Abstract
Background
Recently, production of 16S rRNA methylases by Gram-negative bacilli has emerged as a novel mechanism for high-level resistance to aminoglycosides by these organisms in a variety of geographic locations. Therefore, the spread of high-level aminoglycoside resistance determinants has become a great concern.
Methods
Between January 2006 and July 2008, 680 distinct Escherichia coli clinical isolates were collected from a teaching hospital in Wenzhou, China. PCR and DNA sequencing were used to identify 16S rRNA methylase and extended-spectrum β-lactamase (ESBL) genes, including armA and rmtB, and in situ hybridization was performed to determine the location of 16S rRNA methylase genes. Conjugation experiments were subsequently performed to determine whether aminoglycoside resistance was transferable from the E. coli isolates via 16S rRNA methylase-bearing plasmids. Homology of the isolates harboring 16S rRNA methylase genes was determined using pulse-field gel electrophoresis (PFGE).
Results
Among the 680 E. coli isolates, 357 (52.5%), 346 (50.9%) and 44 (6.5%) isolates were resistant to gentamicin, tobramycin and amikacin, respectively. Thirty-seven of 44 amikacin-resistant isolates harbored 16S rRNA methylase genes, with 36 of 37 harboring the rmtB gene and only one harboring armA. The positive rates of 16S rRNA methylase genes among all isolates and amikacin-resistant isolates were 5.4% (37/680) and 84.1% (37/44), respectively. Thirty-one isolates harboring 16S rRNA methylase genes also produced ESBLs. In addition, high-level aminoglycoside resistance could be transferred by conjugation from four rmtB-positive donors. The plasmids of incompatibility groups IncF, IncK and IncN were detected in 34, 3 and 3 isolates, respectively. Upstream regions of the armA gene contained ISCR1 and tnpU, the latter a putative transposase gene,. Another putative transposase gene, tnpD, was located within a region downstream of armA. Moreover, a transposon, Tn3, was located upstream of the rmtB. Nineteen clonal patterns were obtained by PFGE, with type H representing the prevailing pattern.
Conclusion
A high prevalence of plasmid-mediated rmtB gene was found among clinical E. coli isolates from a Chinese teaching hospital. Both horizontal gene transfer and clonal spread were responsible for the dissemination of the rmtB gene.
Background
Aminoglycosides are clinically effective agents for treating a broad range of life-threatening infections caused by Gram-negative pathogens, usually in combination with β-lactam agents. However, increasing resistance to aminoglycosides is becoming a serious clinical problem [1-3]. Resistance to aminoglycosides is frequently due to the acquisition of modifying enzymes that vary in their substrate ranges, such as acetyltransferases, phosphorylases and adenylyltransferases [1,3]. Unlike modifying enzymes, 16S rRNA methylases have recently emerged as a novel mechanism for high-level resistance to all 4, 6-disubstituted deoxystreptamine aminoglycosides, such as arbekacin, amikacin, tobramycin, and gentamicin [4]. To date, six plasmid-encoded 16S rRNA methylases, including ArmA, RmtA, RmtB, RmtC , RmtD and NpmA, have been identified in clinical isolates of Gram-negative bacilli from multiple geographic locations [4-10]. Escherichia coli producing plasmid-mediated isolates extended-spectrum β lactamases (ESBLs) conferring to resistance to third-generation cephalosporins are mainly associated with hospital-acquired infections [11], but plasmid-mediated ArmA, RmtB and NpmA 16S rRNA methylases have been found in E. coli as well [5,10]. In China, although 16S rRNA methylases have been found among E. coli isolates from animals and humans [12-14], epidemiological data regarding 16S rRNA methylase production in E. coli clinical isolates are lacking. The aim of this study was to investigate the occurrence of 16S rRNA methylase genes in E. coli clinical isolates from a teaching hospital in Wenzhou, China.
Methods
Clinical isolates
Between January 2006 and July 2008, 680 non-duplicate E. coli clinical isolates were consecutively collected from a teaching hospital in Wenzhou, China. The clinical isolates were confirmed as E. coli using Gram stain and GNI cards on the VITEK-60 system (bioMe'rieux, Marcy l'E' toile, France). As plasmid-mediated 16S rRNA methylases can confer high-level resistance to most clinically important aminoglycosides, the isolates with concomitant resistance to gentamicin, amikacin, and tobramycin based on the criteria following the Clinical and Laboratory Standards Institute (CLSI) [15] were first screened for potential 16S rRNA methylase producers.
Antimicrobial susceptibility testing
Initial antimicrobial susceptibilities were determined by using GNS cards on the VITEK-60 system (bioMe'rieux, Marcy l'E' toile, France). The antimicrobial susceptibilities of amikacin-, tobramycin-, or gentamicin-resistant isolates were further determined by the disk diffusion test using commercial disks for cefotaxime, ceftazidime, ciprofloxacin, levofloxacin, cefoxitin, imipenem, tetracycline and trimethoprim/sulfamethoxazole according to the criteria recommended by the CLSI [15]. MICs of amikacin, tobramycin, and gentamicin were further determined by the agar dilution method in accordance with the CLSI guidelines [15]. E. coli ATCC 25922 was used as quality control strain for antimicrobial susceptibility testing.
Extraction of total DNA and plasmid DNA
Total DNA was extracted by boiling. Briefly, a fresh bacterial colony was suspended in 150 μL of sterile distilled water and boiled at 100°C for 10 min. After centrifugation at 15000 rpm for 15 min at 4°C, the supernatant was removed and stored at -20°C for further assays. Plasmid DNA of the donors, transconjugants and transformants was extracted with the QIAGEN (Hilden, Germany) Plasmid Midi kit according to the manufacturer's instructions. Plasmid size was estimated as previously described [16]. E. coli V517 and E. coli J53 containing plasmid R1 were used as a standard for plasmid size.
Screening for 16S rRNA methylase genes
The armA, rmtA, rmtB, rmtC, rmtD , and npmA genes were detected by PCR with total DNA from potential 16S rRNA methylase producers and a series of primers as described previously [5,6,8-10]. The Klebsiella pneumoniae F25 harboring both armA and rmtB described in our previous study [17], was used as positive control in every test for detecting 16S rRNA methylase genes. All PCR products were directly sequenced on an ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster City, CA). The nucleotide sequences were analyzed with software available over the Internet http://www.ncbi.nlm.nih.gov/.
β lactamase characterization
All 16S rRNA methylase gene-positive isolates were tested for ESBL production by the CLSI-recommended confirmatory double disk combination test [15]. K. pneumoniae ATCC 700603 was used for the ESBL positive control. PCR was performed for the detection of β-lactamase genes in 16S rRNA methylase gene-positive isolates with the previously reported oligonucleotide primers, including blaTEM, blaSHV and blaCTX-M [18]. All PCR products were directly sequenced, and the sequences were compared with those in the GenBank nucleotide database at http://www.ncbi.nlm.nih.gov/blast/.
Transfer of 16S rRNA methylase genes
To determine if aminoglycoside resistance was transferable in E. coli isolates with 16S rRNA methylase-bearing plasmids, a conjugation experiment was carried out in Luria-Bertani broth with E. coli J53 as the recipient as previously described [19]. Transconjugants were selected on tryptic soy agar plates containing sodium azide (100 μg/mL) for counterselection and amikacin (30 μg/mL) for plasmid-mediated aminoglycoside resistance selection. The plasmid extracts were electroporated into E. coli DH5α by using a Gene Pulser II apparatus (Bio-Rad, Hercules, CA), and the transformants were selected on Luria-Bertani agar plates containing amikacin (30 μg/mL).
PCR-based replicon typing
All obtained transconjugants and transformants were subjected to typing by a PCR method described previously based on replicons of the major plasmid incompatibility groups among Enterobacteriaceae [20]. The plasmid DNA from the transconjugants and transformants was amplified by five multiplex and three simplex PCRs using 18 pairs of primers that recognized Inc replicons FIA, FIB, FIC, HI1, HI2, I1-I, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA.
Southern hybridization
Southern hybridization was performed by standard methods with armA-and rmtB-specific digoxigenin (DIG)-labeled probes formed by the PCR DIG detection system (Roche Diagnostics GmbH). Briefly, plasmid DNA was subjected to electrophoresis in 0.7% agarose gels. After depurination, denaturation, and neutralization of the agarose gels, DNA was transferred to a positive-charged nylon membrane by capillary action. Plasmid DNA was hybridized successively with armA-specific and rmtB-specific DIG-labeled probes at 42°C under highly stringent conditions.
Determination of flanking regions of the armA gene and the rmtB gene
To investigate the genetic environments of the armA and rmtB genes, the 54-kb plasmid of E31 transformant harboring the armA gene (pE31) and the other 54-kb plasmid of E35 transformant harboring the rmtB gene (pE35) were selected for DNA sequencing. Purified armA-and rmtB-bearing plasmids were directly sequenced by using a series of outwardly-directed primers specific to the locations near to the armA gene and the rmtB gene.
PFGE
Chromosomal DNA was prepared from all 16S rRNA methylase gene-positive isolates and digested with 40 U XbaI (New England Biolabs, Beverly, MA). Electrophoresis was performed on 1.0% agarose gels in 0.5 M Tris/borate/EDTA buffer on a CHEF-Mapper XA PFGE system (Bio-Rad, Hercules, CA) for 22 h at 14°C, with run conditions of 6 V/cm, a pulse angle of 120° and pulse times from 5 to 20 s. A λDNA ladder (Amersham Biosciences, Piscataway, NJ) was used as molecular mass marker, and bands were stained with ethidium bromide (0.5 μg/mL) and photographed under UV light. Band profiles were interpreted by the criteria described previously [21]. The patterns with a difference of no more than three bands were considered to belong to the same type.
Ethical approval was not needed for the present study
Results
Prevalence of 16S rRNA methylase genes and antimicrobial susceptibility
Among tested 680 E. coli isolates, 357 (52.5%), 346 (50.9%) and 44 (6.5%)isolates were resistant to gentamicin, tobramycin and amikacin, respectively. All 44 amikacin-resistant isolates were concomitantly resistant to tobramycin and gentamicin. Thirty-seven of 44 (84.1%) amikacin-resistant isolates were positive for 16S rRNA methylase genes, among which 36 isolates were positive for rmtB gene and only one isolate was positive for armA gene. The armA and rmtB amplicons showed 100% identity with armA in K. pneumoniae BM4536 (GenBank AY220558) and rmtB in Serratia marcescens S-95 (GenBank AB103506). The rmtA, rmtC, rmtD, and npmA genes were not detected in any of the isolates tested. All 16S rRNA methylase gene-positive isolates were highly resistant to gentamicin, amikacin and tobramycin (MICs, ≥256 μg/mL) (Table 1). The positive rates of 16S rRNA methylase genes among overall isolates and amikacin-resistant isolates were 5.4% (37/680) and 84.1% (37/44), respectively. All 37 isolates harboring 16S rRNA methylase genes were resistant to ciprofloxacin, levofloxacin and trimethoprim/sulfamethoxazole but susceptible to imipenem. The sites of infection or colonization by 16S rRNA methylase gene-positive isolates from 37 patients were as follows: urine, 16 patients; pus, 10 patients; blood, 9 patients; sputum, 1 patient; and ascites, 1 patient. (Table 1).
Table 1.
characteristics of the 37 E. coli isolates harboring 16S rRNA methylase genes and their transconjugants and transformants
| Organisma | Origin | Wardb | PFGE | MIC (μg/ml)c | rmtB | armA | ESBL | Inc group | TEM | CTX-M | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | AMK | TOB | ||||||||||
| E1 | Urine | 11 | A | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-14 | |
| E1T | >512 | 512 | 512 | + | + | F | TEM-1 | CTX-M-14 | ||||
| E2 | Pus | 15 | B | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E2T | 512 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E5 | Urine | 28 | C | >512 | 512 | 256 | + | - | + | TEM-1 | CTX-M-15' | |
| E5T | >512 | 256 | 256 | + | - | + | N | TEM-1 | CTX-M-15' | |||
| E6 | Urine | 11 | D | >512 | 512 | >512 | + | - | + | TEM-1 | CTX-M-15 | |
| E6T | 256 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-15 | |||
| E7 | Pus | 15 | E | >512 | >512 | >512 | + | - | - | TEM-1 | - | |
| E7T | 512 | 256 | 256 | + | - | - | F | TEM-1 | - | |||
| E11 | Blood | 28 | F | >512 | 512 | >512 | + | - | - | TEM-1 | - | |
| E11T | >512 | 512 | 512 | + | - | - | F | TEM-1 | - | |||
| E17 | Urine | 10 | G | >512 | >512 | 512 | + | - | + | TEM-1 | CTX-M-14/27 | |
| E17T | >512 | 512 | 512 | + | - | + | N | TEM-1 | CTX-M-14/27 | |||
| E20 | Blood | 17 | H | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-15 | |
| E20C | >512 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-15 | |||
| E21 | Urine | 11 | H | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-24 | |
| E21T | >512 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-24 | |||
| E23 | Urine | 9 | H | >512 | 512 | >512 | + | - | + | TEM-1 | CTX-M-14 | |
| E23T | 512 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E24 | Urine | 11 | H | 512 | 256 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E24T | 512 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E25 | Blood | 4 | H | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E25T | 512 | 512 | 512 | + | - | + | F | TEM-1 | CTX-M-14/15 | |||
| E26 | Pus | 7 | I | >512 | 512 | 512 | + | - | - | TEM-1 | CTX-M-14 | |
| E26T | 512 | 256 | 256 | + | - | - | F | TEM-1 | CTX-M-14 | |||
| E27 | Pus | 8 | I | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E27T | >512 | 512 | 512 | + | - | + | F | TEM-1 | CTX-M-14/15 | |||
| E28 | Urine | 11 | J | 512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E28T | 256 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E29 | Urine | 4 | H | >512 | 512 | >512 | + | - | + | TEM-1 | CTX-M-14/27 | |
| E29T | >512 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-14/27 | |||
| E30 | Blood | 1 | A | >512 | 512 | >512 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E30C | 512 | 256 | 256 | + | - | + | F, K | TEM-1 | CTX-M-14/15 | |||
| E31 | Urine | 15 | K | >512 | >512 | >512 | - | + | + | TEM-1 | CTX-M-14 | |
| E31T | >512 | 256 | 512 | - | + | + | F | TEM-1 | CTX-M-14 | |||
| E32 | Blood | 57 | L | >512 | 256 | 512 | + | - | - | TEM-1 | CTX-M-15 | |
| E32T | >512 | 256 | 256 | + | - | - | F | TEM-1 | CTX-M-15 | |||
| E33 | Pus | 21 | H | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E33T | 512 | 256 | 256 | + | - | + | F, K | TEM-1 | CTX-M-14 | |||
| E34 | Blood | 26 | A | >512 | 512 | >512 | + | - | + | TEM-1 | CTX-M-14 | |
| E34T | >512 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E35 | Pus | 11 | M | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E35T | 512 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E36 | Urine | 11 | N | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-3 | |
| E36T | >512 | 512 | 512 | + | - | + | F | TEM-1 | CTX-M-3 | |||
| E37 | Urine | 15 | O | >512 | 512 | 512 | + | - | - | TEM-1 | - | |
| E37T | 512 | 256 | 256 | + | - | - | F | TEM-1 | - | |||
| E38 | Urine | 28 | N | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E38T | >512 | >512 | >512 | + | - | + | F | TEM-1 | CTX-M-14/15 | |||
| E39 | Urine | 56 | P | 512 | 256 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E39T | 256 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E40 | Pus | 19 | Q | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E40C | 512 | 256 | 256 | + | - | + | N | TEM-1 | CTX-M-14 | |||
| E41 | Blood | 2 | R | 512 | 256 | 512 | + | - | - | TEM-1 | - | |
| E41T | 512 | 256 | 256 | + | - | - | F | TEM-1 | - | |||
| E42 | Pus | 7 | H | >512 | >512 | >512 | + | - | + | TEM-1 | CTX-M-14 | |
| E42T | >512 | 512 | >512 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E43 | Pus | 15 | P | 512 | 512 | 256 | + | - | + | TEM-1 | CTX-M-27 | |
| E43T | 512 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-27 | |||
| E44 | Urine | 9 | H | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E44T | >512 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14/15 | |||
| E45 | Urine | 11 | G | 512 | 256 | 256 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E45T | 256 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14/15 | |||
| E46 | Sputum | 54 | H | >512 | 256 | 512 | + | - | - | TEM-1 | - | |
| E46T | 512 | 256 | 256 | + | - | - | F | TEM-1 | - | |||
| E47 | Blood | 25 | S | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14/15 | |
| E47T | >512 | 256 | 512 | + | - | + | F | TEM-1 | CTX-M-14/15 | |||
| E48 | Blood | 9 | N | >512 | 512 | >512 | + | - | + | TEM-1 | CTX-M-14 | |
| E48C | >512 | 256 | 512 | + | - | + | F, K | TEM-1 | CTX-M-14 | |||
| E49 | Pus | 7 | H | >512 | 512 | 512 | + | - | + | TEM-1 | CTX-M-14 | |
| E49T | >512 | 256 | 256 | + | - | + | F | TEM-1 | CTX-M-14 | |||
| E51 | Ascites | 6 | H | >512 | 512 | 512 | + | - | - | TEM-1 | - | |
| E51T | 512 | 256 | 256 | + | - | - | F | TEM-1 | - | |||
a; T, transformant; C, transconjugant b;11, 15, 28, 10, 17, 9, 4, 7, 8, 1, 57, 21, 26, 56, 19, 2, , 54, 25 and 6 represent different clinical units. C; GEN, gentamicin; AMK, amikacin; TOB, tobramycin
β lactamase characterization
Among 37 isolates harboring 16S rRNA methylase genes, 29 isolates (78.4%, 29/37) were found to coproduce ESBLs. Thirty-one isolates were found to harbor blaCTX-M, among which 24, 11, 3, 1 and 1 isolates were positive for blaCTX-M-14, blaCTX-M-15, blaCTX-M-27, blaCTX-M-3 and blaCTX-M-24, respectively. Seven of 31 blaCTX-M-positive isolates harbored both blaCTX-M-14 and blaCTX-M-15, and two isolates harbored both blaCTX-M-27 and blaCTX-M-14 (Table 1). All 16S rRNA methylase gene-positive isolates were positive for blaTEM, and all blaTEM type genes were identified as the narrow spectrum β-lactamase gene, blaTEM-1.
Transfer of aminoglycoside resistance and plasmid analysis
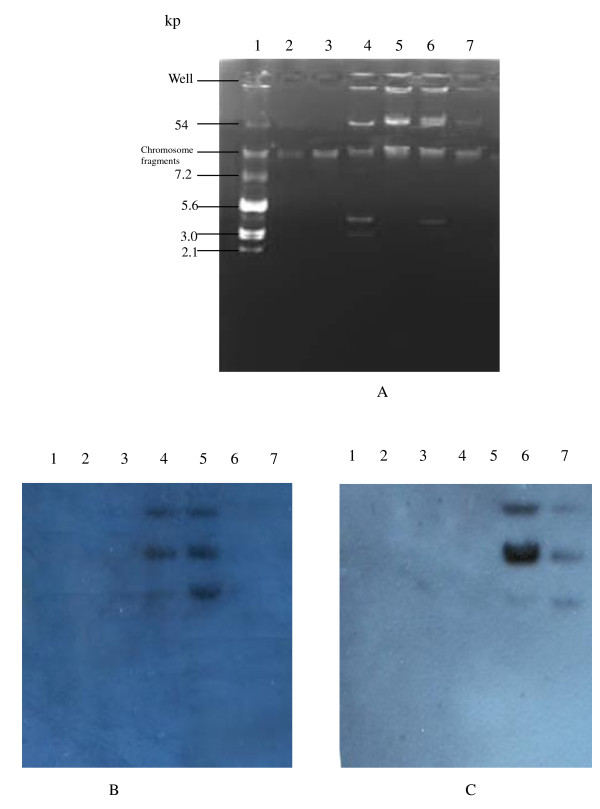
High-level aminoglycoside resistance could be transferred by conjugation with frequencies between 10-4 and 10-6 only from four rmtB-positive donors through repeat attempts including using filter mating. The plasmids of the remaining 33 isolates with 16S rRNA methylase genes were transferred into the recipients, E. coli DH5α, by transformation. The transconjugants and transformants were highly resistant to amikacin, gentamicin and tobramycin (MICs, ≥256 μg/mL). The results of PCR and sequence analyses revealed that blaTEM-1, blaCTX-M-14 and blaCTX-M-15 were co-transferred with rmtB on plasmids to the recipients (Table 1). The plasmids of incompatibility groups IncF, IncK and IncN were detected in 34, 3 and 3 isolates, respectively, among which both IncF and IncK were detected in three isolates. The armA-positive (E. coli E31) and rmtB-positive (E. coli E35) isolates were selected for plasmid analyses. E. coli E31 contained three plasmids including 54-, 3- and 2-kb plasmids whereas E. coli E35 contained a 54-kb plasmid and a 3-kb plasmid (Fig. 1A). The 54-kb plasmids of E. coli E31 and its transformant hybridized with the armA-specific probe (Fig. 1B). The hybridization signals for a rmtB-specific probe were obtained on the 54-kb plasmids of the E. coli E35 harboring rmtB and its transformant (Fig. 1C). The DNA of E. coli V517, V517/R1 and amikacin-susceptible E. coli isolates had no hybridization signals with rmtB- and armA-specific probes.
Figure 1.
Plasmid extractions from reference strains of E. coli and clinical isolates carrying armA or rmtB and their transformants (A), Southern hybridization with an armA-specific probe (B) and with a rmtB-specific probe (C). Lanes:1, E. coli V517; 2. E. coli V517/R1; 3, Amikacin-susceptible E. coli isolate; 4, E31; 5, E31 transformant; 6, E35; 7, E35 transformant.
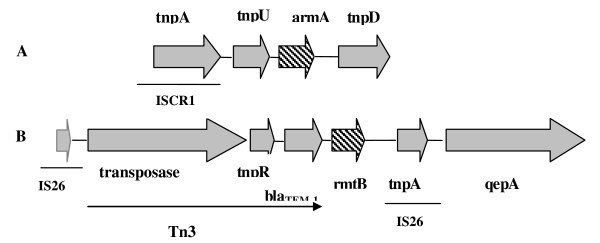
Genetic environments of the armA and rmtB gene
Upstream sequences of the armA gene contained ISCR1 and the putative transposase gene, tnpU. Another putative transposase gene, tnpD, was located downstream of the armA gene (Fig. 2A). The structural gene for the rmtB was preceded by a transposon, Tn3, including tnpA encoding transposase, tnpR encoding resolvase, and blaTEM-1 encoding spectrum beta-lactamase. A transposase gene, tnpA, and a quinolone efflux pump gene, qepA, were located downstream of the rmtB gene. The rmtB gene was flanked by two transposase genes comprising insertion sequence (IS26). Flanking regions of the rmtB gene are exhibited in Fig. 2B.
Figure 2.
Genetic environments of armA (A) and rmtB (B) genes.
PFGE
Thirty-seven 16S rRNA methylase gene-positive isolates distributing 19 different clinical units were grouped into 19 clonal patterns by PFGE, designated PFGE type A to S (Table 1). The predominant PFGE type was type H which accounted for 32.4% (12/37), followed by type A (3/37), type N (3/37), type G (2/37), type I (2/37) and type P (2/37). The remaining PFGE types were represented by a single isolate. The 12 PFGE type H isolates distributed nine different wards, whereas eight isolates isolated from the same ward (ward 11) belonged to seven separate PFGE types.
Discussion
Although resistance rates of the E. coli isolates to gentamicin and tobramycin were more than 50.0% in our study, resistance to amikacin was relatively low. Since the armA gene was first identified in K. pneumoniae BM4536 isolated from a French patient in 2000 and the rmtB gene was initially found in Serratia marcescens S-95 isolated from a Japanese patient in 2002, these two genes have been found in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii in many areas and particularly in Asia [4-6,14,17,22-24]. In the present study, the overall prevalence (5.4%) of 16S rRNA methylase genes in E. coli clinical isolates is higher than those previously reported in a Taiwanese study (0.4%) and a study from Shanghai, China (3.4%) [5,14]. Although rmtA, rmtC, and npmA have been detected in P. aeruginosa, Proteus mirabilis, and E. coli, respectively, in Japan [7,8,10], these genes were not detected in other Asian countries. Previous reports indicate a higher prevalence of armA relative to rmtB for Gram-negative bacilli [5,22,25,26]. One study reported that only armA was found in imipenem-resistant A. baumannii isolates from China [27]. However, armA was only detected in one E. coli isolate from our study, and our data are consistent with other studies indicating that rmtB may be more prevalent than armA among Enterobacteriaceae isolates and, in fact, that rmtB is the most prevalent 16S rRNA methylase gene among Enterobacteriaceae isolates in China [14,17]. This report further highlights the wide dissemination of 16S rRNA methylase genes among Enterobacteriaceae, and the spread of 16S rRNA methylase genes has become a great concern for hospitalized patients.
16S rRNA methylase genes have been linked to other resistance determinants, such as blaTEM-1, blaCTX-M-3, blaCTX-M-14, sul1, and dfrXII, and co-transferred with other resistance determinants on self-transferable plasmids to recipients by conjugation [5,28]. Similar to previous studies[4,5,22], most of 16S rRNA methylase gene-positive E. coli isolates reported in the present study were found to harbor blaCTX-M genes encoding ESBLs and were, therefore, resistant to multiple antibiotics. However, aminoglycoside resistance was not transferred by conjugation from 33 of 37 isolates harboring 16S rRNA methylase genes. Galimand et al. reported that the armA gene was part of functional composite transposon Tn1548 in plasmid pIP1204 [28]. The rmtB gene was found in the flanking region of the Tn3-like structure [6]. The genetic environment of the armA gene reported here was similar to that in K. pneumoniae reported in our previous study in which IS6100 and ISCR1 were first found to be located upstream of the armA gene [17], indicating that a similar mechanism may be responsible for the transfer of armA between K. pneumoniae and E. coli isolates in our hospital. The flanking regions of the rmtB gene in the present study were similar to that in previous reports in which qepA encoding quinolone efflux was located downstream of rmtB [29,30], but were different from those among in K. pneumoniae isolates in our previous report in which traI encoding conjugal nickase and helicase involved in conjugation were located downstream of rmtB [17]. The armA gene was usually carried by conjugative plasmids of replicon groups IncL/M and the rmtB gene was mainly associated with IncA/C and IncF conjugative plasmids [22,23,28]. In the present study, rmtB was borne by IncF plasmids in the majority of isolates. A small clonal dissemination was found, which accounted for 12 PFGE type H isolates. Based on our PFGE data, most 16S rRNA methylase gene-positive isolates exhibited only distantly related patterns and were from separate clinical units, suggesting considerable molecular heterogeneity amongst these isolates.
Conclusion
High prevalence of plasmid-mediated rmtB gene was found among clinical E. coli isolates from a teaching hospital in China. The majority of 16S rRNA methylase gene-positive isolates also produced CTX-M type ESBLs. Both horizontal gene transfer and clonal spread were responsible for the dissemination of the rmtB gene.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FYY, DY, CC, LHY and QQL performed the laboratory measurements. DQ and LXW made substantial contributions to conception and design. CP and DQ revised the manuscript critically for important intellectual content. ZQQ performed the analysis and interpretation of data. ZQQ, JYP and XQZ participated in design and coordination. FYY drafted the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Fang-you Yu, Email: wzjxyfy@163.com.
Dan Yao, Email: zdyaodan@163.com.
Jing-ye Pan, Email: panjingye@hosp1.ac.cn.
Chong Chen, Email: cky2000@163.com.
Zhi-qiang Qin, Email: qinzh@musc.edu.
Chris Parsons, Email: parsonch@musc.edu.
Le-he Yang, Email: lyanglehe@163.com.
Qiao-qiao Li, Email: litong.xxx@163.com.
Xue-qing Zhang, Email: xueqingzhang@hosp1.ac.cn.
Di Qu, Email: dqu@shmu.edu.cn.
Liang-xing Wang, Email: wzyxywlx@163.com.
Acknowledgements
We are grateful to Dr George A. Jacoby for providing reference strains E. coli J53AzR and E. coli V517 and Dr Xiaogan Xu and Dr Minggui Wang, Huashan Hospital, Fudan University, for the determination of PFGE. This work was supported by the grants from Medical Science Fund of Zhejiang province, China (2008A116) and Wenzhou Municipal Science and Technology Bureau, China (H20080064).
References
- Yamane K, Wachino J, Doi Y, Kurokawa H, Arakawa Y. Global spread of multiple aminoglycoside resistance genes. Emerg Infect Dis. 2005;11(6):951–953. doi: 10.3201/eid1106.040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45(1):88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- Kotra LP, Haddad J, Mobashery S. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother. 2000;44(12):3249–3256. doi: 10.1128/AAC.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003;47(8):2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JJ, Wu JJ, Ko WC, Tsai SH, Chuang CL, Wu HM, Lu YJ, Li JD. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J Antimicrob Chemother. 2004;54(6):1007–1012. doi: 10.1093/jac/dkh455. [DOI] [PubMed] [Google Scholar]
- Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother. 2004;48(2):491–496. doi: 10.1128/AAC.48.2.491-496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, Yagi T, Kato H, Arakawa Y. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet. 2003;362(9399):1888–1893. doi: 10.1016/S0140-6736(03)14959-8. [DOI] [PubMed] [Google Scholar]
- Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, Doi Y, Kimura K, Ike Y, Arakawa Y. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother. 2006;50(1):178–184. doi: 10.1128/AAC.50.1.178-184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother. 2007;51(3):852–856. doi: 10.1128/AAC.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother. 2007;51(12):4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TY, Ng LS, He J, Hsu LY. CTX-M and ampC beta-lactamases contributing to increased prevalence of ceftriaxone-resistant Escherichia coli in Changi General Hospital, Singapore. Diagn Microbiol Infect Dis. 2009. [DOI] [PubMed]
- Liu JH, Deng YT, Zeng ZL, Gao JH, Chen L, Arakawa Y, Chen ZL. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6')-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob Agents Chemother. 2008;52(8):2992–2993. doi: 10.1128/AAC.01686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du XD, Wu CM, Liu HB, Li XS, Beier RC, Xiao F, Qin SS, Huang SY, Shen JZ. Plasmid-mediated ArmA and RmtB 16S rRNA methylases in Escherichia coli isolated from chickens. J Antimicrob Chemother. 2009;64(6):1328–1330. doi: 10.1093/jac/dkp354. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Han L, Sun J, Ni Y. Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob Agents Chemother. 2009;53(1):271–272. doi: 10.1128/AAC.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute CaLS. Performance standards for antimicrobial susceptibility testing, 18th informational supplement (M100-S18) Clinical and Laboratory Standards Institute, Wayne, PA, USA; 2008. [Google Scholar]
- Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. 2003;47(7):2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Wang L, Pan J, Yao D, Chen C, Zhu T, Lou Q, Hu J, Wu Y, Zhang X. Prevalence of 16S rRNA methylase genes in Klebsiella pneumoniae isolates from a Chinese teaching hospital: coexistence of rmtB and armA genes in the same isolate. Diagn Microbiol Infect Dis. 2009;64(1):57–63. doi: 10.1016/j.diagmicrobio.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ji S, Chen Y, Zhou W, Wei Z, Li L, Ma Y. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect. 2007;54(1):53–57. doi: 10.1016/j.jinf.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Wang M, Sahm DF, Jacoby GA, Hooper DC. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother. 2004;48(4):1295–1299. doi: 10.1128/AAC.48.4.1295-1299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts P, Galimand M, Bauraing C, Deplano A, Vanhoof R, De Mendonca R, Rodriguez-Villalobos H, Struelens M, Glupczynski Y. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J Antimicrob Chemother. 2007;59(3):459–464. doi: 10.1093/jac/dkl527. [DOI] [PubMed] [Google Scholar]
- Kang HY, Kim KY, Kim J, Lee JC, Lee YC, Cho DT, Seol SY. Distribution of conjugative-plasmid-mediated 16S rRNA methylase genes among amikacin-resistant Enterobacteriaceae isolates collected in 1995 to 1998 and 2001 to 2006 at a university hospital in South Korea and identification of conjugative plasmids mediating dissemination of 16S rRNA methylase. J Clin Microbiol. 2008;46(2):700–706. doi: 10.1128/JCM.01677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche TR, Castanheira M, Miller GH, Jones RN, Armstrong ES. Detection of methyltransferases conferring high-level resistance to aminoglycosides in enterobacteriaceae from Europe, North America, and Latin America. Antimicrob Agents Chemother. 2008;52(5):1843–1845. doi: 10.1128/AAC.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HY, Kim J, Seol SY, Lee YC, Lee JC, Cho DT. Characterization of conjugative plasmids carrying antibiotic resistance genes encoding 16S rRNA methylase, extended-spectrum beta-lactamase, and/or plasmid-mediated AmpC beta-lactamase. J Microbiol. 2009;47(1):68–75. doi: 10.1007/s12275-008-0158-3. [DOI] [PubMed] [Google Scholar]
- Sabtcheva S, Saga T, Kantardjiev T, Ivanova M, Ishii Y, Kaku M. Nosocomial spread of armA-mediated high-level aminoglycoside resistance in Enterobacteriaceae isolates producing CTX-M-3 beta-lactamase in a cancer hospital in Bulgaria. J Chemother. 2008;20(5):593–599. doi: 10.1179/joc.2008.20.5.593. [DOI] [PubMed] [Google Scholar]
- Yu YS, Zhou H, Yang Q, Chen YG, Li LJ. Widespread occurrence of aminoglycoside resistance due to ArmA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J Antimicrob Chemother. 2007;60(2):454–455. doi: 10.1093/jac/dkm208. [DOI] [PubMed] [Google Scholar]
- Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005;49(7):2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51(9):3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perichon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob Agents Chemother. 2008;52(7):2581–2592. doi: 10.1128/AAC.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]