2 for function. When we treated Hepa1c1c7 cells with BaP, we observed the formation of ROS and BaP metabolites, which in turn might modify the Keap1 protein. Thus, the downregulation of the Keap1 protein by BaP in Hepa1c1c7 cells can very well be caused, in part, by modification of the Keap1 protein. In the case of Jurkat cells, it appears more straightforward: BaP causes a decrease of the Keap1 protein by downregulating the Keap1 message, and this process does not involve ROS or the formation of the oxidized BaP metabolites (as redox cycling molecules or electrophiles). We observed that this downregulation of the Keap1 protein in Jurkat cells by 2.5μM BaP occurred as early as 12 h after treatment (data not shown), and the amount of the Keap1 protein was suppressed to 72% of the vehicle control after 24 h of BaP treatment. Lack of ROS and BaP metabolism in Jurkat cells is probably the reason for no observable cell death, in contrast to apparent cell death in Hepa1c1c7 cells after treatment with 2.5μM BaP for up to 48 h. Downregulation of the Keap1 message is likely caused by BaP but not its metabolites in Jurkat cells. However, we cannot rule out the possibility that the minute amount of a BaP metabolite in Jurkat cells is responsible for the Keap1 regulation.
Interestingly, the amount of the nuclear Nrf2 protein after BaP treatment is significantly more in Jurkat than in Hepa1c1c7 cells (Fig. 6C). We were able to detect the BaP-induced NQO1 protein expression in Hep1c1c7 cells, showing that the increased amount of Nrf2 induces NQO1 expression. However, no NQO1 protein was detected in BaP-treated Jurkat cells, even though NQO1 was clearly induced at the message level. Other researchers had reported that NQO1 was inducible in Jurkat cells by t-BHQ; however, this induction was only examined at the message level (Sakamoto et al., 2009). We were unable to detect the NQO1 protein in Jurkat cells up to 200 μg, with or without BaP treatment. As a comparison, as little as 2 μg of MCF-7 WCE gave a definite NQO1 band (data not shown). In addition, absolutely no NQO1 activity was detected up to 200 μg of the Jurkat lysate, whereas 1 μg of the Hepa1c1c7 lysate gave measureable NQO1 activity (data not shown). We believe that Jurkat cells do not express the NQO1 protein. More importantly, accumulation of the nuclear Nrf2 protein appears to induce the gstp1 gene expression in Jurkat cells, and this induction does not involve ROS and any reactive electrophile. It is tempting to speculate that there might be a feedback mechanism to limit the amount of the Nrf2 protein accumulation by increasing the NQO1 activity, which subsequently lowers the stimuli that cause the Keap1 protein modification. Therefore, the lack of NQO1 expression in Jurkat cells may allow a higher amount of the Nrf2 protein accumulation.
Although it is clear that the net effect of BaP is carcinogenic but not cancer preventive, the downregulation of the Keap1 message by BaP reveals a novel mechanism in regulating the Nrf2 function, which has not been previously reported. Regulation of the Keap1 gene transcription may very well be playing a vital role in controlling the cellular Nrf2 function. This regulation must be explored to fully understand our cellular response to oxidative stress and bioactivated molecules.
FUNDING
National Institutes of Health (R01 ES014050).
References
- Adachi T, Nakagawa H, Chung I, Hagiya Y, Hoshijima K, Noguchi N, Kuo MT, Ishikawa T. Nrf2-dependent and -independent induction of ABC transporters ABCC1, ABCC2, and ABCG2 in HepG2 cells under oxidative stress. J. Exp. Ther. Oncol. 2007;6:335–348. [PubMed] [Google Scholar]
- Aguirre P, Valdes P, Aracena-Parks P, Tapia V, Nunez MT. Upregulation of gamma-glutamate-cysteine ligase as part of the long-term adaptation process to iron accumulation in neuronal SH-SY5Y cells. Am. J. Physiol. Cell Physiol. 2007;292:C2197–C2203. doi: 10.1152/ajpcell.00620.2006. [DOI] [PubMed] [Google Scholar]
- Ahmad A, Kikuchi H, Takami Y, Nakayama T. Different roles of N-terminal and C-terminal halves of HIRA in transcription regulation of cell cycle-related genes that contribute to control of vertebrate cell growth. J. Biol. Chem. 2005;280:32090–32100. doi: 10.1074/jbc.M501426200. [DOI] [PubMed] [Google Scholar]
- Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Alexandrov K, Cascorbi I, Rojas M, Bouvier G, Kriek E, Bartsch H. CYP1A1 and GSTM1 genotypes affect benzo[a]pyrene DNA adducts in smokers’ lung: comparison with aromatic/hydrophobic adduct formation. Carcinogenesis. 2002;23:1969–1977. doi: 10.1093/carcin/23.12.1969. [DOI] [PubMed] [Google Scholar]
- Baird WM, Hooven LA, Mahadevan B. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ. Mol. Mutagen. 2005;45:106–114. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- Bostrom CE, Gerde P, Hanberg A, Jernstrom B, Johansson C, Kyrklund T, Rannug A, Tornqvist M, Victorin K, Westerholm R. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health. Perspect. 2002;110(Suppl. 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab. Dispos. 2009;37:847–856. doi: 10.1124/dmd.108.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conney AH, Chang RL, Jerina DM, Wei SJ. Studies on the metabolism of benzo[a]pyrene and dose-dependent differences in the mutagenic profile of its ultimate carcinogenic metabolite. Drug Metab. Rev. 1994;26:125–163. doi: 10.3109/03602539409029788. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato MT, Gomez-Lechon MJ, Castell JV. A microassay for measuring cytochrome P450IA1 and P450IIB1 activities in intact human and rat hepatocytes cultured on 96-well plates. Anal. Biochem. 1993;213:29–33. doi: 10.1006/abio.1993.1381. [DOI] [PubMed] [Google Scholar]
- Ellard S, Mohammed Y, Dogra S, Wolfel C, Doehmer J, Parry JM. The use of genetically engineered V79 Chinese hamster cultures expressing rat liver CYP1A1, 1A2 and 2B1 cDNAs in micronucleus assays. Mutagenesis. 1991;6:461–470. doi: 10.1093/mutage/6.6.461. [DOI] [PubMed] [Google Scholar]
- Faustman EM, Omenn GS. Casaret and Doull's Toxicology: The Basic Science of Poisons. New York, NY: McGraw-Hill; 2008. [Google Scholar]
- Hattemer-Frey HA, Travis CC. Benzo-a-pyrene: environmental partitioning and human exposure. Toxicol. Ind. Health. 1991;7:141–157. doi: 10.1177/074823379100700303. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem. Soc. Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- Hossain A, Tsuchiya S, Minegishi M, Osada M, Ikawa S, Tezuka FA, Kaji M, Konno T, Watanabe M, Kikuchi H. The Ah receptor is not involved in 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated apoptosis in human leukemic T cell lines. J. Biol. Chem. 1998;273:19853–19858. doi: 10.1074/jbc.273.31.19853. [DOI] [PubMed] [Google Scholar]
- Hu L, Miao W, Loignon M, Kandouz M, Batist G. Putative chemopreventive molecules can increase Nrf2-regulated cell defense in some human cancer cell lines, resulting in resistance to common cytotoxic therapies. Cancer Chemother. Pharmacol. 2009 doi: 10.1007/s00280-009-1182-7. Advance Access published on November 26, 2009; doi: 10.1007/s00280-009-1182-7. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- Jensen KA, Luu TC, Chan WK. A truncated Ah receptor blocks the hypoxia and estrogen receptor signaling pathways: a viable approach for breast cancer treatment. Mol. Pharm. 2006;3:695–703. doi: 10.1021/mp0600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DC, LaFrance R, Su ZZ, Fisher PB. Reciprocal subtraction differential RNA display: an efficient and rapid procedure for isolating differentially expressed gene sequences. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13788–13793. doi: 10.1073/pnas.95.23.13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum YS, Chang PP, Kwon KH, Yuan X, Li W, Hu L, Kong AN. 3-Morpholinopropyl isothiocyanate is a novel synthetic isothiocyanate that strongly induces the antioxidant response element-dependent Nrf2-mediated detoxifying/antioxidant enzymes in vitro and in vivo. Carcinogenesis. 2008;29:594–599. doi: 10.1093/carcin/bgm208. [DOI] [PubMed] [Google Scholar]
- Knize MG, Salmon CP, Pais P, Felton JS. Food heating and the formation of heterocyclic aromatic amine and polycyclic aromatic hydrocarbon mutagens/carcinogens. Adv. Exp. Med. Biol. 1999;459:179–193. doi: 10.1007/978-1-4615-4853-9_12. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Ahmed S, Ishida M, Kasai S, Kikuchi H. Calcium/calmodulin signaling elicits release of cytochrome c during 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced apoptosis in the human lymphoblastic T-cell line, L-MAT. Toxicology. 2009;258:25–32. doi: 10.1016/j.tox.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Loignon M, Miao W, Hu L, Bier A, Bismar TA, Scrivens PJ, Mann K, Basik M, Bouchard A, Fiset PO, et al. Cul3 overexpression depletes Nrf2 in breast cancer and is associated with sensitivity to carcinogens, to oxidative stress, and to chemotherapy. Mol. Cancer Ther. 2009;6:2432–2440. doi: 10.1158/1535-7163.MCT-08-1186. [DOI] [PubMed] [Google Scholar]
- Matkar SS, Wrischnik LA, Hellmann-Blumberg U. Production of hydrogen peroxide and redox cycling can explain how sanguinarine and chelerythrine induce rapid apoptosis. Arch. Biochem. Biophys. 2008;477:43–52. doi: 10.1016/j.abb.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Miller KP, Ramos KS. Impact of cellular metabolism on the biological effects of benzo[a]pyrene and related hydrocarbons. Drug Metab. Rev. 2001;33:1–35. doi: 10.1081/dmr-100000138. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Puga A, Vasiliou V. Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann. N.Y. Acad. Sci. 1993;685:624–640. doi: 10.1111/j.1749-6632.1993.tb35928.x. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Im H, Oh E, Lee J, Khim JY, Mun J, Kim Y, Lee E, Kim J, Sul D. Effects of benzo[a]pyrene on protein expression in Jurkat T-cells. Proteomics. 2004;4:3514–3526. doi: 10.1002/pmic.200400981. [DOI] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Parsanejad R, Fields WR, Morgan WT, Bombick BR, Doolittle DJ. The time course of expression of genes involved in specific pathways in normal human bronchial epithelial cells following exposure to cigarette smoke. Exp. Lung Res. 2008;34:513–530. doi: 10.1080/01902140802271826. [DOI] [PubMed] [Google Scholar]
- Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat. Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Pi J, Bai Y, Reece JM, Williams J, Liu D, Freeman ML, Fahl WE, Shugar D, Liu J, Qu W, et al. Molecular mechanism of human Nrf2 activation and degradation: role of sequential phosphorylation by protein kinase CK2. Free Radic. Biol. Med. 2007;42:1797–1806. doi: 10.1016/j.freeradbiomed.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi J, Qu W, Reece JM, Kumagai Y, Waalkes MP. Transcription factor Nrf2 activation by inorganic arsenic in cultured keratinocytes: involvement of hydrogen peroxide. Exp. Cell Res. 2003;290:234–245. doi: 10.1016/s0014-4827(03)00341-0. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Iwasaki K, Sugiyama H, Tsuji Y. Role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription and associated histone modifications. Mol. Biol. Cell. 2009;20:1606–1617. doi: 10.1091/mbc.E08-07-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- Shimada T, Sugie A, Yamada T, Kawazoe H, Hashimoto M, Azuma E, Nakajima T, Inoue K, Oda Y. Dose-response studies on the induction of liver cytochromes P4501A1 and 1B1 by polycyclic aromatic hydrocarbons in arylhydrocarbon-responsive C57BL/6J mice. Xenobiotica. 2003;33:957–971. doi: 10.1080/0049825031000140896. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Nakatsuru Y, Ichinose M, Takahashi Y, Kume H, Mimura J, Fujii-Kuriyama Y, Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U. S. A. 2000;97:779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D9. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarx EJ, Tabone EK, Osmond MJ, Anderson HJ, Kunz BA. Arabidopsis homologue of human transcription factor IIH/nucleotide excision repair factor p44 can function in transcription and DNA repair and interacts with AtXPD. Plant J. 2006;46:512–521. doi: 10.1111/j.1365-313X.2006.02705.x. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willinger T, Freeman T, Herbert M, Hasegawa H, McMichael AJ, Callan MF. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J. Immunol. 2006;176:1439–1446. doi: 10.4049/jimmunol.176.3.1439. [DOI] [PubMed] [Google Scholar]