Abstract
Histamine N-methyltransferase (HNMT) is the sole enzyme responsible for inactivating histamine in the mammalian brain. The human HNMT gene contains a common threonine-isoleucine polymorphism at residue 105, distal from the active site. The 105I variant has decreased activity and lower protein levels relative to the 105T protein. Crystal structures of both variants have been solved, but reveal little regarding how the T105I polymorphism affects activity. We performed molecular dynamics simulations of both 105T and 105I at 37°C to explore the structural and dynamic consequences of the polymorphism. The simulations indicate that replacing Thr with the larger Ile residue leads to greater burial of residue 105 and heightened packing interactions between residue105 and residues within helix α3 and strand β3. This altered packing is directly translated to the active site resulting in the reorientation of several co-substrate-binding residues. The simulations also show that the hydrophobic histamine-binding domain in both proteins undergoes a large-scale breathing motion that exposes key catalytic residues and lessens the hydrophobicity of the substrate-binding site.
Introduction
Histamine is a neurotransmitter and neuromodulator that plays important roles in regulating inflammatory and allergic responses (1, 2), gastric acid secretion (3, 4), memory and learning (5-7). Defects in the histaminergic system have been linked with cognitive deficiencies in neurodegenerative diseases, including Alzheimer's disease and Down syndrome (8-11).
Histamine N-methyltransferase (HNMT, EC.2.1.1.8) is the sole enzyme responsible for histamine metabolism in the mammalian brain (12). The human HNMT gene contains a common single nucleotide polymorphism (SNP) that codes for either threonine (T, Thr) or isoleucine (I, Ile) at residue 105 (rs1801105) (13). The 105I allele is present in 10-15% of the Caucasian population and ∼6% of the Han Chinese population (13-16). Replacing T105 with isoleucine results in lower enzymatic activity and decreased levels of immunoreactive protein in vivo (13, 17-20). The apparent KM values for histamine and co-substrate S-adenosylmethionine (SAM) are 1.3 and 1.8 fold higher, respectively, in the 105I variant than in the 105T protein (18). The T105I polymorphism was initially thought to be associated with increased risk for asthma and inflammation (21). However, recent studies have shown no link between the 105I allele and asthma (22-24) or gastric inflammation (25). Interestingly, it is the more active105T protein that appears to be associated with disease. Two recent studies suggest that the 105I allele confers protection against alcoholism (26, 27). The elevated levels of histamine present in individuals carrying the 105I allele may also provide protection against infectious agents as well as affect anxiety levels, cognition and sedation (9, 10, 26).
Several crystal structures of human 105T and 105I HNMT with bound S-adenosylhomocysteine (SAH) and a diverse set of inhibitors have been solved (18, 28), revealing a two-domain structure (Figure 1A). The larger domain consists of the highly conserved SAM-binding fold present in many SAM-dependent methyltransferases (Figure 1B). Elements from both the amino and carboxy termini form the hydrophobic substrate-binding domain, which buries histamine substrates in a pocket lined with 14 aromatic residues (Figure 1C). The T105I polymorphism is ∼16 Å from the active site in a loop between helix α4 and strand β3, both of which have SAM-binding residues at their distal ends (Figure 1). The root-mean-square-deviation of Cα atoms (Cα-RMSD) between the 105T and 105I structures is 0.4 Å. The largest deviations are in the polymorphic loop region (residues 104-110), which has larger crystallographic B-factors in105I HNMT. Residue 105 interacts with the same residues in both variants, the sole exception being the loss of a hydrogen bond between the hydroxyl group of T105 and the backbone carbonyl L101 in the 105I protein. Both proteins interact with co-substrate SAH in an identical manner.
Figure 1. Bi-domain structure of histamine N-methyltransferase.
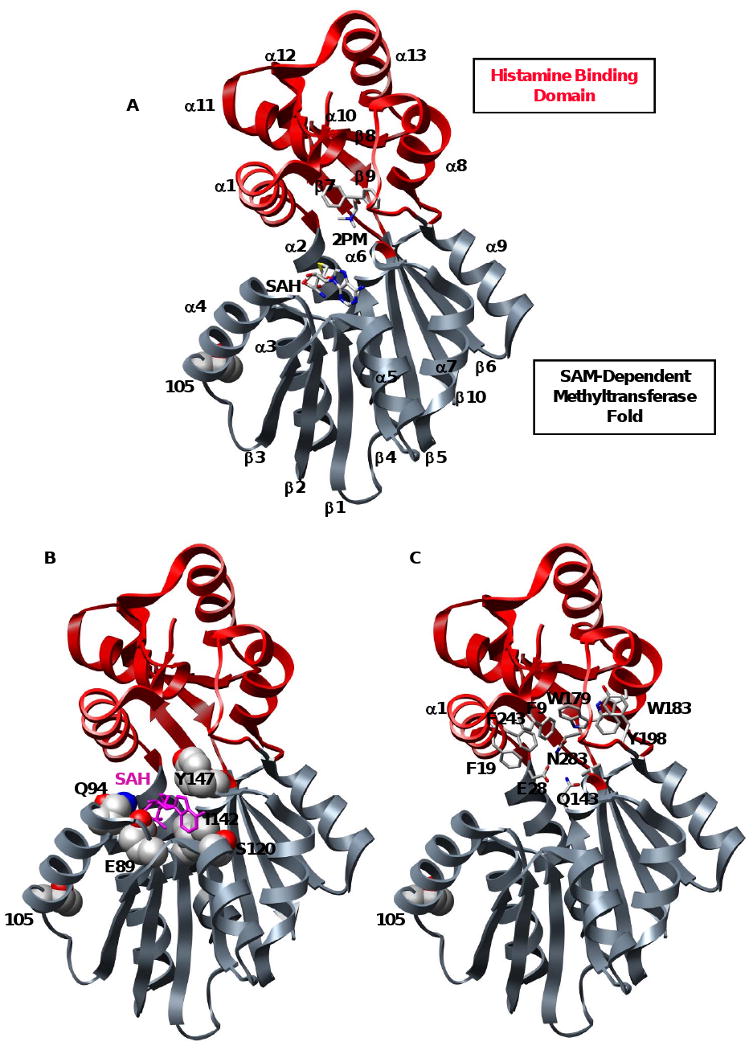
(A) Ribbon diagram of HNMT (2AOT) with the SAM and histamine binding domains colored gray and red, respectively. Product S-adenosylhomocysteine (SAH), inhibitor diphenhydramine (2PM), and polymorphic residue 105 are colored by atom. (B) Key residues in the highly conserved SAM-dependent methyltransferase domain. SAM interacts with residues from α4 (Q94), α5 (S120), β2 (E89, P90), β3 (T119) and β4 (I142, M144). SAH is colored magenta and shown in licorice representation, residues are colored by atom and shown in space-filling representation. (C) Key residues in the histamine-binding domain. A trio of catalytic residues (E28, Q143 and N283) is involved in the N-methylation of HNMT substrates. The substrate is buried in a hydrophobic domain lined with aromatic residues (shown in licorice representation).
The static crystal structures of 105T and 105I HNMT reveal little in regards to how the T105I polymorphism affects substrate binding or catalysis. Therefore, we have performed multiple molecular dynamics (MD) simulations of 105T and 105I HNMT at 37°C to examine the dynamic and structural consequences of the T105I polymorphism. The simulations show that the hydrophobic histamine-binding domains of both proteins undergo a breathing motion, exposing the catalytic residues and consequently providing a less hydrophobic site for substrate docking. Overall, this motion is more restricted in I105 HNMT, and the Ile variant remains more compact. The larger Ile residue is buried to a greater extent than T105, and interacts with more of the surrounding residues in α3 and β4. This altered packing is transmitted directly to the SAM-binding site through α3 and β4, resulting in the reorientation of several key catalytic residues at their distal ends.
Methods
Protein Preparation
Chain A of a 1.9 Å crystal structure of human 105T HNMT (2AOT.PDB (28), residues 5-292) bound with S-adenosylhomocysteine (SAH) and diphenhydramine (2PM) was the starting structure for all of the simulations. The substrate molecules were removed, and S-hydroxy-cysteine was replaced by cysteine at positions 82, 217 and 248. The T105I variant was generated by replacing Thr 105 with Ile and minimizing the torsional, electrostatic and van der Waals potential energy of the resultant structure in vacuo (29).
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations of the 105T and 105I HNMT proteins were preformed with the in lucem molecular mechanics (ilmm) simulation package (30) using protocols described elsewhere (30-33). The simulations included all hydrogen atoms and explicit flexible three-centered waters (33). The proteins were solvated in a rectangular box with walls extending at least 10 Å from any protein atom. Solvent densities were set to 0.997 and 0.933 g/ml for simulations performed at 25 and 37°C, respectively (34). After setting the densities, the box volume was held fixed and the NVE microcanonical ensemble was employed. A 10Å non-bonded cut off was used and it was updated every 2 steps. The potential energy function and associated methods have been described (29-32). A time step of 2 fs was used in all calculations. All simulations were run for 21 ns and structures were saved every 1 ps for analysis. One 25 °C simulation and three independent 37°C simulations were performed for each system.
Analyses
Average Cα-RMSDs, solvent-accessible surface areas (SASA), and contact-distances were calculated using structures from the last 5 ns (5,000 structures) of each simulation. SASA was determined using in-house software implementing the NACCESS algorithm (35). A contact was defined as a C-C atom distance within 5.4Å, or a heavy atom (O, N, S) distance within 4.6Å between two non-neighboring residues. The data described in Tables I, II, and III are based on sets of three simulations at 37°C for both 105T and 105I HNMT. The errors are the standard deviations in the average values of the ensembles for each property reported. In Figure 2, Cα-RMSF values for the HNMT crystal structure (2AOT (28)) were calculated using crystallographic B-factors via the equation: Cα–RMSF = (3B-factor/(8π2))1/2 (36). Figure images were created using CHIMERA (37).
Table I. General Properties of the Simulations.
| Property | 105T HNMT | 105I HNMT | ||
|---|---|---|---|---|
| Initial | 37°C | Initial | 37°C | |
| Cα–RMSD of Methyltransferase Domain (Å)a | NAg | 3.4 ± 0.6 | NA | 3.0 ± 0.8 |
| Cα-RMSD of Histamine Binding Domain (Å)b | NA | 5.2 ± 0.7 | NA | 3.7 ± 1.3 |
| Total SASA (Å2)c | 13618 | 16745 ± 609 | 13596 | 16064 ± 754 |
| SAM Site SASA (Å2)d | 289 | 501 ± 102 | 288 | 455 ± 93 |
| Histamine Binding Site SASA (Å2) e | 359 | 645 ± 70 | 354 | 613 ± 92 |
| Residue 105 SASA (%) | 41.2 | 43.0 ± 1.7 | 52.0 | 29.8 ± 19.9 |
| Polymorphic Site SASA (Å2)f | 498 | 411 ± 2 | 514 | 460 ± 43 |
Cα-RMSD values were calculated using structures from the last 5 ns (5,000 structures) of each simulation. The Cα–RMSD of the methyltransferase domain was calculated using the Cα-atoms of all residues colored black in Figure 1. All values are expressed as means and standard deviations of the means from three independent simulations at 37°C.
The Cα–RMSD of the histamine-binding domain was calculated using the Cα-atoms of all residues colored red in Figure 1.
The total solvent-accessible surface area (SASA, Å2) was determined using the NACCESS algorithm (35).
The following residues were used to calculate the SASA of the SAM binding-site: M32, G60, E89, P90, Q94, T119, S120, I142, M144, and Y147.
The following residues were used to calculate the histamine binding-site SASA: F9, F19, F22, Y147, V173, W179, W183, C196, Y198, and F243.
The following residues were used to calculate the polymorphic site SASA: L68, L71, S72, L101, K104, S106, N107, L108, F113, V111, and residue 105.
NA: not applicable.
Table II.
Active-Site Distances (Å)
| Contact Paira | 105T HNMT | 105I HNMT | ||
|---|---|---|---|---|
| SAM-Binding Domain | Initial | 37°C | Initial | 37°C |
| M32 CE – I142 CG2 | 3.6 | 8.5 ± 4.3 | 3.6 | 7.1 ± 2.4 |
| P90 CD –T119 CB | 5.3 | 5.3 ± 0.4 | 5.3 | 5.2 ± 0.6 |
| P90 CD – M144 CD | 8.2 | 13.1 ± 2.1 | 8.2 | 13.3 ± 2.7 |
| Q94 CG – I142 CB | 12.7 | 13.9 ± 1.6 | 12.7 | 14.7 ± 2.1 |
| S120 CB – M144 CG | 7.1 | 9.1 ± 1.6 | 7.1 | 8.5 ± 2.0 |
| Histamine-Binding Domain | ||||
| E28 OE1-Q143 NE2 | 3.5 | 12.4 ± 7.1 | 3.0 | 9.5 ± 4.1 |
| E28 OE2 – N283 ND2 | 2.7 | 9.2 ± 4.6 | 2.7 | 8.8 ± 2.7 |
| Q143 NE2 – N283 ND2 | 5.7 | 8.6 ± 1.5 | 5.8 | 11.2 ± 1.7 |
| F9 CB – C196 CB | 6.7 | 19.1 ± 6.0 | 6.7 | 11.7 ± 3.8 |
| F19 CG – Y147 OH | 7.4 | 17.6 ± 2.8 | 7.4 | 11.4 ± 3.6 |
| V173 CG1 – F243 CZ | 8.6 | 16.5 ± 2.2 | 8.6 | 12.4 ± 1.9 |
| W183 CG – Y198 CG | 3.7 | 4.7 ± 0.4 | 3.7 | 4.7 ± 0.1 |
Contact distances were calculated over the last 5 ns (5,000 structures) of each simulation. All values are expressed as means and standard deviations of the means of three independent simulations at 37°C.
Table III.
Active-Site Residue Solvent Exposure
| Side-Chain SASA (Å2) a | 105T HNMT | 105I HNMT | ||
|---|---|---|---|---|
| SAM-Binding Domain | Initial | 37°C | Initial | 37°C |
| E89 | 10 | 36 ± 4 | 10 | 40 ± 18 |
| P90 | 56 | 47 ± 5 | 56 | 43 ± 13 |
| Q94 | 60 | 41 ± 25 | 60 | 57 ± 5 |
| T119 | 47 | 52 ± 4 | 47 | 49 ± 5 |
| I142 | 12 | 57 ± 40 | 12 | 41 ± 29 |
| M144 | 20 | 54 ± 23 | 20 | 57 ± 22 |
| Histamine-Binding Domain | ||||
| F9 | 34 | 112 ± 42 | 34 | 131 ± 32 |
| E28 | 5 | 72 ± 40 | 5 | 77 ± 36 |
| Q143 | 30 | 68 ± 23 | 30 | 82 ± 17 |
| N283 | 1 | 16 ± 12 | 1 | 10 ± 5 |
| Y147 | 78 | 114 ± 30 | 78 | 102 ± 22 |
| C196 | 14 | 30 ± 16 | 14 | 34 ± 13 |
| Y198 | 58 | 63 ± 24 | 58 | 57 ± 15 |
| E242 | 18 | 79 ± 24 | 18 | 73 ± 30 |
| F243 | 25 | 43 ± 17 | 25 | 42 ± 9 |
| L285 | 0 | 25 ±18 | 0 | 27 ± 18 |
Side-chain solvent-accessible surface areas (SASA, Å2) were determined using the NACCESS algorithm (35). Calculations used structures from the last 5 ns (5,000 structures) of each simulation. All values are expressed as means and standard deviations of the means of three independent simulations at 37°C.
Figure 2. Mobility of HNMT during MD.
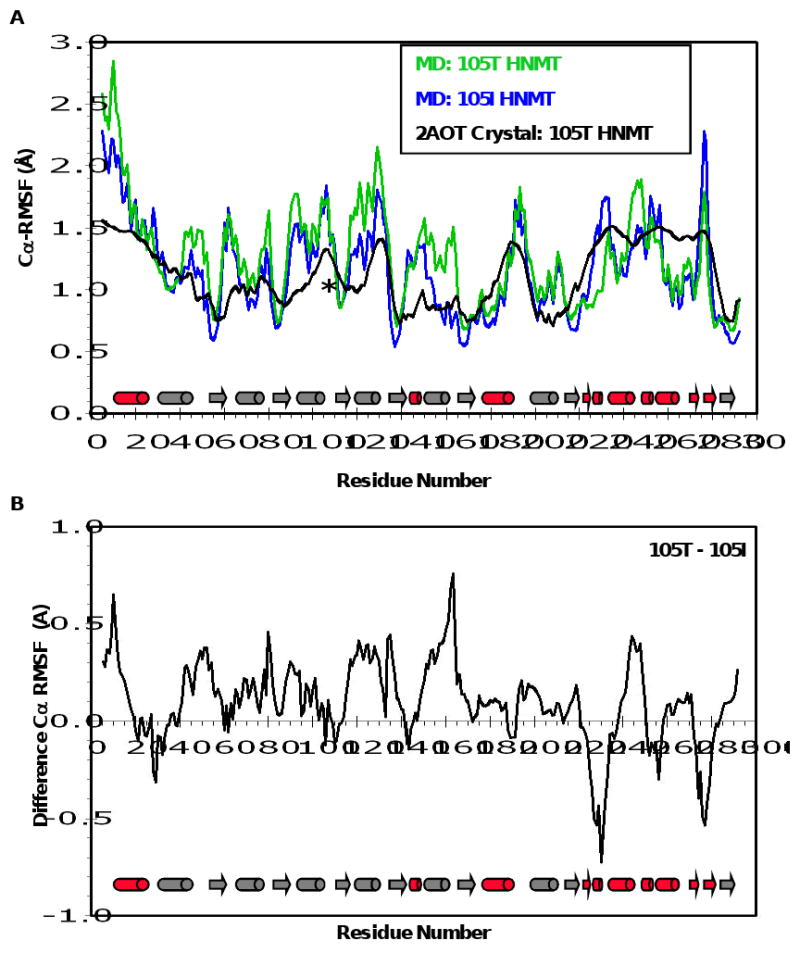
(A) Cα-RMS fluctuations (Å) per residue from the 105T (green) and 105I (blue) MD simulations at 37°C. Cα-RMSFs were calculated relative to the average structure over the last 10 ns of each simulation. Experimental B-factors of the 105T HNMT crystal structure (2AOT, (28)) are colored in black. (B) Cα-RMSF difference plot for the HNMT simulations. Positive and negative values indicate greater overall fluctuations in the 105T and 105I HNMT proteins, respectively. Secondary structural elements are depicted as  for α-helices, and
for α-helices, and  for β-strands, and are colored to match the SAM- and histamine-binding domains shown in Figure 1. * = residue 105.
for β-strands, and are colored to match the SAM- and histamine-binding domains shown in Figure 1. * = residue 105.
Results and Discussion
We ran multiple simulations of both 105T and 105I HNMT to examine the effects of the T105I polymorphism on the structure and dynamics of the protein. The root-mean-square deviation of the Cα atoms (Cα-RMSD) between the 105T and 105I HNMT starting structures after energy minimization was 0.4 Å. The overall HNMT structure was maintained throughout all of the simulations, and the polymorphism did not grossly disrupt the protein. However, the structures of both variants expanded and became slightly flattened during the simulations, while their overall solvent-accessible surface area (SASA) increased. This was most likely due to the removal of both the SAH and 2PM substrates (Figure 1). At 37°C, the SAM- and histamine-binding domains both became more exposed to solvent, and the Cα-RMSD of the SAM-binding domain reached ∼3Å in both proteins (Table 1). However, the histamine-binding domain becomes slightly more disrupted in the 105T HNMT simulations, reaching a Cα-RMSD of 5.2 ± 0.7 Å compared to 3.7 ± 1.3 Å in the 105I protein.
Figure 2 shows the average Cα-root-mean square fluctuations (Cα-RMSF) about the mean structure for the 37°C simulations, along with the crystallographic B-factors of HNMT (2AOT, (28)). The fluctuations during the simulations are of the same magnitude and follow a pattern similar to the B-factors. The largest Cα–RMSFs were in the helices and loops of both proteins, especially α1, which makes up one face of the histamine-binding domain, α5 (residues 120-130), and the solvent exposed E276. β-strands positioned within the core of the SAM-binding domain demonstrated the smallest fluctuations (Figure 2A). The Cα-RMSF values differ significantly between the105T and 105I HNMT simulations at several places within the protein structure. Residues on the surface of the histamine-binding domain (β7, α10) fluctuate more in the105I HNMT simulations. 105T HNMT exhibits larger fluctuations for residues 145-163 and 243-248, which contain buried core SAM (I142, M144, Y147) and histamine (Y146, Y147, F243, E246) binding residues. The Cα-RMSD and Cα-RMSF values suggest that the histamine-binding domain of HNMT is quite flexible (Figure 2B, Table I).
Substrate Binding and the Inherent Flexibility of the Histamine-Binding Domain
The histamine-binding domain has a mixed α/β structure and is comprised of residues from both the amino and carboxy termini of HNMT. The interior of the domain is lined with 14 aromatic residues, providing a very hydrophobic pocket for substrate docking and burial. Three polar residues (E28, Q143, N283) and several water molecules form a hydrogen-bonded network at the base of the pocket (Figure 1C). These residues interact directly with the substrate's charged residues to both orient the substrate and catalyze its N-methylation (18, 28). In addition, residues C196 and E246 positioned at opposite ends of the pocket are important for substrate binding (28). Because HNMT is inhibited by a variety of rigid, aromatic compounds that share little structural similarity, it has been hypothesized that the histamine-binding domain exhibits an inherent flexibility that allows tight binding of diverse compounds (18, 28). More importantly, because the bound substrate or inhibitor is completely buried in the crystal structures, the histamine-binding domain must open up in order for the substrate to enter the active site.
The Cα–RMSD of the histamine-binding domain reached 5.2 Å in simulations of 105T HNMT (Table I). This deviation was due mainly to the flexibility of α1 (Figures 1, 2). Several aromatic residues (F9, Y15, F19, F22) align along one side of α1 to form part of the histamine-binding pocket (Figure 1C). α1 reorients and extends during the simulations, separating the domain into two clusters of aromatic residues (Figure 3). Throughout the simulations, α1 and α11 remain in contact via multiple salt-bridges and a hydrophobic interaction between F22 and F243, and the two helices to move in concert (Figure 4B). A hydrophobic patch (V173, W179, W183, F190, C196, F243) remains available for substrate binding at the back of the pocket (Figure 3). The motions of α1 open up the histamine-binding domain, increasing the solvent exposure of the active site by ∼ 200 Å2 (Table I). There also is a large increase in the solvent accessibility of residues E28, Q143 and N283 at the base of the active site (Figure 3, Table III). Overall, this provides a more accessible site for docking an amphipathic substrate than the initial, tightly packed cluster of hydrophobic residues.
Figure 3. Snapshots from a MD simulation of 105T HNMT at 37°C.
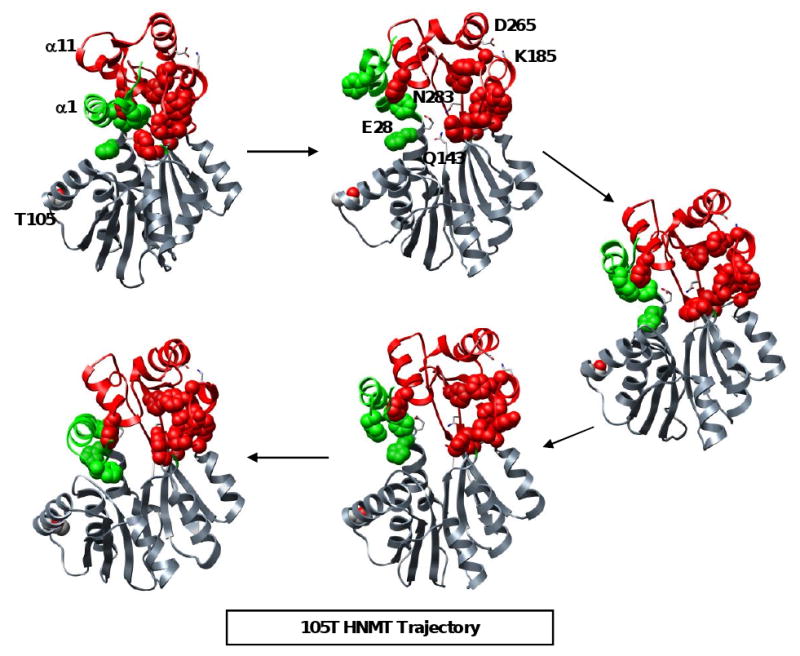
α1 elongates, breaking apart the aromatic cluster within the histamine-binding domain, and exposing E28, Q143 and N283 to the solvent. Hydrophobic interactions between α1 and α11, and salt bridges throughout the domain allow the hydrophobic cluster to reform. This breathing motion may facilitate the entry of charged substrates in a highly hydrophobic environment. Hydrophobic residues within the histamine-binding domain are shown in space-filling representation and colored in green (α1) and red. The catalytic trio (E28, Q143, N283) and the K185-D265 salt bridge are colored by atom and shown in licorice representation.
Figure 4. Distances between core residues within the histamine-binding domains of 105T and 105I HNMT.
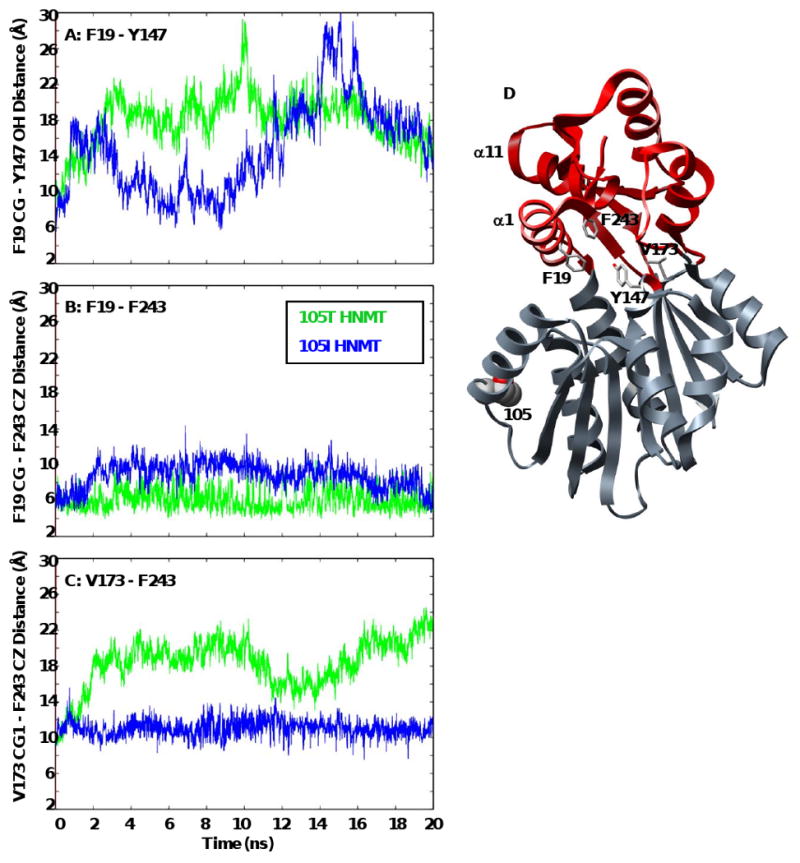
The contact distances between core residues within the histamine-binding domain (F19-Y147, V173-F243) fluctuate greatly with time, reflecting the periodic breathing motion of HNMT that may facilitate substrate binding. The distance between F19 and F243 is more constant throughout the simulations, as α1 and α11 remain in contact and move together. The figures shows plots of (A) F19 CG (α1) – Y147 OH (α6), (B) F19 CG (α1) – F243 CZ (α11), and (C) V173 CG1 (β5) – F243 CZ (α11) contact distances with time for the 105T (green) and 105I (blue) HNMT simulations at 37°C. (D) Ribbon diagram of HNMT showing the positions of residues F19, Y147, V173 and F243 within the histamine binding domain (red). Residue side-chains are shown in licorice representation and colored by atom.
Interestingly, α1 appeared to move back towards its starting orientation, in several simulations, resulting in a partial repacking of the histamine-binding domain's hydrophobic core (Figures 3, 4). This movement was facilitated by the close association of helices α1 and α11 (Figure 4). It is possible that SAM binding affects the motion of α1, as the co-substrate interacts with several residues (M32, M144, Y147) positioned near the histamine-binding domain (Figure 1). This periodic opening and closing, or breathing, of the active site would expose both a hydrophobic patch and the catalytic residues necessary for substrate-docking and orientation, allowing substrates of various sizes and polarities to bind. The α1-α11 arm could then close off the active site, burying the substrate.
Effects of the T105I Polymorphism
Horton et al. (2001) reported larger crystallographic B-factors for the polymorphic loop of 105I HNMT than for the 105T protein (18). However, increased flexibility in the residues immediately surrounding I105 was not observed in the simulations (Figure 2). In the initial structures of both 105T and 105I HNMT, residue 105 formed backbone hydrogen bonds with residues in α4 (L101, V102) and hydrophobic contacts with residues in α3 (L68), α4 (L101, A103, V102, K104) and the adjacent loop (S106, N107, L108) (Figure 5A). The additional hydrogen bond formed between the hydroxyl group of T105 and the backbone carbonyl of L101 was maintained throughout the 105T simulations.
Figure 5. Snapshots of polymorphic packing taken from the 20 ns structures of the 105T and 105I HNMT MD simulations at 37°C.
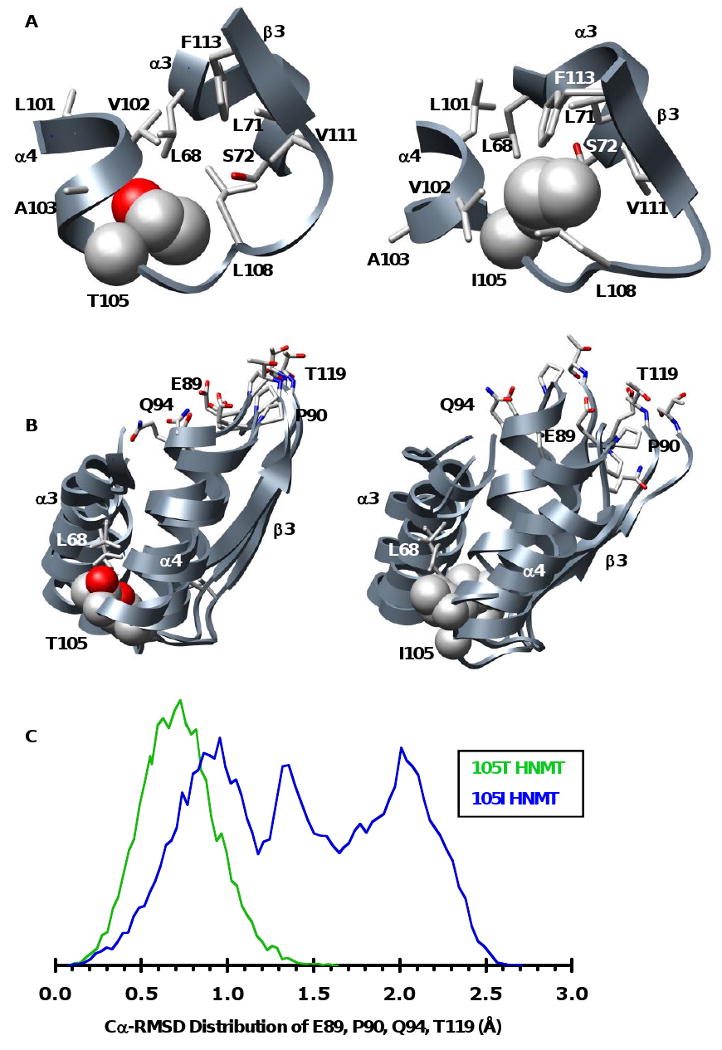
(A) Packing at the polymorphic site. The larger Ile is more buried, forming side-chain contacts with residues from α3 (L71, S72) and β3 (V111, F113) that are not present in the 105T simulations. (B) The polymorphic packing affects the SAM-binding site. Structural overlay of the polymorphic and SAM binding-sites from the 20ns structures of three independent simulations of 105T and 105I HNMT. The more tightly packed Ile acts as a pivot point between α3, α4 and β3 disrupting the orientation of key SAM-binding residues (E89, P90, Q94, T119). The side chain of the polymorphic residue and its contacts are colored by atom and shown in space-filling and licorice representations, respectively. (C) Distributions of the Cα-RMSDs for residues E89, P90, Q94 and T119 during the last 10ns of all three of the 105T (green) and 105I (blue) HNMT simulations at 37°C. The SAM-binding residues are more mobile in 105I HNMT, existing in a large ensemble of conformations that differ greatly from their respective positions in the starting structure. The distribution for 105T HNMT is narrower, indicating that the active site structure of 105T HNMT is maintained throughout the simulations. The vertical scales are arbitrary.
The overall solvent exposure of the polymorphic site was similar in all simulations (Table I); however, large variations in the solvent accessibility of both I105 and its surroundings occurred at 37°C (Table I). Residue 105 was buried to a greater extent in the 105I protein and the larger Ile formed side-chain contacts with additional residues in α3 (L71, S72) and β3 (V111, F113) that were absent in all of the 105T simulations (Table 1, Figures 5A, 6B). These packing differences suggest that any changes in the orientation of the smaller Thr side-chain are buffered by the additional space present in the polymorphic site and have little effect on the overall protein structure (Figure 5B). In contrast, the more tightly interacting Ile appears to act as a pivot point, affecting the orientation of nearby α3, α4 and β3 through its direct side-chain contacts and transmitting these changes to the SAM –binding site (Figure 5). Several residues at the distal ends of α4 (E89, P90, Q94) and β3 (T119) interact with the adenosine ring of SAM (Figure 1B). Although some of the contact distances and solvent accessibilities of residues within the SAM-binding site remain similar in both proteins (Tables I, II, III), many of the SAM-binding residues adjacent to the polymorphic site become much more flexible and disordered in the 105I simulations (Figures 3B, 3C, Tables II, III). This may explain the increase in the apparent KM of 105I HNMT for SAM (18).
Figure 6. Translation of polymorphic packing effects to the histamine-binding site.
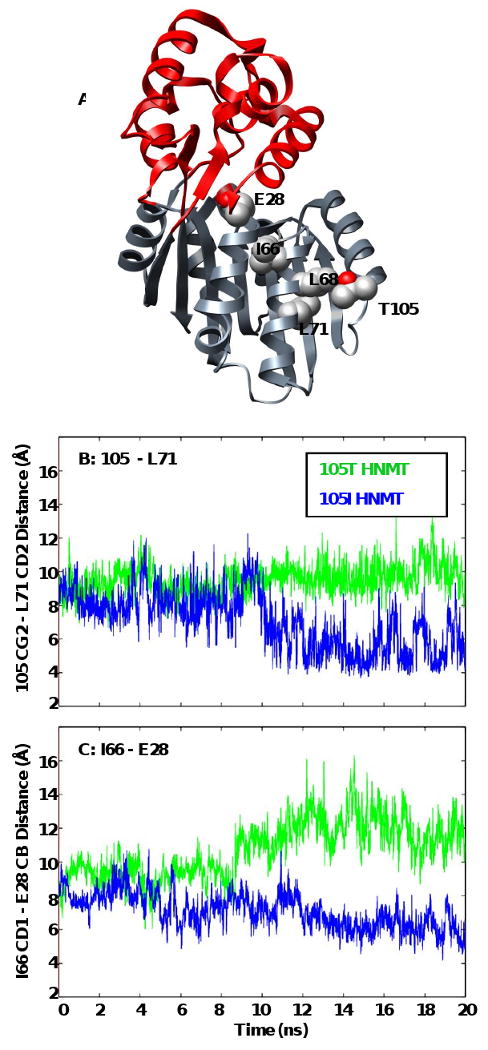
Secondary structures surrounding the polymorphic residue are more susceptible to changes in orientation brought on by altered contacts between L71 and S72 with the larger Ile side-chain. These structural changes are translated to the histamine-binding site via a new contact between α3 (I66) and α2 (E28). (A) Ribbon diagram of HNMT showing the location of residues 105, L71, L72, I66 and E28. Side-chains are shown in space-filling representation and colored by atom. Plots of (B) 105 CG2 – L71 CD2 (α3), and (C) I66 (α3) – E28 (α2, catalytic trio) contact distances for the 105T (green) and 105I (blue) HNMT simulations at 37°C.
In contrast to the effects on the SAM-binding site, the active-site distances and the solvent exposure of residues within the histamine-binding domain are larger and show greater fluctuations in the 105T HNMT simulations than those of the 105I protein (Tables II, III). The additional contacts of I105 that disorder the SAM-binding site may play a role in stabilizing the histamine-binding domain. The larger Ile forms side-chain contacts with L68, L71 and S72 in α3 (Figure 5). The reorientation of α3 in the 105I HNMT simulations allows these residues to interact with E28 (α2), a catalytic residue, through I66 (Figure 6C). Neither the I105-L71 nor the E28-I66 interactions occur in the T105 simulations (Figure 6). The restricted flexibility of this region in 105I HNMT could possibly impair substrate binding, accounting for the slight increase in its apparent KM for histamine (Table 3) (18).
Interestingly, catechol O-methyltransferase (COMT), a fellow member of the SAM-dependent methyltransferase fold family, also has a common polymorphism (V108M) that occupies an almost identical position in the COMT tertiary structure as T105I in HNMT (18, 28, 38). However, unlike T105I in HNMT, the COMT V108M polymorphism appears to result in decreased protein stability (39-41) and to have little effect on substrate binding (40, 42). MD simulations of the COMT variants showed that the larger Met formed closer side-chain contacts with residues within the polymorphic site, resulting in an increased sensitivity to structural changes in nearby helices and a distortion of the SAM-binding site (38). These changes were propagated throughout the protein, resulting in an increase in the overall SASA of 108M COMT and destabilization of the protein.
Conclusions
The HNMT active site undergoes a breathing motion, periodically exposing a trio of catalytic residues and a core hydrophobic patch important for substrate docking and orientation. Aromatic residues lining the highly flexible α1 then bury the substrate in the enzyme-substrate complex. The identity of residue 105 had a significant effect on active site structure and dynamics. I105 was buried to a greater extent and consequently contacted more residues within α3 and β3 than did T105. This altered packing reoriented α3 and β3, disordering several key SAM-binding residues on their distal ends while slightly stabilizing the histamine-binding domain.
Acknowledgments
Financial support for this work was provided by the National Institutes of Health (GM 50789 to VD) and the Canadian Institutes of Health (DRA MD-75910 to KR). Figures were produced using the USCF Chimera package from the Computer Graphics Laboratory, University of California, San Francisco (supported by NIH P41 RR-01081) (37). We thank Amanda Jonsson for running the simulations.
- HNMT
histamine N-methyltransferase
- SNP
single nucleotide polymorphism
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- 2PM
diphenhydramine
- COMT
catechol O-methyltransferase
- MD
molecular dynamics
- SASA
solvent accessible surface area
- Cα-RMSD
Cα-root-mean-square deviation from the starting structure
- Cα-RMSF
Cα-root-mean-square fluctuation about the mean structure
Footnotes
Financial support for this work was provided by the National Institutes of Health (GM 50789 to VD) and the Canadian Institutes of Health (DRA MD-75910 to KR).
References
- 1.Rosenwasser L. New insights into the pathophysiology of allergic rhinitis. Allergy Asthma Proc. 2007;28:10–5. doi: 10.2500/aap.2007.28.2977. [DOI] [PubMed] [Google Scholar]
- 2.Zhang M, Thurmond RL, Dunford PJ. The histamine H(4) receptor: a novel modulator of inflammatory and immune disorders. Pharmacol Ther. 2007;113:594–606. doi: 10.1016/j.pharmthera.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Schubert ML. Regulation of gastric acid secretion. Curr Opin Gastroenterol. 1999;15:457. doi: 10.1097/00001574-199911000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, Friis-Hansen L, Hakanson R, Zhao CM. Genetic dissection of the signaling pathways that control gastric acid secretion. Inflammopharmacology. 2005;13:201–7. doi: 10.1163/156856005774423872. [DOI] [PubMed] [Google Scholar]
- 5.Bacciottini L, Passani MB, Mannaioni PF, Blandina P. Interactions between histaminergic and cholinergic systems in learning and memory. Behav Brain Res. 2001;124:183–94. doi: 10.1016/s0166-4328(01)00230-3. [DOI] [PubMed] [Google Scholar]
- 6.Passani MB, Bacciottini L, Mannaioni PF, Blandina P. Central histaminergic system and cognition. Neurosci Biobehav Rev. 2000;24:107–13. doi: 10.1016/s0149-7634(99)00053-6. [DOI] [PubMed] [Google Scholar]
- 7.Yanai K, Tashiro M. The physiological and pathophysiological roles of neuronal histamine: an insight from human positron emission tomography studies. Pharmacol Ther. 2007;113:1–15. doi: 10.1016/j.pharmthera.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Schneider C, Risser D, Kirchner L, Kitzmuller E, Cairns N, Prast H, Singewald N, Lubec G. Similar deficits of central histaminergic system in patients with Down syndrome and Alzheimer disease. Neurosci Lett. 1997;222:183–6. doi: 10.1016/s0304-3940(97)13379-1. [DOI] [PubMed] [Google Scholar]
- 9.Kim SH, Cairns N, Fountoulakisc M, Lubec G. Decreased brain histamine-releasing factor protein in patients with Down syndrome and Alzheimer's disease. Neurosci Lett. 2001;300:41–4. doi: 10.1016/s0304-3940(01)01545-2. [DOI] [PubMed] [Google Scholar]
- 10.Kim SH, Krapfenbauer K, Cheon MS, Fountoulakis M, Cairns NJ, Lubec G. Human brain cytosolic histamine-N-methyltransferase is decreased in Down syndrome and increased in Pick's disease. Neurosci Lett. 2002;321:169–72. doi: 10.1016/s0304-3940(02)00051-4. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Novoa L, Cacabelos R. Histamine function in brain disorders. Behav Brain Res. 2001;124:213–33. doi: 10.1016/s0166-4328(01)00215-7. [DOI] [PubMed] [Google Scholar]
- 12.Takemura M, Kitanaka N, Kitanaka J. Signal transduction by histamine in the cerebellum and its modulation by N-methyltransferase. Cerebellum. 2003;2:39–43. doi: 10.1080/14734220310015601. [DOI] [PubMed] [Google Scholar]
- 13.Preuss CV, Wood TC, Szumlanski CL, Raftogianis RB, Otterness DM, Girard B, Scott MC, Weinshilboum RM. Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity. Mol Pharmacol. 1998;53:708–17. doi: 10.1124/mol.53.4.708. [DOI] [PubMed] [Google Scholar]
- 14.Chen GL, Wang H, Wang W, Xu ZH, Zhou G, He F, Zhou HH. Histamine N-methyltransferase gene polymorphisms in Chinese and their relationship with enzyme activity in erythrocytes. Pharmacogenetics. 2003;13:389–97. doi: 10.1097/00008571-200307000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Chen GL, Wang W, Xu ZH, Zhu B, Wang LS, Zhou G, Wang D, Zhou HH. Genotype-phenotype correlation for histamine N-methyltransferase in a Chinese Han population. Clin Chim Acta. 2003;334:179–83. doi: 10.1016/s0009-8981(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 16.Chen GL, Xu ZH, Wang W, Wang GP, Zhou G, Wang D, Zhou HH. Analysis of the C314T and A595G mutations in histamine N-methyltransferase gene in a Chinese population. Clin Chim Acta. 2002;326:163–7. doi: 10.1016/s0009-8981(02)00299-1. [DOI] [PubMed] [Google Scholar]
- 17.Scott MC, Van Loon JA, Weinshilboum RM. Pharmacogenetics of N-methylation: heritability of human erythrocyte histamine N-methyltransferase activity. Clin Pharmacol Ther. 1988;43:256–62. doi: 10.1038/clpt.1988.30. [DOI] [PubMed] [Google Scholar]
- 18.Horton JR, Sawada K, Nishibori M, Zhang X, Cheng X. Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons. Structure. 2001;9:837–49. doi: 10.1016/s0969-2126(01)00643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girard B, Otterness DM, Wood TC, Honchel R, Wieben ED, Weinshilboum RM. Human histamine N-methyltransferase pharmacogenetics: cloning and expression of kidney cDNA. Mol Pharmacol. 1994;45:461–8. [PubMed] [Google Scholar]
- 20.Price RA, Scott MC, Weinshilboum RM. Genetic segregation analysis of red blood cell (RBC) histamine N-methyltransferase (HNMT) activity. Genet Epidemiol. 1993;10:123–31. doi: 10.1002/gepi.1370100205. [DOI] [PubMed] [Google Scholar]
- 21.Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000;10:261–6. doi: 10.1097/00008571-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki Y, Ihara K, Ahmed S, Yamawaki K, Kusuhara K, Nakayama H, Nishima S, Hara T. Lack of association between atopic asthma and polymorphisms of the histamine H1 receptor, histamine H2 receptor, and histamine N-methyltransferase genes. Immunogenetics. 2000;51:238–40. doi: 10.1007/s002510050037. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S, Mann D, Singh TP, Ghosh B. Lack of association of histamine-N-methyltransferase (HNMT) polymorphisms with asthma in the Indian population. J Hum Genet. 2005;50:611–7. doi: 10.1007/s10038-005-0302-4. [DOI] [PubMed] [Google Scholar]
- 24.Deindl P, Peri-Jerkan S, Deichmann K, Niggemann B, Lau S, Sommerfeld C, Sengler C, Muller S, Wahn U, Nickel R, Heinzmann A. No association of histamine- N-methyltransferase polymorphism with asthma or bronchial hyperresponsiveness in two German pediatric populations. Pediatr Allergy Immunol. 2005;16:40–2. doi: 10.1111/j.1399-3038.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen GL, Zhu B, Nie WP, Xu ZH, Tan ZR, Zhou G, Liu J, Wang W, Zhou HH. Single nucleotide polymorphisms and haplotypes of histamine N-methyltransferase in patients with gastric ulcer. Inflamm Res. 2004;53:484–8. doi: 10.1007/s00011-004-1290-0. [DOI] [PubMed] [Google Scholar]
- 26.Oroszi G, Enoch MA, Chun J, Virkkunen M, Goldman D. Thr105Ile, a functional polymorphism of histamine N-methyltransferase, is associated with alcoholism in two independent populations. Alcohol Clin Exp Res. 2005;29:303–9. doi: 10.1097/01.alc.0000156128.28257.2e. [DOI] [PubMed] [Google Scholar]
- 27.Reuter M, Jeste N, Klein T, Hennig J, Goldman D, Enoch MA, Oroszi G. Association of THR105Ile, a functional polymorphism of histamine N-methyltransferase (HNMT), with alcoholism in German Caucasians. Drug Alcohol Depend. 2006 doi: 10.1016/j.drugalcdep.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Horton JR, Sawada K, Nishibori M, Cheng X. Structural basis for inhibition of histamine N-methyltransferase by diverse drugs. J Mol Biol. 2005;353:334–44. doi: 10.1016/j.jmb.2005.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levitt, M. (1990), Molecular Applications Group, Palo Alto, CA, and Yeda, Rehovot, Israel.
- 30.Beck, D. A. C., Alonso, D.O.V., Daggett, V. (2006), University of Washington, Seattle, WA.
- 31.Beck DA, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34:112–20. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Computer Physics Communications. 1995;91:215–231. [Google Scholar]
- 33.Levitt M, Hirshberg M, Sharon R, Laidig KE, Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. Journal of Physical Chemistry B. 1997;101:5051–5061. [Google Scholar]
- 34.Kell GS. Precise representation of volume properties of water at one atmosphere. Journal of Chemical Engineering Data. 1967;12:66–69. [Google Scholar]
- 35.Hubbard, S. J., Thorton, J.M. (1993), Department of Biochemicstry and Molecular Biology, University College, London.
- 36.Hunenberger PH, Mark AE, van Gunsteren WF. Fluctuation and cross-correlation analysis of protein motions observed in nanosecond molecular dynamics simulations. J Mol Biol. 1995;252:492–503. doi: 10.1006/jmbi.1995.0514. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Rutherford K, Bennion BJ, Parson WW, Daggett V. The 108M polymorph of human catechol O-methyltransferase is prone to deformation at physiological temperatures. Biochemistry. 2006;45:2178–88. doi: 10.1021/bi051988i. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, Kolachana BS, Hyde TM, Herman MM, Apud J, Egan MF, Kleinman JE, Weinberger DR. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–21. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, Taskinen J. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34:4202–10. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- 41.Shield AJ, Thomae BA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatry. 2004;9:151–60. doi: 10.1038/sj.mp.4001386. [DOI] [PubMed] [Google Scholar]
- 42.Goodman JE, Jensen LT, He P, Yager JD. Characterization of human soluble high and low activity catechol-O-methyltransferase catalyzed catechol estrogen methylation. Pharmacogenetics. 2002;12:517–28. doi: 10.1097/00008571-200210000-00003. [DOI] [PubMed] [Google Scholar]


