Abstract
DNA microarrays are invaluable tools for biosensing applications such as diagnostic detection of DNA and analysis of gene expression. Surface plasmon resonance imaging (SPRI) can detect unlabeled oligonucleotide targets adsorbed to the array elements. The variety of biosensing applications can be expanded by enzymatic manipulation of DNA microarray elements and the sensitivity of detection can be enhanced with the use of oligonucleotide immobilized onto a gold nanoparticle surface (detector-NP). We describe a novel method that couples a template-directed polymerase extension of a surface array element with nanoparticle-enhanced detection of the reaction product. Using this technique, it is possible to see as little as 10–100 attomoles of polymerase product representing as little as 0.25% of a monolayer. This sensitivity would allow for the detection of a specific DNA target that is present in low amounts in a sample and with partially unknown sequence. One application of this method would be to identify the presence of the aberrantly recombined DNA sequences, such as those found in the fragile sites of chromosomes.
DNA microarrays have become an invaluable tool for biosensing applications, such as diagnostic detection of DNA1 and analysis of gene expression.2 These arrays are used to adsorb a specific target oligonucleotide onto an array element, typically consisting of a complementary DNA sequence that has been covalently attached to the surface. Surface plasmon resonance imaging (SPRI)3,4 is increasingly used for the label-free detection of adsorbed strands. Enzymatic manipulation of DNA microarray elements offers another level of specificity for target oligonucleotide detection.5–7 Enzymes are used to either build the surface array elements (using RNA or DNA ligase)8–10 or detect target RNA or DNA binding to the array elements using RNase H5,6 and Exonuclease III8, respectively. Moreover, the sensitivity of biomolecule detection can be increased using nanoparticle-enhanced SPRI.11–14
Microarrays have previously been constructed to capture oligonucleotides14 and oligonucleotide-nanoparticle complexes have been used for detection of both surface-attached DNA probes13 and surface-adsorbed RNA oligonucleotides.11 Here we show that 10–100 attomoles of enzymatically extended surface-attached probe DNA can be detected with SPRI using oligonucleotide-coupled gold nanoparticles (Figure 1). This array-based method is an extremely sensitive means of detecting longer single-stranded DNA (>40 nts), but more importantly it is sequence specific at two different 20 nt locations that do not have to be contiguous. This allows for the analysis of longer DNA molecules with partially unknown sequence by providing separate sequences to anneal to a surface-bound oligonucleotide and a detector sequence. One application of this method will be in sensing aberrantly recombined DNA sequences of common and uncommon fragile sites of chromosomes. Fragile sites are specific areas of chromosomes where rearrangements, deletions, or breaks are found to occur reproducibly in the human genome and can cause human genetic diseases such as fragile X syndrome mental retardation and cancer.15 Our method allows highly sensitive detection of the sequences surrounding the fragile sites, e.g. in cancer samples with low concentration of recombined chromosomes, in which one of the flanking sequences is replaced by another sequence through recombination.
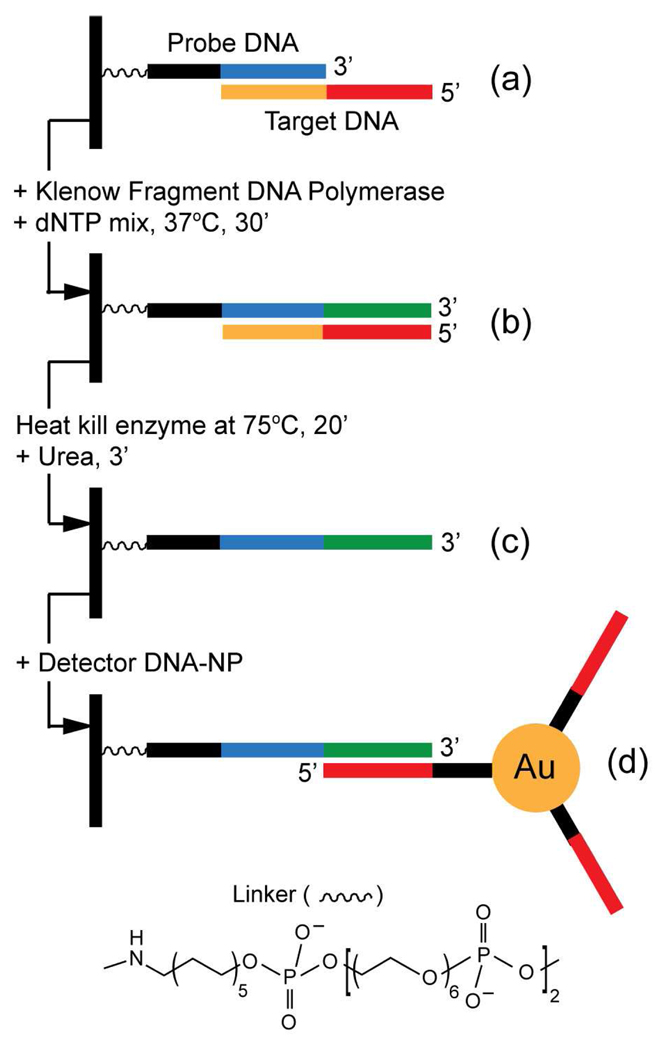
Figure 1.
The reaction scheme showing binding of a target DNA to the surface-bound probe DNA with a 5´ overhang. This overhang is used as a template for Klenow Fragment DNA polymerase. The slide is then heated and SPRI is used to detect the binding of a complementary DNA-associated gold nanoparticle.
Our process employs a primer attached to a glass surface that binds to an overhanging DNA template, which is then extended by DNA polymerase. This newly polymerized sequence is detected by base-pairing to a complementary DNA coupled to a gold nanoparticle (Figure 1). We began by performing the mesophilic DNA polymerase reaction on commercial CodeLink slides and detecting the reaction product using a fluorescently labeled oligodeoxynucleotide (general reaction scheme in Figure 1). An excess of aminolated DNA (250 µM) was covalently attached to the surface in areas where the NHS esters of the CodeLink slide had been protected from UV radiation by a chromium quartz mask. When the surface-bound DNA was annealed to the complementary target strand of DNA with a 5′ overhang, the Klenow Fragment of E. coli DNA polymerase I extended the probe. Following the reaction, the slide was heated to 75°C to denature both the enzyme and double-stranded DNA. The slide was placed in urea solution to prevent the re-annealing of complementary strands of DNA. The extended sequence of the DNA probe was detected by annealing to a Cy3-labeled complementary DNA (see Supporting Information). The fluorescent images of the reaction slide exhibited bright fluorescence and low background (Figure S1).
These fluorescence results indicated that the slides needed to be heated to remove non-specific binding of the enzyme to the surface. To study the Klenow Fragment reaction using SPRI, this same amino-modified DNA was to be attached to a gold spot on a glass surface using a method that would require the preliminary formation of a gold-sulfur bond.16 Since the gold-sulfur bond cannot withstand temperatures greater than 50°C, gold-spotted slides were glassified using a modified version of the method developed by Phillips et al. (see Supporting Information).17 The thickness of the glassified surfaces was 23 nm as measured by SPR, and SEM showed that the morphology was smooth (Figure S2).
DNA was attached to the calcinated surface using a combination of chemistries.12,16 A solution of 50 mM 3-aminopropyl-trimethoxysilane in ethanol/water (95%/5%) was prepared and allowed to stand for 20 minutes before filtering with a 0.22-µm cut-off syringe filter. The slide was immersed in the ethanol/water solution for 45 minutes, rinsed with water and reacted with a 2 mg/mL polyglutamic acid (pGlu) solution for 30 minutes. Two amine-modified oligonucleotides, probe DNA and T30 control (in 10 mM phosphate, 0.1 M NaCl, 5 mM MgCl2, pH 7.2) were mixed with NHS and EDC to form a covalent linkage between the 5′ terminal amine group of the DNA and the carboxylate of pGlu. Varying ratios (0:100 to 100:0) of DNA probe to control T30 DNA were reacted with the slide above the 1-mm gold spots (Figure 2, right inset).
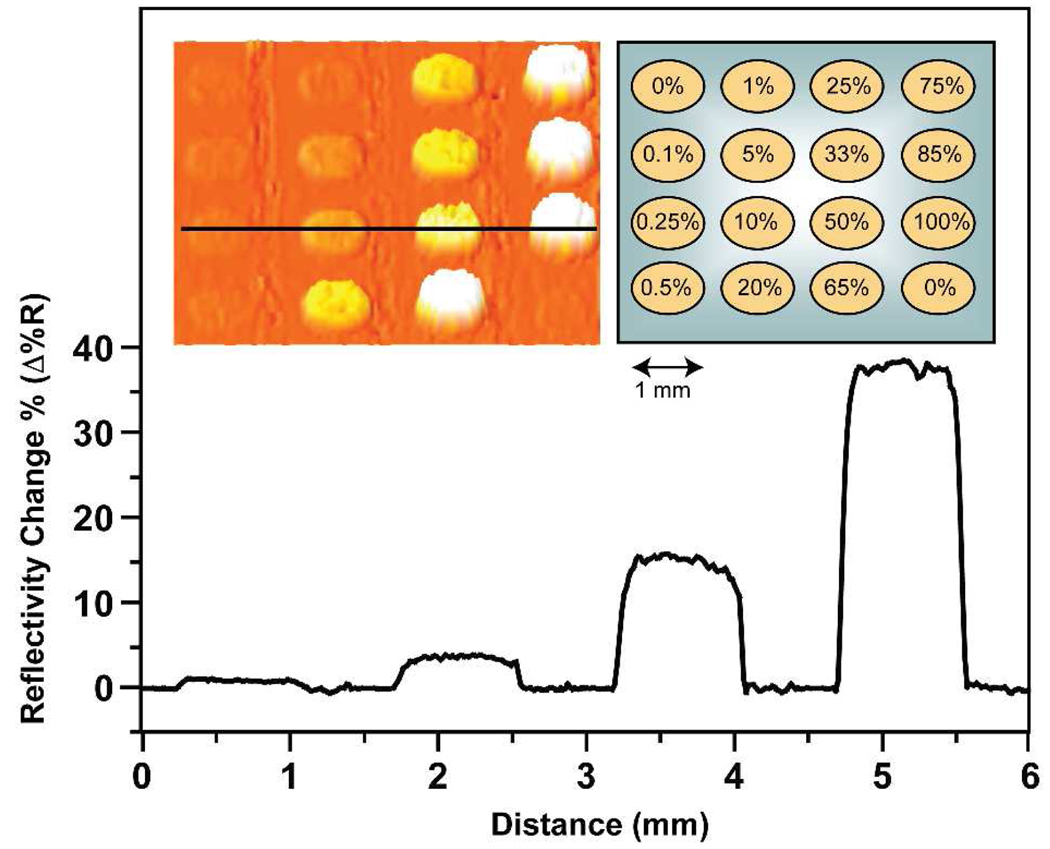
Figure 2.
Nanoparticle-enhanced SPR Imaging of the Klenow Fragment reaction products on glassified gold-spotted slides. Varying ratios of probe DNA:T30 were spotted onto the glassified surface above the gold spots (inset, right). The line profile corresponds to the area indicated by the black line in the left inset and shows the percent reflectivity change (Δ%R) due to the binding of nanoparticles to the surface-bound DNA.
The Klenow Fragment (0.5 U/µL) of the E. coli DNA polymerase was used to elongate the surface-bound probe using 16 µM target DNA as the template. The reaction took place in a Frame-Seal chamber and proceeded for up to 20 minutes at 37°C. The slide was heated to 75°C to denature the protein and DNA and placed in the SPR Imager. Gold nanoparticles coated with DNA (detector-NP) complementary to the newly synthesized part of the probe DNA were flowed into the SPR cell and allowed to bind for 15 minutes.
The SPR image obtained shows that the detector-NP binding has saturated the signal on all spots with ratios greater than 65:35 probe DNA:control DNA (Fig 2., left inset; Figure S3). The third row of the slide contains the full range of percent reflectivity change observed. Line profile analysis of this row revealed that the spot containing 100:0 probe:control DNA has an approximately 40% reflectivity change. The lowest signal was seen in the 0.25:99.75 probe:control DNA spot, or 0.25% probe DNA (Figure 2).
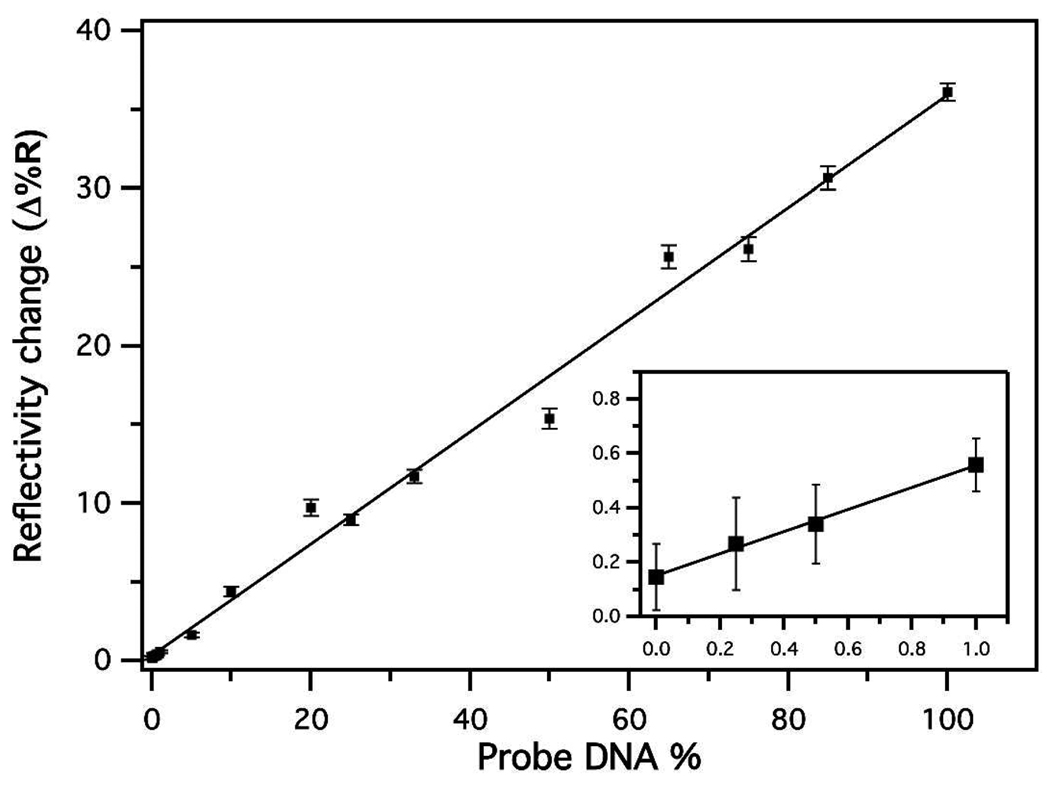
SPR data were collected at several different time points and used to calculate the change in the percent reflectivity due to detector-NP binding the enzymatically extended surface-bound DNA. The graph of the data gathered at 7 minutes indicates that the binding of detector-NP is increasing in a linear fashion, from the spots containing the lowest concentration of probe DNA (Figure 3, inset) to the spots comprised entirely of probe DNA (Figure 3).
Figure 3.
Adsorption of the detector-NPs to the enzymatically extended surface bound probe DNA. The change in percent reflectivity after detector-NPs were allowed to bind for 7 minutes increases linearly as a function of probe DNA surface density.
There is a detectable difference in reflectivity even at small concentrations of target DNA if the nanoparticles are allowed to bind for longer times. Using enzymatic amplification coupled with nanoparticle-enhanced SPRI, it is possible to see as little as 0.25% of the surface-bound DNA reaction product (Figure 3, Figure S4). The gold spot is approximately 1 mm2 and can hold 1010 DNA molecules. We can detect binding to 0.25% of a monolayer. This is 107–108 Klenow reaction products, or 10–100 attomoles of DNA. Assuming 10 amoles of DNA in a total reaction volume of 25 µL, the target concentration is calculated to be 400 fM. The advantage of the enzymatic detection method is that the traditional bioaffinity binding constant, Kads, does not have to be used in this calculation.14
While the binding of detector-NP to smaller amounts of surface DNA is still increasing linearly at 15 minutes, the plot of % reflectivity change for all concentrations of the surface polymerase product at 15 minutes fits to a first-order exponential curve, indicating saturation of binding to the spots containing higher amounts of probe DNA (Figure S5). Allowing the detector-NP to bind to the lower concentrations for longer periods of time could result in detection of even smaller amounts of enzyme product.
This method takes advantage of the innate template-directed polymerization function of DNA polymerase to extend the sequence of our surface attached-DNA probe in the 5′ to 3′ direction. The presence of a specific DNA sequence in solution can be determined by using the enzyme to create a DNA sequence that is: 1) covalently attached to the surface and 2) detectable by base-pairing to a complementary detector DNA. The coupling of nanoparticles to the detector DNA increases the sensitivity of this method so that attomole amounts of reaction product can be detected. The ability to identify a specific DNA present in low concentrations in a sample containing many other DNA sequences without thermal cycling will be a useful tool in diagnostic testing.
Supplementary Material
Acknowledgement
This work was supported by grants to RMC from the National Institute of Health (GM-059622) and the National Science Foundation (CHE-0551935) and University of California-Irvine (to AL). The authors are grateful to the California Institute for Telecommunications and Information Technology at UCI for help in the use of SEM instrumentation, Reginald Penner for the kind use of the furnace, and Aaron Halpern for useful discussions. RMC and AL are members of the Chao Family Comprehensive Cancer Center. RMC has a financial interest in GWC Technologies.
Footnotes
Supporting Information Available: Experimental section, Figures S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Kimmel A, Oliver B, editors. DNA Microarrays Part A: Array Platforms and Wet-Bench Protocols (Methods in Enzymology) Vol. 410. San Diego, CA: Elsevier Academic Press; 2006. [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Nelson BP, Grimsrud TE, Liles MR, Goodman RM, Corn RM. Anal. Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 4.Smith EA, Corn RM. Applied Spec. 2003;57:320A–332A. doi: 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- 5.Goodrich TT, Lee HJ, Corn RM. Anal. Chem. 2004;76:6173–6178. doi: 10.1021/ac0490898. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich TT, Lee HJ, Corn RM. J. Am. Chem. Soc. 2004;126:4086–4087. doi: 10.1021/ja039823p. [DOI] [PubMed] [Google Scholar]
- 7.Li YA, Wark AW, Lee HJ, Corn RM. Anal. Chem. 2006;78:3158–3164. doi: 10.1021/ac0600151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HJ, Li Y, Wark AW, Corn RM. Anal. Chem. 2005;77:5096–5100. doi: 10.1021/ac050815w. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Lee HJ, Corn RM. Nucleic Acids Res. 2006;34:6416–6424. doi: 10.1093/nar/gkl738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Lee HJ, Corn RM. Anal. Chem. 2007;79:1082–1088. doi: 10.1021/ac061849m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang SP, Lee HJ, Wark AW, Corn RM. J. Am. Chem. Soc. 2006;128:14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sendroiu IE, Warner ME, Corn RM. Langmuir. 2009;25:11282–11284. doi: 10.1021/la902675s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wark AW, Lee HJ, Qavi AJ, Corn RM. Anal. Chem. 2007;79:6697–6701. doi: 10.1021/ac071062b. [DOI] [PubMed] [Google Scholar]
- 14.Lee HJ, Wark AW, Corn RM. Langmuir. 2006;22:5241–5250. doi: 10.1021/la060223o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland GR. In: Genetic Translocations and Other Chromosome Aberrations. Leydon GT, editor. Hauppauge, NY: Nova Science Publishers, Inc.; 2008. pp. 209–222. [Google Scholar]
- 16.Chen YL, Nguyen A, Niu LF, Corn RM. Langmuir. 2009;25:5054–5060. doi: 10.1021/la804021t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips KS, Han JH, Martinez M, Wang ZZ, Carter D, Cheng Q. Anal. Chem. 2006;78:596–603. doi: 10.1021/ac051644y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.