Abstract
We compute the absolute binding affinities for two ligands bound to the FKBP protein using nonequilibrium unbinding simulations. The methodology is straightforward requiring little or no modification to many modern molecular simulation packages. The approach makes use of a physical pathway, eliminating the need for complicated alchemical decoupling schemes. We compare our nonequilibrium results to those obtained via a fully equilibrium approach and to experiment. The results of this study suggest that to obtain accurate results using nonequilibrium approaches one should use the stiff-spring approximation with the second cumulant expansion. From this study we conclude that nonequilibrium simulation could provide a simple means to estimate protein-ligand binding affinities.
INTRODUCTION
The accurate estimation of binding affinities for protein-ligand systems (ΔG) remains one of the most challenging tasks in computational biophysics and biochemistry.1 Due to the high computational cost of such free energy computation, it is of interest to understand the advantages and limitations of various ΔG methods.
Many previous studies2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 have calculated protein-ligand binding affinities using equilibrium free energy methods such as thermodynamic integration,24 free energy perturbation,25, 26 and weighted histogram analysis method (WHAM).27 Due to the introduction of the novel Jarzynski approach28 it is also possible to estimate ΔG from nonequilibrium simulations. However, the estimation of ΔG for protein-ligand binding using nonequilibrium approaches remains largely untested. Two recent studies used nonequilibrium simulations to test unbinding pathways.29, 30 Studies by other groups found that estimating ΔG via nonequilibrium simulations resulted in a large error compared to experiment.31, 32 A recent study by Bastug et al.33 demonstrated that use of nonequilibrium simulation required longer simulation times than umbrella sampling.
The importance of pursuing nonequilibrium methods such as used in this report is threefold. (i) The approach is trivially parallelizeable since each nonequilibrium unbinding simulation is performed independently. (ii) The method is simple to implement in many existing simulation packages such as GROMACS (Ref. 34) used here; little or no modification to the code is necessary. (iii) Since a physical pathway is utilized, there is no need to use alchemical decoupling schemes.
In this report, apparently for the first time, we demonstrate the ability to compute accurate (as compared to experimental data) protein-ligand binding ΔG estimates following a nonequilibrium methodology. The approach relies on performing multiple nonequilibrium unbinding simulations using a physical pathway, i.e., pulling the ligand out of the binding pocket, and then uses the Hummer–Szabo approach35 or the Jarzynski method28 to estimate ΔG. The system is an FKBP protein complexed with 4-hydroxy-2-butanone (BUQ) and dimethyl sulfoxide (DMSO). The motivation for using this system is that comparison to experiment is possible36 and many previous computational studies have been performed.12, 13, 18, 20, 21 This study represents the first stage of a project comparing the efficiencies of various free energy methods for protein-ligand ΔG computation. We note that efficiency studies have been carried out for other nonprotein systems.37, 38, 39, 40, 41, 42
THEORY
In general, the absolute binding affinity is defined as the free energy difference between the unbound (apo) and bound (holo) states of the protein-ligand system. We define the apo state as when the protein and ligand are not interacting due to a large separation between them. The holo state is defined to be when the ligand is in the binding pocket of the protein. Experimentally, the binding affinity is measured by determining the equilibrium constant Keq=[PL]∕[P][L], where [PL] denotes the concentration of the protein-ligand complex, and [P] and [L] are the concentrations of the apo protein and free ligand, respectively. Then the absolute binding affinity is given by ΔGbind=−kBT ln(Keq∕V0), where kB is the Boltzmann constant, T is the system temperature, and V0 is the standard volume used for the experiment (typically V0=1.661 nm2 corresponding to 1.0 mol∕liter concentration).
Following the notation of Roux and co-workers16, 18 (also see discussion in Refs. 1, 4, 10) the equilibrium constant is given by a ratio of integrals over the apo and holo regions of configurational space
| (1) |
where β=1∕kBT is the inverse temperature, represents the configurational coordinates of the ligand, are the coordinates of the protein and solvent, and is the potential energy of the system. The vector defines the location of the center of mass of the ligand relative to the center of mass of the protein (see Fig. 1) and is some arbitrary reference value taken to be when the ligand and protein are not interacting. The subscript holo indicates that the integral only includes configurations for which the ligand is bound; this corresponds to a range of . The subscript apo indicates that the integral includes only configurations for which the ligand is unbound, i.e., when . The expression Eq. 1 assumes that the concentration is low enough that no ligand-ligand or receptor-receptor interactions take place.
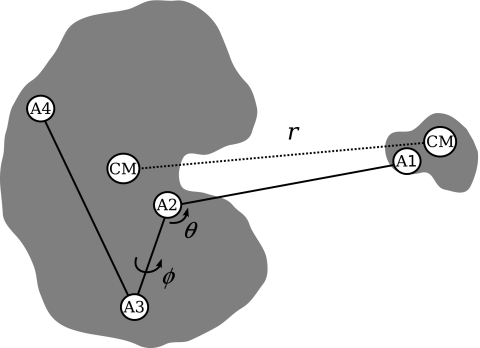
Figure 1.
The coordinate system used for the restraints Ua(θ,ϕ) and Ur(r,t). The value of r is given by the center of mass separation between the protein and ligand. (A1)–(A4) are the heavy atoms used to define the coordinate system for Ua. For the FKBP-DMSO system A1=DMSO:S1, A2=TRP59:N, A3=HIS25:N, and A4=ALA64:N. For the FKBP-BUQ system A1=BUQ:C2 and (A2)–(A4) are the same as FKBP-DMSO.
Equation 1 can be used to compute binding affinities using various computational strategies. In our study, we restrain the ligand relative to the protein so that the ligand remains along the binding axis. The potential energy of this axial restraint is given by , where ka is the force constant and θ0, ϕ0 are reference values of the coordinates, see Fig. 1. With this restraint defined, Eq. 1 can be written as a product of dimensionless ratios of integrals
| (2) |
where strategies for computing the terms I1 and I2 will be discussed below.
The first term in Eq. 2 corresponds to the free energy difference associated with restraining the protein to the binding axis while in the holo state. This free energy can be computed using any standard technique by performing simulations for a range of force constants from 0 to ka. For the current study we chose to compute this free energy difference via a multistage Bennett approach.43
To determine the second term I2 in Eq. 2 we define the potential of mean force (PMF) W with the restraint potential Ua present as a function of the scalar distance r,
| (3) |
To obtain I2 we first multiply Eq. 3 by and then integrate over both apo and holo regions
| (4) |
where we have used the fact that for the apo integral since the PMF is independent of the direction of when the ligand is not interacting with the protein. Using the relationship shown in Eq. 4 it is now possible to evaluate I2 via
| (5) |
Below the apo integral will be evaluated analytically and the holo integral will be evaluated using quadrature.
With our approximations above the absolute binding free energy can now be estimated via the relation
| (6) |
This is our central theoretical result. Thus, estimating ΔGbind in the current framework requires three computations: the PMF W must be calculated (detailed below), must be approximated, and the apo integral must be analytically evaluated.
One may note that another viable nonequilibrium approach would be to set ka=0, i.e., remove the axial restraint. This would simplify the ΔGbind calculation since and the apo integral would be 4π in Eq. 6, and thus only the PMF would be needed. Further, the binding axis would not need to be defined by the researcher, which would allow the ligand more flexible unbinding routes. While this may simplify the computation, there are two important advantages to using the axial restraint. Most important is that since the axial restraint limits the allowable configurational space for the ligand, the PMF converges much more quickly than with no restraint. This observation is in agreement with some of our preliminary studies (data not shown). Also, in cases where the binding pocket is not at the protein surface, or when more than one viable pathway exists, it may be advantageous or even necessary to define the binding path to obtain meaningful results.
Estimating the PMF
The PMF was computed using two different approaches: the Hummer–Szabo method and the stiff-spring approximation with the second cumulant expansion. Below we summarize these techniques.
In the Hummer–Szabo approach the PMF is estimated by performing multiple nonequilibrium pulling simulations along the reaction coordinate r. A time-dependent biasing potential Ur(r,t)=kr[r−(r0+vt)]2 is utilized, where kr is the force constant, r=r(t) is the protein-ligand center of mass separation, r0 is an initial reference separation, which is constant for all pulling simulations [i.e., r0≠r(0)], and v is the speed at which the biasing center is moved. The PMF is then estimated via35, 44
| (7) |
where the sum is over time slices t and the 2 ln(r) term is the Jacobian correction, which is necessary since r is a radial distance.45 The ⟨…⟩ indicates an ensemble average for pulling simulations drawn from the Boltzmann distribution corresponding to the initial system potential energy . The work for a given time slice is given by35
| (8) |
Note that Wt is the accumulated work minus the initial t=0 biasing energy.
The stiff-spring approximation utilizes the well-known Jarzynski equality28, 46, 47 to estimate the PMF. The approximation is that for a sufficiently large force constant kr that the protein-ligand separation closely follows the biasing center, i.e., ξ≡r0+vt≈r. Park and co-workers48, 49 thus concluded that the accumulated work along the reaction coordinate r is approximately equal to the accumulated work for a given time slice
| (9) |
where the 2 ln(r) term is necessary due to the Jacobian correction45 and the work is determined by integrating the biasing force over the location of the bias center
| (10) |
where we have used the fact that ∂r∕∂ξ≈1. Applying the cumulant expansion to Eq. 9, we obtain the final expression used to estimate the PMF for the stiff-spring approach48, 49
| (11) |
We note two aspects of relationships embodied in Eqs. 7, 11. (i) The equality in Eq. 7 holds only in the case of obtaining all possible pulling trajectories. The approximation in Eq. 11 is an equality for the case that the work value distribution is perfectly Gaussian. Thus, it is important to calculate uncertainty estimates for the PMF, and if possible, to compare results to an independent computational measure. Below we will compare our results to use of equilibrium umbrella sampling. (ii) The relation is independent of the speed at which the system is forced, i.e., the unbinding speed. In practice, however, it has been found that the speed chosen can dramatically affect the bias of the resulting estimates.39, 42, 50
For the results given in this report the ligand is pulled out of the binding pocket, and the reverse process of pulling the ligand into the pocket is not considered. Future studies will include the reverse process since the use of bidirectional simulation has been shown to be an effective approach to accurate ΔG estimation.40, 42, 43, 51, 52, 53, 54, 55
Use of a physical pathway
It is useful to consider the advantages and disadvantages of using a physical (rather than alchemical) pathway. The regions of configurational space corresponding to apo and holo in Eq. 1 are well separated with no overlap, thus a pathway connecting them is typically created. For our discussion below, we will assume that this pathway is parametrized using the variable λ. In the case of a physical pathway, such as in the current study, λ=r represents the protein-ligand separation. By contrast, for an alchemical pathway λ is generally a parameter that scales the strength of the interactions between the ligand and the rest of the system.
Our use of a physical pathway is motivated by several factors. Alchemical pathways are typically much more difficult to implement than physical pathways since interactions must be scaled carefully. In addition, restraints must often be employed such that the noninteracting parts do not drift away from the region of interest.
We note that there are disadvantages to using physical pathways. Physical pathways may require the researcher to determine the pulling direction such that the ligand exits the binding pocket, i.e., determined by choice of Ua in this report. Alchemical pathways do not require such a choice. Perhaps most importantly, physical pathways require larger system sizes when explicit solvent is used, as in the current report. The size of the system must be large enough that the ligand can be pulled to a distance such that interactions between the ligand and protein are negligible.
In cases where the binding site is buried deep within the protein, alchemical methods should be much more efficient than physical approaches. However, when the binding pocket is close to the protein surface, as for the current study, it is not clear where alchemical or physical approaches are more efficient and∕or accurate.
Another important consideration is that the use of a physical pathway allows the researcher to determine the PMF along the pathway. This PMF can give insights into binding which are simply not possible when using alchemical methods, e.g., determining the preferred binding pathway when multiple pathways are present.29, 30
Use of a nonequilibrium approach
Nonequilibrium approaches, such as used in the current study, rely on computing the work required to force the system from one state to the other rapidly enough that equilibrium is not attained at any value of λ. This process is repeated many times and the resulting distribution of work values is used to estimate ΔG.28 By contrast, equilibrium free energy methodologies such as thermodynamic integration,24 free energy perturbation,25, 26 and WHAM (Ref. 27) share the common strategy of generating equilibrium ensembles of configurations for multiple values of the scaling parameter λ. It is important when performing such ΔG computation that enough simulation time is spent to equilibrate at each value of λ such that the resulting ensemble is valid for the current λ.
It is not currently known whether equilibrium or nonequilibrium methodologies offer an efficiency advantage for typical protein-ligand binding affinity computation. Equilibrium methods have been widely used to generate accurate ΔG estimates for protein-ligand binding.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 However, if equilibrium is not attained the resulting ΔG estimate can be heavily biased. With a very few recent exceptions29, 30, 31, 32, 33 nonequilibrium methods are largely untested on protein-ligand systems. In previous calculations of relative solvation free energy non-equilibrium methods were proven to be equal or superior in efficiency to commonly used equilibrium methods.42
A key advantage of nonequilibrium methodologies is the ease that one can parallelize the ΔG calculation. Since each work value must necessarily be generated independently, the corresponding simulations can be run in parallel with no loss of accuracy to the final ΔG estimate. Equilibrium ΔG computations, by contrast, are not trivially parallelizeable. One can imagine performing each λ simulation in parallel; however one must be very careful about the configurations used to start each λ simulation. In typical cases it is necessary to start the current λ simulation using the final snapshot from the previous λ simulation; thus, the λ simulations are performed in a serial fashion. If this is not done, the amount of time needed to equilibrate at each value of λ could be heavily dependent on the chosen starting structures. The ΔG estimate could be heavily biased if the time spent for equilibration at each λ value is inadequate.
METHODS
Computational details
The initial coordinates for the FKBP-ligand complexes were obtained from the Protein Data Bank:56 1D7H for FKBP-DMSO and 1D7J for FKBP-BUQ. The topologies for DMSO and BUQ were then generated by the PRODRG server57 with partial charges modified by the author.
The GROMACS simulation package version 3.3.3 (Ref. 34) was used with the default GROMOS-96 43A1 forcefield.58 The software was slightly modified to provide the biasing potential Ur, which depends only on the center of mass separation between the ligand and the protein. Protonation states for the histidine residues were selected by the GROMACS program pdb2gmx: HIS25 was protonated at Nδ1 and HIS87 and HIS94 were protonated at Nϵ2. The protein-ligand complexes were then solvated in a cubic box of simple point charge (SPC) water59 with approximate initial size of 6.8 nm a side. A single chloride ion was randomly placed in each water box to give a net neutral charge, and then each system was minimized using steepest decent for 500 steps. To allow for equilibration of water, each system was then simulated for 1.0 ns with the positions of all heavy atoms in the ligand and protein harmonically restrained with a force constant of 1000 kJ∕mol∕nm2. The temperature was maintained at 300 K using Langevin dynamics60 with a friction coefficient of 1.0 amu∕ps. The pressure was maintained at 1.0 atm using the Berendsen algorithm.61 We note that the Berendsen algorithm does not produce canonically distributed structures; however, none of the resulting simulation frames was used for generating ΔG estimates, as will be seen below. The LINCS algorithm62 was used to constrain hydrogens to their ideal lengths and heavy hydrogens were used—the hydrogen mass was increased by a factor of 4 and this increase was subtracted from the bonded heavy atom so that the mass of the system remained unchanged—allowing the use of a 4.0 fs time step. Particle mesh Ewald63 was used for electrostatics with a real-space cutoff of 1.0 nm and a Fourier spacing of 0.1 nm. van der Waals interactions used a cutoff with a smoothing function such that the interactions smoothly decayed to zero between 0.75 and 0.9 nm. Dispersion corrections for energy and pressure were utilized.64
After the position restrained simulation, a 4.0 ns equilibrium simulation at constant temperature and volume with restraints was used to generate starting configurations for use in the PMF calculations. Each FKBP-protein complex was simulated with parameters chosen identical to the position restrained simulation above except that the volume was fixed at the value of the final configuration from the position restrained simulations. Importantly, for Eqs. 7, 11 to be used these equilibrium simulations must include the restraints, i.e., both Ua and Ur were present. For both the DMSO and BUQ systems the axial restraint used a force constant ka=1000 kJ∕mol∕nm2, and θ0, ϕ0 were chosen to be equal to the values from the final snapshot of the position restrained simulation. For both systems the biasing potential Ur used a force constant kr=3000 kJ∕mol∕nm2 and r0=0.5 nm and a speed v=0.
Starting structures for the unbinding simulations were chosen to be equally spaced within the 4.0 ns equilibrium simulation. So, if 100 starting structures were desired, then the spacing between snapshots was 400 ps. The pulling simulations were performed using identical parameters to the 4.0 ns equilibrium simulation, except that the bias center was moved at a constant speed v ranging from 1.0×10−4 to 8.0×10−4 nm∕ps. The pulling simulations were discontinued when the bias center was at a position of 2.5 nm.
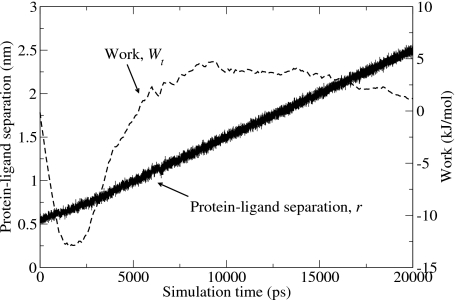
The nonequilibrium unbinding simulations provided us with the protein-ligand separation r at every time step, which we used to compute the work and thus the resulting PMF. An example of this is shown in Fig. 2 where we have used Eq. 8 to compute the work as a function of simulation time Wt.
Figure 2.
Results of a single nonequilibrium pulling simulation performed on the FKBP-DMSO system using a pulling speed of 1.0×10−4 nm∕ps. The solid line shows the protein-ligand center of mass separation as a function of simulation time. The dashed line shows the accumulated work Wt as computed by Eq. 8 as a function of simulation time.
Computing
We used the Bennett acceptance ratio approach to compute the free energy differences associated with the axial restraints.43 With the ligand bound to the protein we performed 1.0 ns equilibrium simulations for each of the values ka=0,25,40,60,90,150,200,300,450,700,1000. The first 0.5 ns of each simulation was discarded for equilibration and the remaining 0.5 ns was used to compute . We did not attempt to optimize the efficiency of the computations. Our only concern was accurate values, so it may be possible to reduce the total computational time from that described above.
Nonequilibrium uncertainty estimation
We estimated the uncertainty in our nonequilibrium ΔGbind estimates using the bootstrap approach applied to the PMF. (i) The reference value of the PMF given by W(r∗) was computed via Eq. 11 using N work values chosen at random (with replacement) from a data set containing N values. (ii) The above step was repeated until the mean and standard deviation of the free energy estimates converged; we used 100 000 trials in our study. (iii) The uncertainty is given by the converged standard deviation of the free energy estimates.
For comparison, we also used the uncertainty analysis obtained by Zuckerman and Woolf65 and Gore et al.50 These uncertainty estimates are reported to be accurate when the variance in the estimate dominates over the bias (as in the case of large N).
Generating an independent PMF estimate via WHAM
Since the purpose of the current study was to test the effectiveness of nonequilibrium strategies it is important to have an independent estimate of the PMF. Thus, we computed the PMF using umbrella sampling and WHAM.27 Simulations were performed using the same restraints as for the nonequilibrium simulations, Ua and Ur. For the umbrella sampling simulations the speed was set to v=0. All other parameters were identical to the nonequilibrium simulations and 41 windows were used r0=0.50, 0.55, 0.60, … 2.45, 2.50. Each window was simulated for a total time of 12 ns; 6 ns was discarded for equilibration and 6 ns was used for the WHAM analysis. The total simulation time of 492 ns was chosen to be nearly the same as the 505 ns used for the nonequilibrium simulations detailed above. No attempt was made to test the efficiency since the goal was to generate an accurate PMF. Note that the 2 ln(r) Jacobian correction from Eqs. 7, 11 was also used for the WHAM PMF.
RESULTS AND DISCUSSION
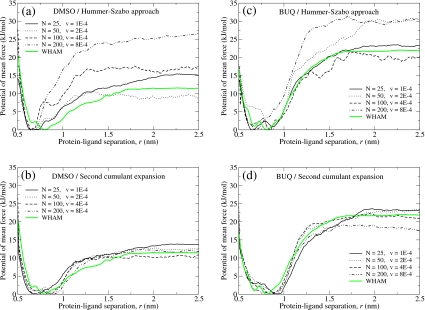
Figure 3 shows the PMF as a function of protein-ligand separation for both DMSO and BUQ systems with the pulling speeds indicated on each plot. Note that the same amount of total simulation time was spent on each nonequilibrium PMF estimate, and that the WHAM estimates utilized approximately the same amount of time as the nonequilibrium estimates. The plots show that there is a tendency for the Hummer–Szabo nonequilibrium approach to overestimate the PMF compared to WHAM and to underestimate the broadness of the PMF minimum. This effect is more pronounced at faster pulling speeds, suggesting that the pull rate must be slow enough to properly sample the PMF. Use of stiff-spring approximation with the second cumulant expansion significantly improves the nonequilibrium PMF estimates and does not appear to be correlated with pulling speed.
Figure 3.
The PMF as a function of the protein-ligand center of mass separation. These PMF curves were numerically integrated for use in Eq. 6 and utilized to generate the ΔGbind estimates shown in Table 1. All nonequilibrium and WHAM results used approximately the same total amount of simulation time. For all plots the light colored solid curve shows the PMF generated via equilibrium umbrella sampling and WHAM. (a) FKBP-DMSO system using the Hummer–Szabo approach of Eq. 7. (b) FKBP-DMSO system using the stiff-spring second cumulant expansion approximation of Eq. 11. (c) FKBP-BUQ system using the Hummer–Szabo approach of Eq. 7. (d) FKBP-BUQ system using the stiff-spring second cumulant expansion approximation of Eq. 11.
To compute ΔGbind estimates the reference distances were chosen as r∗=2.45 nm for both DMSO and BUQ. The value of the restraint free energy was found to be for FKBP-DMSO and for FKBP-BUQ. The value of the apo integral in Eq. 6 was computed analytically to be 10.6 kJ∕mol for FKBP-DMSO and 10.7 kJ∕mol for FKBP-BUQ.
Table 1 shows the binding affinity results obtained via Eq. 6 with the PMF computed using both the Hummer–Szabo approach of Eq. 7 and the stiff-spring second cumulant approximation of Eq. 11. The computational estimates of ΔGbind for the PMF generated via umbrella sampling and WHAM are in excellent agreement with experimental data. The nonequilibrium estimates using the second cumulant expansion are typically within less than 4.0 kJ∕mol of the WHAM and experimental values.
Table 1.
Comparison of binding affinity results obtained via nonequilibrium simulation, equilibrium umbrella sampling and WHAM, and experiment. All energy values are shown in units of kJ∕mol. The first column describes the ligand used. The second column contains the number of work values N used in the nonequilibrium estimate, and the third and fourth columns are, respectively, the corresponding speed of the restraint attached to the ligand and the standard deviation of the work values. The rightmost column gives the experimental results reported in Ref. 36.
| Ligand | N | Speed (nm∕ps) | σW | ΔGbinda | ΔGbindb | Unctyc | Unctyd | WHAM | Expt. |
|---|---|---|---|---|---|---|---|---|---|
| DMSO | 25 | 1.0×10−4 | 5.8 | −13.3 | −12.2 | 1.3 | 1.3 | −9.4 (1.1) | −9.7 |
| 50 | 2.0×10−4 | 7.3 | −7.3 | −10.9 | 3.6 | 2.5 | |||
| 100 | 4.0×10−4 | 10.4 | −13.3 | −8.8 | 3.4 | 2.0 | |||
| 200 | 8.0×10−4 | 10.9 | −23.0 | −11.0 | 0.9 | 0.9 | |||
| BUQ | 25 | 1.0×10−4 | 7.2 | −19.9 | −19.8 | 1.8 | 1.5 | −18.4 (1.4) | −18.9 |
| 50 | 2.0×10−4 | 9.9 | −26.3 | −19.7 | 3.0 | 2.0 | |||
| 100 | 4.0×10−4 | 10.5 | −16.3 | −17.6 | 4.4 | 2.6 | |||
| 200 | 8.0×10−4 | 12.8 | −26.3 | −15.1 | 1.9 | 1.5 |
Binding affinity estimate obtained using the stiff-spring second cumulant expansion approximation using Eqs. 6, 11.
Uncertainty estimate computed via the bootstrap method.
Binding affinity estimate obtained using umbrella sampling and WHAM. Uncertainty estimate shown in parentheses was obtained via bootstrapping using Alan Grossfield’s software (Ref. 67).
Table 1 includes the standard deviation of the work values σW measured at the reference distance r∗=2.45 nm. Previous studies have suggested that the optimal efficiency for use of the Jarzynski relation is when the speed is slow enough that σW≈1.0kBT≈2.5 kJ∕mol.39, 42, 47 Apparently the speeds attempted for the current study were not slow enough to generate work values with such a small σW. For the current study, the bias of the Hummer–Szabo results tends to be larger for faster pulling speeds, but use of the second cumulant expansion appears to eliminate this trend. Future studies would be necessary to determine if there is an optimal pulling speed for protein-ligand systems.
The results from Table 1 suggest that use of the Hummer–Szabo approach, while exact in the limit of infinite sampling, is not feasible for estimating ΔGbind for the systems studied here. This is likely due to the fact that the pulling speeds used were too fast to generate work values with σW≈1.0kBT. However, use of the stiff-spring second cumulant expansion, while approximate, does significantly improve the ΔGbind estimates. This is consistent with the recent study by Minh and McCammon,66 which determined that for the case of utilizing a narrow range of speeds the stiff-spring second cumulant expansion method performed better than the other tested methods.
We realize that the use of larger more flexible ligands may lead to difficulties in using the method suggested here. This is due to the large number of possible conformations the ligand may adopt in the apo state; all of which must be sampled adequately to obtain an accurate PMF. However, the method may be modified by including an additional restraint to the root mean square deviation (RMSD) of the ligand, thus restricting the conformational freedom of the ligand. The free energy of release from this RMSD restraint must then be included in the binding affinity estimate.16, 18, 22
Note on simulation time
Each nonequilibrium estimate in Table 1 was generated using a total simulation time of 516.0 ns (1.0 ns position restrained a +4.0 ns equilibrium to generate starting configurations of +500.0 ns for unbinding simulations of +11.0 ns for estimation). The WHAM estimate was generated using a total simulation time of 504.0 ns (1.0 ns position restrained a +492.0 ns total for the 41 windows of +11.0 ns for estimation). Note however, that the nonequilibrium unbinding simulations were performed in parallel. So, for example, at a speed of 2.0×10−4 nm∕ps, 50 independent 10.0 ns simulations were performed in parallel. Therefore, all the simulation data needed to compute ΔGbind can be obtained in around 1–2 days of wall clock time with the use of a computer cluster and fast computer software such as GROMACS used here.
CONCLUSIONS
We have demonstrated that nonequilibrium unbinding simulations utilizing a physical pathway can be used to generate estimates of the binding affinity for the FKBP-DMSO and FKBP-BUQ systems studied here. We utilized the Hummer–Szabo nonequilibrium approach,35, 44 the stiff-spring second cumulant expansion nonequilibrium approach,48, 49 and equilibrium umbrella sampling with the WHAM.27
Our results suggest that when the standard deviation of the work values is larger than the optimal σW≈1.0kBT, as for our study, that the stiff-spring second cumulant expansion approximation provides a more accurate ΔGbind estimate than the Hummer–Szabo method. Estimates of ΔGbind using both WHAM and the second cumulant expansion were within 4.0 kJ∕mol of the experimental results.
The importance of pursuing methods such as described here is that such nonequilibrium approaches are trivially parallelizeable since each unbinding simulation is performed independently. Also, due to the use of a physical pathway, the method is simple to implement in many existing simulation packages with little or no modification to the software.
We note that the method described here is not expected to produce accurate binding affinities when the ligand is large and flexible. In this case, it is necessary to extend the approach to include additional restraints to the ligand during the unbinding simulation to prevent large-scale fluctuations. The contribution to the binding affinity from these additional restraints must then be taken into account.16, 18, 22
ACKNOWLEDGMENTS
Funding for this research was provided by the University of Idaho, Idaho NSF-EPSCoR, and BANTech. Computing resources were provided by IBEST at University of Idaho and by the TeraGrid Advanced Support Program. F.M.Y. would like to thank Ronald White, David Mobley, and Daniel Zuckerman for helpful discussions.
References
- Chipot C. and Pohorille A., Free Energy Calculations: Theory and Applications in Chemistry and Biology (Springer, Berlin, 2007). [Google Scholar]
- Bash P. A., Singh U. C., Brown F. K., Langridge R., and Kollman P. A., Science 235, 574 (1987). 10.1126/science.3810157 [DOI] [PubMed] [Google Scholar]
- Hermans J. and Wang L., J. Am. Chem. Soc. 119, 2707 (1997). 10.1021/ja963568+ [DOI] [Google Scholar]
- Gilson M. K., Given J. A., Bush B. L., and McCammon J. A., Biophys. J. 72, 1047 (1997). 10.1016/S0006-3495(97)78756-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Chang C. -E., and Gilson M. K., Biophys. J. 87, 3035 (2004). 10.1529/biophysj.104.049494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms V. and Wade R. C., J. Am. Chem. Soc. 120, 2710 (1998). 10.1021/ja9738539 [DOI] [Google Scholar]
- Oostenbrink B. C., Pitera J. W., van Lipzip M. M., Meerman J. H. N., and van Gunsteren W. F., J. Med. Chem. 43, 4594 (2000). 10.1021/jm001045d [DOI] [PubMed] [Google Scholar]
- Dixit S. B. and Chipot C., J. Phys. Chem. A 105, 9795 (2001). 10.1021/jp011878v [DOI] [Google Scholar]
- Banavali N. K., Im W., and Roux B., J. Chem. Phys. 117, 7381 (2002). 10.1063/1.1507108 [DOI] [Google Scholar]
- Boresch S., Tettinger F., Leitgeb M., and Karplus M., J. Phys. Chem. B 107, 9535 (2003). 10.1021/jp0217839 [DOI] [Google Scholar]
- Oostenbrink C. and van Gunsteren W. F., J. Comput. Chem. 24, 1730 (2003). 10.1002/jcc.10304 [DOI] [PubMed] [Google Scholar]
- Swanson J. M. J., Henchman R. H., and McCammon J. A., Biophys. J. 86, 67 (2004). 10.1016/S0006-3495(04)74084-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani H., Tanida Y., Ito M., Jayachandran G., Snow C. D., Shirts M. R., Sorin E. J., and Pande V. S., J. Chem. Phys. 123, 084108 (2005). 10.1063/1.1999637 [DOI] [PubMed] [Google Scholar]
- Pearlman D. A., J. Med. Chem. 48, 7796 (2005). 10.1021/jm050306m [DOI] [PubMed] [Google Scholar]
- Carlsson J. and Aqvist J., J. Phys. Chem. B 109, 6448 (2005). 10.1021/jp046022f [DOI] [PubMed] [Google Scholar]
- Woo H. -J. and Roux B., Proc. Natl. Acad. Sci. U.S.A. 102, 6825 (2005). 10.1073/pnas.0409005102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. and Roux B., J. Chem. Theory Comput. 2, 1255 (2006). 10.1021/ct060037v [DOI] [PubMed] [Google Scholar]
- Wang J., Deng Y., and Roux B., Biophys. J. 91, 2798 (2006). 10.1529/biophysj.106.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley D. L., Chodera J. D., and Dill K. A., J. Chem. Phys. 125, 084902 (2006). 10.1063/1.2221683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran G., Shirts M. R., Park S., and Pande V. S., J. Chem. Phys. 125, 084901 (2006). 10.1063/1.2221680 [DOI] [PubMed] [Google Scholar]
- Lee M. S. and Olson M. A., Biophys. J. 90, 864 (2006). 10.1529/biophysj.105.071589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley D. L., Chodera J. D., and Dill K. A., J. Chem. Theory Comput. 3, 1231 (2007). 10.1021/ct700032n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S. and Olson M. A., J. Phys. Chem. B 112, 13411 (2008). 10.1021/jp802460p [DOI] [PubMed] [Google Scholar]
- Kirkwood J. G., J. Chem. Phys. 3, 300 (1935). 10.1063/1.1749657 [DOI] [Google Scholar]
- Zwanzig R. W., J. Chem. Phys. 22, 1420 (1954). 10.1063/1.1740409 [DOI] [Google Scholar]
- Valleau J. P. and Card D. N., J. Chem. Phys. 57, 5457 (1972). 10.1063/1.1678245 [DOI] [Google Scholar]
- Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H., and Kollman P. A., J. Comput. Chem. 13, 1011 (1992). 10.1002/jcc.540130812 [DOI] [Google Scholar]
- Jarzynski C., Phys. Rev. Lett. 78, 2690 (1997). 10.1103/PhysRevLett.78.2690 [DOI] [Google Scholar]
- Zhang D., Gullingsrud J., and McCammon J. A., J. Am. Chem. Soc. 128, 3019 (2006). 10.1021/ja057292u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisth H. and Abrams C. F., Biophys. J. 95, 4193 (2008). 10.1529/biophysj.108.139675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräter F., de Groot B. L., Jiang H., and Grubmüller H., Structure 14, 1567 (2006). 10.1016/j.str.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Cuendet M. A. and Michielin O., Biophys. J. 95, 3575 (2008). 10.1529/biophysj.108.131383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastug T., Chen P. -C., Patra S. M., and Kuyucak S., J. Chem. Phys. 128, 155104 (2008). 10.1063/1.2904461 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., and Berendsen H. J. C., J. Comput. Chem. 26, 1701 (2005). 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Hummer G. and Szabo A., Proc. Natl. Acad. Sci. U.S.A. 98, 3658 (2001). 10.1073/pnas.071034098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P., Taylor P., and Walkinshaw M. D., J. Mol. Biol. 295, 953 (2000). 10.1006/jmbi.1999.3411 [DOI] [PubMed] [Google Scholar]
- Radmer R. J. and Kollman P. A., J. Comput. Chem. 18, 902 (1997). [DOI] [Google Scholar]
- Kofke D. A. and Cummings P. T., Mol. Phys. 92, 973 (1997). 10.1080/002689797169600 [DOI] [Google Scholar]
- Hummer G., J. Chem. Phys. 114, 7330 (2001). 10.1063/1.1363668 [DOI] [Google Scholar]
- Shirts M. R. and Pande V. S., J. Chem. Phys. 122, 144107 (2005). 10.1063/1.1873592 [DOI] [PubMed] [Google Scholar]
- Oostenbrink C. and van Gunsteren W. F., Chem. Phys. 323, 102 (2006). 10.1016/j.chemphys.2005.08.054 [DOI] [Google Scholar]
- Ytreberg F. M., Swendsen R. H., and Zuckerman D. M., J. Chem. Phys. 125, 184114 (2006). 10.1063/1.2378907 [DOI] [PubMed] [Google Scholar]
- Bennett C. H., J. Comput. Phys. 22, 245 (1976). 10.1016/0021-9991(76)90078-4 [DOI] [Google Scholar]
- Hummer G. and Szabo A., Acc. Chem. Res. 38, 504 (2005). 10.1021/ar040148d [DOI] [PubMed] [Google Scholar]
- Trzesniak D., Kunz A. -P. E., and van Gunsteren W. F., ChemPhysChem 8, 162 (2007). 10.1002/cphc.200600527 [DOI] [PubMed] [Google Scholar]
- Jarzynski C., Phys. Rev. E 56, 5018 (1997). 10.1103/PhysRevE.56.5018 [DOI] [Google Scholar]
- Crooks G. E., Phys. Rev. E 61, 2361 (2000). 10.1103/PhysRevE.61.2361 [DOI] [Google Scholar]
- Park S., Khalili-Araghi F., Tajkhorshid E., and Schulten K., J. Chem. Phys. 119, 3559 (2003). 10.1063/1.1590311 [DOI] [Google Scholar]
- Park S. and Schulten K., J. Chem. Phys. 120, 5946 (2004). 10.1063/1.1651473 [DOI] [PubMed] [Google Scholar]
- Gore J., Ritort J., and Bustamante C., Proc. Natl. Acad. Sci. U.S.A. 100, 12564 (2003). 10.1073/pnas.1635159100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Singh J. K., and Kofke D. A., J. Chem. Phys. 118, 2977 (2003). 10.1063/1.1537241 [DOI] [Google Scholar]
- Shirts M. R., Bair E., Hooker G., and Pande V. S., Phys. Rev. Lett. 91, 140601 (2003). 10.1103/PhysRevLett.91.140601 [DOI] [PubMed] [Google Scholar]
- Lu N., Kofke D. A., and Woolf T. B., J. Comput. Chem. 25, 28 (2004). 10.1002/jcc.10369 [DOI] [PubMed] [Google Scholar]
- Lu N., Wu D., Woolf T. B., and Kofke D. A., Phys. Rev. E 69, 057702 (2004). 10.1103/PhysRevE.69.057702 [DOI] [PubMed] [Google Scholar]
- Minh D. D. L. and Adib A. B., Phys. Rev. Lett. 100, 180602 (2008). 10.1103/PhysRevLett.100.180602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., and Bourne P. E., Nucleic Acids Res. 28, 235 (2000). 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttelkopf A. W. and van Aalten D. M. F., Acta Crystallogr. D Biol. Crystallogr. 60, 1355 (2004). 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- van Gunsteren W. F., Billeter S. R., Eising A. A., Hünenberger P. H., Krüger P., Mark A. E., Scott W. R. P., and Tironi I. G., Biomolecular Simulation: The GROMOS96 manual and user guide (Hochschulverlag, Zürich, 1996). [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., and Hermans J., Intermolecular Forces (Reidel, Dordrecht, 1981). [Google Scholar]
- van Gunsteren W. F., Berendsen H. J. C., and Rullmann J. A. C., Mol. Phys. 44, 69 (1981). 10.1080/00268978100102291 [DOI] [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., and Haak J. R., J. Chem. Phys. 81, 3684 (1984). 10.1063/1.448118 [DOI] [Google Scholar]
- Hess B., Bekker H., Berendsen H. J. C., and Fraaije J. G. E. M., J. Comput. Chem. 18, 1463 (1997). [DOI] [Google Scholar]
- Darden T., York D., and Pedersen L., J. Chem. Phys. 98, 10089 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- Allen M. P. and Tildesley D. J., Computer Simulation of Liquids (Oxford University Press, New York, 1989). [Google Scholar]
- Zuckerman D. M. and Woolf T. B., Phys. Rev. Lett. 89, 180602 (2002). 10.1103/PhysRevLett.89.180602 [DOI] [PubMed] [Google Scholar]
- Minh D. D. L. and McCammon J. A., J. Phys. Chem. B 112, 5892 (2008). 10.1021/jp0733163 [DOI] [PubMed] [Google Scholar]
- Grossfield A., http://membrane.urmc.rochester.edu.