Abstract
Epigenetic changes are among the most common alterations observed in cancer cells, yet the mechanism by which cancer cells acquire and maintain abnormal DNA methylation patterns is not understood. Cancer cells have an altered distribution of DNA methylation and express aberrant DNA methyltransferase 3B transcripts, which encode truncated proteins, some of which lack the C-terminal catalytic domain. To test if a truncated DNMT3B isoform disrupts DNA methylation in vivo, we constructed two lines of transgenic mice expressing DNMT3B7, a truncated DNMT3B isoform commonly found in cancer cells. DNMT3B7 transgenic mice exhibit altered embryonic development, including lymphopenia, craniofacial abnormalities, and cardiac defects, similar to Dnmt3b-deficient animals, but rarely develop cancer. However, when DNMT3B7 transgenic are bred with Eμ-Myc transgenic mice, which model aggressive B cell lymphoma, DNMT3B7 expression increases the frequency of mediastinal lymphomas in Eμ-Myc animals. Eμ-Myc/DNMT3B7 mediastinal lymphomas have more chromosomal rearrangements, increased global DNA methylation levels, and more locus-specific perturbations in DNA methylation patterns compared to Eμ-Myc lymphomas. These data represent the first in vivo modeling of cancer-associated DNA methylation changes and suggest that truncated DNMT3B isoforms contribute to the re-distribution of DNA methylation characterizing virtually every human tumor.
Introduction
Epigenetic changes, including DNA methylation and histone modifications, play an important role in cellular gene regulation (1). Highly methylated DNA is typically transcriptionally inactive, whereas hypomethylated DNA is associated with active, open chromatin (2). Repetitive DNA is usually highly methylated and transcriptionally silenced, effectively inactivating transposable elements that could mediate genomic rearrangements. The distribution of DNA methylation is highly regulated during development and can become altered during aging and malignant transformation (1, 3). Repetitive DNA sequences and some gene promoters are hypomethylated in tumors compared to normal cells, and the promoters of many tumor suppressor genes are hypermethylated (1). These changes contribute to the abnormalities in genetic/chromosomal stability, growth control, and apoptosis observed in tumor cells.
Three DNA methyltransferase (DNMT) enzymes have been identified in eukaryotic cells (4). DNMT1 is generally considered to be a maintenance methyltransferase, although it also has de novo methyltransferase activity (4). Both DNMT3A and DNMT3B catalyze de novo methylation of DNA sequences (5).
The mechanism(s) by which cancer cells acquire abnormal DNA methylation is not understood completely, but aberrant transcription of the DNMT3B gene is widespread across all human cancer cells studied to date (6). Most of the DNMT3B transcripts found in cancer cells encode catalytically active DNMT3B proteins, but aberrant splicing also produces DNMT3B transcripts containing premature stop codons, which encode truncated proteins lacking the catalytic domain (7). Expression of one of the most commonly expressed aberrant transcripts, DNMT3B7, within 293 cells resulted in hypermethylation of some promoters (e.g. CDH1) and hypomethylation of others (e.g. MAGEA3) in a manner similar to that seen in cancer cells (7). Other transcripts identified in non-small cell lung cancer, the ΔDNMT3B1-7 transcripts, are generated from a promoter internal to the DNMT3B gene (8–10). Additionally, another splice variant termed DNMT3B3Δ5 is highly expressed in induced pluripotent stem cells and displays variable levels of expression in human tumor cells lines depending on the tissue of origin (11).
Altered expression of DNMT3B can also play a role in disease states other than cancer. About half of patients with a rare autosomal recessive syndrome, the Immunodeficiency; Centromere instability; Facial anomalies (ICF) Syndrome, have detectable germline mutations in DNMT3B (12) and display hypomethylation of pericentromeric repetitive DNA. Additionally, mice lacking each of the Dnmts (13, 14) and mice expressing ICF mutations (15) demonstrate hypomethylation of repetitive elements and embryonic lethality and/or developmental abnormalities, all of which substantiate the central role of DNA methylation during embryonic and early post-natal development.
The widespread expression of truncated DNMT3B proteins in human cancers as well as the alteration in DNA methylation seen in vitro with forced expression of DNMT3B7 (7) suggested that truncated DNMT3B proteins could alter DNA methylation within cancer cells. Furthermore, the identification of truncated DNMT3B isoforms consisting of the N-terminal domain within the human ICF Syndrome gave additional support to the hypothesis that truncated DNMT3B proteins have a physiologic effect. The work we present here tests whether DNMT3B7, one of the truncated DNMT3B isoforms found in human tumors, can disrupt mouse development and alter tumorigenesis by inducing changes in DNA methylation.
Materials and Methods
Generation of transgenic mice and monitoring of tumors
The DNMT3B7 transgenic mice were generated using standard techniques (see Supplementary Materials and Methods).
Reverse transcription and PCR amplification
Total RNA was made using STAT-60 (Tel-Test) or Trizol (Invitrogen). Reverse transcription was performed using SuperScriptII (Invitrogen), and PCR amplifications were performed using AmpliTaq Gold polymerase (Applied Biosystems), using primers listed in Supplementary Table S1.
Western blotting
Protein extracts were made as previously described (7), and Western blots were performed with the following antibodies: anti-Dnmt3b: T-16, sc-10236, Santa Cruz Biotechnology; or ab2851, Abcam; anti-UBC9: N-15, sc-5231, Santa Cruz Biotechnology; anti-lamin B: M-20, sc-6217, Santa Cruz Biotechnology; anti-α-tubulin: T9026, Sigma-Aldrich; anti-FLAG: F3165, Sigma-Aldrich. Immunoprecipitation was performed with anti-HA antibody: clone 3F10, 11867423001, Roche Applied Science.
Flow cytometry and serum immunoglobulin level analysis
Flow cytometry using a BD FACSCanto was performed using antibodies from BD Pharmingen: PE anti-mouse B220 (553089), APC anti-mouse IgM (550676), PE anti-mouse CD4 (553730), and APC anti-mouse CD8 (17-0081-81). Serum immunoglobulin levels were performed in triplicate using the SBA Clonotyping™ System/HRP (Southern Biotech).
Generation and growth curve analysis of mediastinal lymphoma cell lines
Primary B cell cultures were generated as described (16).
Cytogenetic analysis
Cytogenetic analysis was performed on fresh single cell suspensions from mediastinal lymphomas as described (17).
Analysis of 5-methylcytosine levels
Genomic DNA was hydrolyzed and analyzed in triplicate for the relative levels of 5-methyldeoxycytidine by liquid chromatography-electrospray ionization tandem mass spectrometry (18).
Gene expression studies
RNA was isolated using Trizol (Invitrogen), purified by the RNeasy Mini Kit (Qiagen), and hybridized to Affymetrix Mouse Genome 430 2.0 gene expression microarrays at The University of Chicago Functional Genomics Facility. For details regarding data analysis, see Supplementary Materials and Methods.
DNA methylation analysis
Genomic DNA was isolated using the AquaPure Genomic DNA Isolation Kit (Bio-Rad). The HELP assay was performed (19), with details provided in Supplementary Materials and Methods. Statistical analysis was performed using the statistical software R 2.8.1 (20). These data can be found at the NCBI Gene Expression Omnibus (GEO) website under accession number GSE21273 (21). PCR primers used to amplify sodium bisulfite-treated DNA are given in Supplementary Table S2.
Results
DNMT3B7 expression in a transgenic mouse model
To test our hypothesis that a cancer-related DNMT3B isoform could disrupt DNA methylation in vivo, we engineered transgenic mice to express DNMT3B7, an isoform consisting of the first 360 N-terminal amino acids of DNMT3B (Fig. 1A). Because cancer cells overexpress full-length DNMT3B transcripts in addition to the aberrant forms, we chose to express DNMT3B7 using transgenic mice to avoid disrupting endogenous murine Dnmt3b expression. We were concerned that widespread DNMT3B7 expression might disrupt embryonic development or cause reproductive defects, since treatment of mice with 5-aza-2′-deoxycytidine, a hypomethylating agent, renders them infertile (22). Therefore, we designed the transgenic construct (Fig. 1A) using the immunoglobulin heavy chain enhancer and promoter with the goal of achieving lymphocyte-specific expression in adult animals. Lymphocytes are very sensitive to changes in DNA methylation, since mice expressing a hypomorphic Dnmt1 allele develop aggressive T cell lymphomas (23, 24).
Figure 1.
Phenotypic abnormalities of DNMT3B7 transgenic mice. A, Top, Schematic diagram of DNMT3B, and the DNMT3B7 isoform. Numbers indicate positions of amino acids (aa). N, N-terminus; C, C-terminus; NLS, nuclear localization signal; PWWP domain, conserved proline-tryptophan-tryptophan-proline motif; PHD domain, plant homeodomain. The asterisks represent the five novel amino acids present in DNMT3B7. Bottom, Schematic diagram of the DNMT3B7 transgenic construct. B, Coronal sections of E15.5 wild-type and DNMT3B7 transgenic mice. Red arrowheads indicate the right eye, and green arrowheads indicate the left eye. Black arrowheads indicate the location of the hard palate. C, Hemotoxylin and eosin-stained sections of the ventricles from E14.5 wild-type and DNMT3B7 animals. A black arrowhead indicates the location of a sub-aortic ventricular septal defect in a homozygous transgenic animal, and yellow arrows indicate the presence of a thin myocardium. RV, right ventricle; LV, left ventricle. D, FACS analysis of B lymphocytes and serum immunoglobulin analysis from Line A transgenic animals. The percentage of total B lymphocytes (top) and of mature IgM+B220+ B lymphocytes (middle) present in the peripheral blood of Line A homozygous versus wild-type animals at P0. Quantitation of IgA levels measured from serum of Line A homozygous versus wild-type animals (bottom). *** denotes P≤0.0003, ** denotes P≤0.009, and * denotes P≤0.01 between wild-type and homozygous animals using the two-tailed Student’s t-test. Values represent mean ± s.e.m.
From our transgenic construct, we obtained eight independent founder animals, six of which were sterile. From the remaining two founders, we generated two lines of DNMT3B7 transgenic mice, Line A and Line C. We used fluorescence in situ hybridization to map the Line A transgene insertion site to chromosome 5, band G1, and the Line C transgene insertion site to chromosome 16, band C1–C2 (data not shown). The endogenous genes Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l are located on other chromosomes, so the transgene insertions could not have resulted directly in Dnmt hypomorphic alleles.
DNMT3B7 transgenic mice show developmental abnormalities
Each line of DNMT3B7 mice displayed developmental abnormalities, the severity of which depended on whether the transgene array was present in the hemizygous or homozygous state. DNMT3B7 mice were smaller than their littermates (Supplementary Fig. S1), and had craniofacial, cardiac, and immune defects (Fig. 1A–D), described in more detail below. The most severely affected Line C homozygotes died between E8.5–E9.5, and Line A homozygotes died within hours of birth (Supplementary Table S3). We observed similar phenotypes in both lines, although they were generally more severe in Line C, suggesting that the phenotypes are a direct result of transgene expression, rather than the result of position effects.
DNMT3B7 is expressed dynamically during embryogenesis
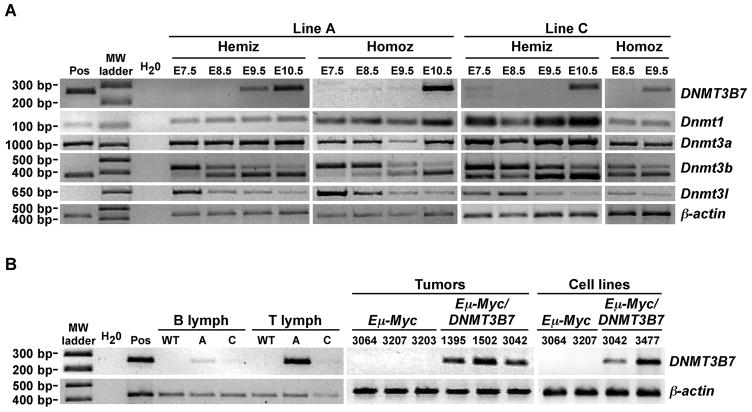
RNA in situ hybridizations demonstrated DNMT3B7 expression during embryogenesis, with widespread expression at E9.5, becoming more restricted with advanced development (Supplementary Fig. S2A–D). We also compared the expression of DNMT3B7 to endogenous levels of Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l by RT-PCR in embryos from E7.5–E10.5 (Fig. 2A). Overall, we observed very low levels of DNMT3B7 expression beginning at E7.5, with a sharp increase in expression at E10.5. Each of the endogenous Dnmts was expressed at much higher levels than the transgene and at levels comparable to those seen in wild-type embryos at the same ages (Supplementary Fig. S2E). DNMT3B7 expression was also observed in adult animals in lymphoid tissues, as expected from the use of the immunoglobulin heavy chain enhancer and promoter, as well as in brain and testis (Fig. 2B; Supplementary Fig. S2F–H).
Figure 2.
DNMT3B7 expression in Line A and Line C by RT-PCR. A, RT-PCR amplification in DNMT3B7 transgenic embryos from E7.5–E10.5. The positive control (Pos) is from a Line A hemizygous embryo at E15.5. Molecular weight markers are indicated to the left. Hemiz, hemizygous; Homoz, homozygous. The identities of each amplicon are given to the right. Amplification of β-actin served as the loading control. B, RT-PCR amplification of DNMT3B7 in: B and T lymphocytes from DNMT3B7 transgenic adults at 10–20 weeks of age; Eμ-Myc/DNMT3B7 mediastinal tumors; and Eμ-Myc/DNMT3B7 mediastinal tumor cell lines. Lymph, lymphocytes; WT, wild-type; A, Line A; C, Line C. Mouse identification numbers are given along the top.
DNMT3B7 transgenic mice have craniofacial, cardiac, and immune defects
The craniofacial abnormalities that were observed in the DNMT3B7 mice included cleft palate and severe microphthalmia on the right, with a normal appearing eye on the left (Fig. 1B; Supplementary Table S4). Cardiac abnormalities, including side-by-side great vessels, thin myocardium, and sub-aortic ventricular septal defects (VSDs), the same type of VSD observed in Dnmt3b−/− mice (15), were also observed in both lines of DNMT3B7 mice (Fig. 1C; Supplementary Table S5). Some animals with severe craniofacial defects had a normal heart (Supplementary Table S6), suggesting some variation in the effects on the cells giving rise to these structures. Similar to what has been observed in humans with ICF syndrome and ICF mutant mice, Line A homozygous animals exhibited lymphopenia and immunodeficiency, with lower numbers of total and mature B lymphocytes and lower IgA levels in the peripheral blood (Fig. 1D).
DNMT3B7 interacts with DNMT3B in 293 cells and leads to chromosomal rearrangements
Since the phenotypes of DNMT3B7 mice are similar to some of the defects seen in Dnmt3b−/− embryos (15), we wanted to test the interaction of DNMT3B7 with full-length DNMT3B. We found that DNMT3B7 can co-immunoprecipitate with DNMT3B, suggesting that DNMT3B7 may be binding to Dnmt3b and disrupting its normal function in transgenic mice (Supplementary Fig. S3) (see Discussion). DNMT3B7 expression led to increased numbers of chromosomal rearrangements of the pericentromeric region of chromosome 15 as well as hypomethylation of a repetitive element located in the region of the chromosomal rearrangements (Supplementary Fig. S4).
DNMT3B7 transgenic mice rarely develop spontaneous cancer
Given the frequent expression of truncated DNMT3B isoforms in human cancers, we followed the DNMT3B7 mice closely for the development of hematopoietic tumors. Out of approximately sixty animals followed over one year, we observed only one spontaneous tumor, an erythroleukemia that developed in a 43 week old Line A DNMT3B7 hemizygous female (Supplementary Fig. S5). We estimate the frequency of leukemia development at 1–2% at one year, which is greater than the rate of spontaneous development of erythroleukemia in C57Bl/6J mice, estimated at <1% (25).
DNMT3B7 expression alters the kinetics of mediastinal lymphoma formation in Eμ-Myc transgenic mice
Given the rarity of cancer development in DNMT3B7 mice, we tested whether DNMT3B7 expression could alter tumorigenesis in Eμ-Myc transgenic mice, in which 94% of the transgenic mice develop B cell lymphomas by 4 months of age (26). Although the majority of Eμ-Myc mice develop peripheral lymphomas, about 7% develop mediastinal lymphomas (27). Since we observe aberrant transcription of DNMT3B in all human tumors tested, we first investigated whether Eμ-Myc-induced lymphomas themselves expressed aberrant Dnmt3b transcripts. RT-PCR for Dnmt3b within Eμ-Myc transgenic tumors demonstrated no aberrant Dnmt3b transcription (data not shown).
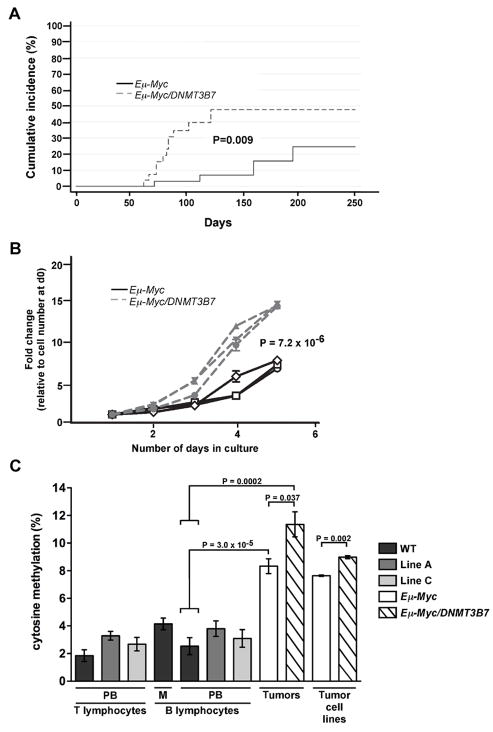
We introduced DNMT3B7 expression into Eμ-Myc mice by crossing them to the DNMT3B7 Line A transgenic mice. Approximately 30 Eμ-Myc and 30 Eμ-Myc/DNMT3B7 mice were followed longitudinally for tumor development. We found that the incidence of mediastinal lymphomas in Eμ-Myc/DNMT3B7 mice was much higher than in Eμ-Myc mice (Fig. 3A), with more than 50% of these mice developing mediastinal lymphomas within the first 120 days. Eμ-Myc/DNMT3B7 mice generated with the Line C insertion also developed mediastinal tumors early, although a large cohort was not followed (data not shown). In contrast, only about 20% of Eμ-Myc mice developed mediastinal lymphomas, the majority of which occurred after 120 days. Both Eμ-Myc and Eμ-Myc/DNMT3B7 mice developed peripheral lymphomas at similar rates (data not shown). RT-PCR confirmed expression of DNMT3B7 in Eμ-Myc/DNMT3B7 transgenic tumors (Fig. 2B).
Figure 3.
Kinetics and DNA methylation levels of mediastinal lymphomas from Eμ-Myc versus Eμ-Myc/DNMT3B7 transgenic mice. A, Cumulative incidence curves for the development of mediastinal lymphomas in Eμ-Myc (solid line) versus Eμ-Myc/DNMT3B7 mice (dashed line). Curves were compared using a K-sample test (Gray statistics=6.80, P=0.009). B, Growth curves of cell lines established from mediastinal lymphomas of Eμ-Myc versus Eμ-Myc/DNMT3B7 mice. Curves represent average growth obtained from triplicate cultures of three independently derived cell lines for each genotype. Curves were compared using the two-tailed Student’s t-test (P=7.2 × 10−6). C, Quantitation of total 5-methylcytosine levels in normal compared to malignant lymphocytes, n ≥ 3 for each genotype. PB, peripheral blood; M, mediastinal cells. Mouse genotypes are distinguished by color: black, wild-type; dark grey, DNMT3B7, Line A; light grey, DNMT3B7, Line C; white, Eμ-Myc; hatched, Eμ-Myc/DNMT3B7. Results shown are mean ± s.e.m. P values were calculated using the two-tailed Student’s t-test.
Cell lines generated from mediastinal lymphomas (Fig. 2B; Supplementary Fig. S2I) showed that cells derived from Eμ-Myc/DNMT3B7 mediastinal lymphomas grew three times more rapidly than those from Eμ-Myc tumors (Fig. 3B). Furthermore, there was no difference in the number of total circulating lymphocytes (Supplementary Table S7) or in the percentage of B lymphocytes in DNMT3B7 mice (Supplementary Fig. S6). Taken together, these results suggest that the expression of DNMT3B7 alters the process of tumorigenesis in the Eμ-Myc/DNMT3B7 mice rather than increasing the susceptible cell population.
Mediastinal lymphomas from Eμ-Myc/DNMT3B7 mice have increased genetic instability
Tumor histology demonstrated that the lymphomas, bone marrows, and spleens from Eμ-Myc and Eμ-Myc/DNMT3B7 mice were indistinguishable, showing sheets of large, blast-like cells with large diffuse nuclei (data not shown). Likewise, the tumors of Eμ-Myc and Eμ-Myc/DNMT3B7 both expressed pre-B cell (B220) and mature B cell markers (IgM) (data not shown). Spectral karyotyping (SKY) analysis demonstrated that Eμ-Myc/DNMT3B7 tumors had an average of 5.6 clonal abnormalities, significantly higher than the 1.75 average clonal abnormalities in the Eμ-Myc tumors (Table 1), suggesting that DNMT3B7 expression promotes chromosomal instability.
Table 1.
Cytogenetic analysis from Eμ-Myc and Eμ-Myc/DNMT3B7 transgenic mediastinal lymphomas.
 |
Only clonal abnormalities are listed. Tetraploidization was scored as a single abnormality. For tumor 3042, we inferred the existence of a primary diploid clone with loss of chromosomes 6, 7, and 19, followed by polyploidization, and the subsequent acquisition of additional numerical abnormalities.
P = 0.03
DNMT3B7 affects global DNA methylation levels in a tumor background
Using mass spectrometry to quantitate total 5-methylcytosine levels, there was little difference in the amount of methylated cytosine in genomic DNA from peripheral blood B and T lymphocytes derived from wild-type and DNMT3B7 transgenic mice (Fig. 3C). However, there was a significant increase in the amount of DNA methylation in Eμ-Myc-induced tumors, with a further increase seen in mediastinal tumors from Eμ-Myc/DNMT3B7 mice. Cell lines generated from mediastinal tumors of those mice also recapitulated this difference.
Mediastinal lymphomas from Eμ-Myc/DNMT3B7 mice demonstrate gene expression changes associated with corresponding DNA methylation alterations
Gene expression changes in mediastinal tumors from two Eμ-Myc and three Eμ-Myc/DNMT3B7 mice were measured by the Affymetrix Mouse Genome 430 2.0 microarray. A P value of 0.05, and a false discovery rate of 0.01 were used as thresholds to perform supervised clustering, identifying 328 genes that distinguished Eμ-Myc/DNMT3B7 tumors from Eμ-Myc tumors (Supplementary Table S8 and Supplementary Fig. S7).
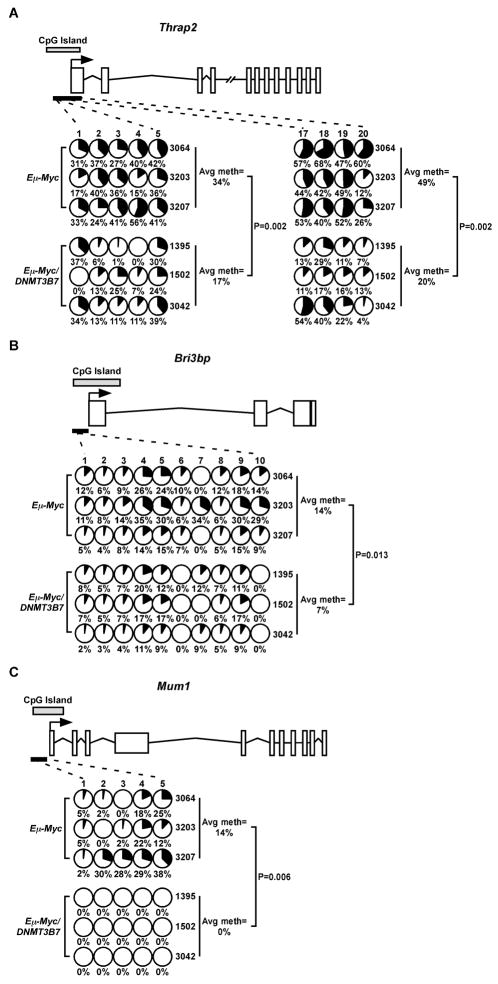
To test if the differential expression of genes in the lymphomas of Eμ-Myc/DNMT3B7 mice was the result of an alteration in their DNA methylation patterns, we performed bisulfite sequencing of selected genes demonstrating a >1.2 fold change that also had a CpG island in the region surrounding their transcriptional start site. Percent methylation at each CpG position was determined by sequencing PCR amplicons in bulk and calculating the ratio of C to T. Thrap2, Bri3bp, and Mum1 had between a 1.8–3.5-fold increase in gene expression in Eμ-Myc/DNMT3B7 lymphomas (Supplementary Table S8; confirmed by quantitative RT-PCR shown in Supplementary Fig. S8), and each gene had regions of hypomethylation within their respective CpG islands/promoters (Fig. 4).
Figure 4.
DNA methylation changes in specific genes from gene expression profiling of mediastinal lymphomas. Methylation changes were assessed by bisulfite sequencing in the CpG islands of A, Thrap2; B, Bri3bp; and C, Mum1. Schematic diagrams for each gene are shown (not to scale). Exons are represented by vertical rectangles, and the location of the CpG island is shown with a horizontal, shaded rectangle. The black arrow indicates the location of the transcriptional start site (TSS). The smaller, black horizontal rectangle indicates the location of the CpGs analyzed for changes in DNA methylation. Each row represents methylation data from a single lymphoma, indicated to the right. Numbers across the top indicate specific CpG dinucleotides in a region of the CpG island. Changes in methylation of specific CpG dinucleotides are indicated by small, shaded circles. Shading indicates the average amount of DNA methylation at each CpG, and the number below the circles represents percent methylated cytosine. Average percent methylation is indicated to the right, and P values were calculated using the two-tailed Student’s t-test.
We also measured DNA methylation by performing a microarray-based assay, HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) (19, 28–30) on mediastinal lymphomas from Eμ-Myc versus Eμ-Myc/DNMT3B7 mice. We validated several genes in Eμ-Myc/DNMT3B7 mediastinal lymphomas that demonstrated changes in their DNA methylation profiles, one of which was hypomethylated (Irs4), and others that were hypermethylated (Galnt3 and Sfrs8) (Supplementary Fig. S9 and data not shown). We also performed the HELP assay on DNA from the brain, to measure DNA methylation perturbations in a non-tumor tissue. In contrast to the mediastinal lymphomas, we observed fewer genes with DNA methylation changes in the brains of DNMT3B7 mice. We found only moderate changes at CpG dinucleotides that are part of HpaII sites in Ubfd1, as well as in its CpG island (Supplementary Fig. S10).
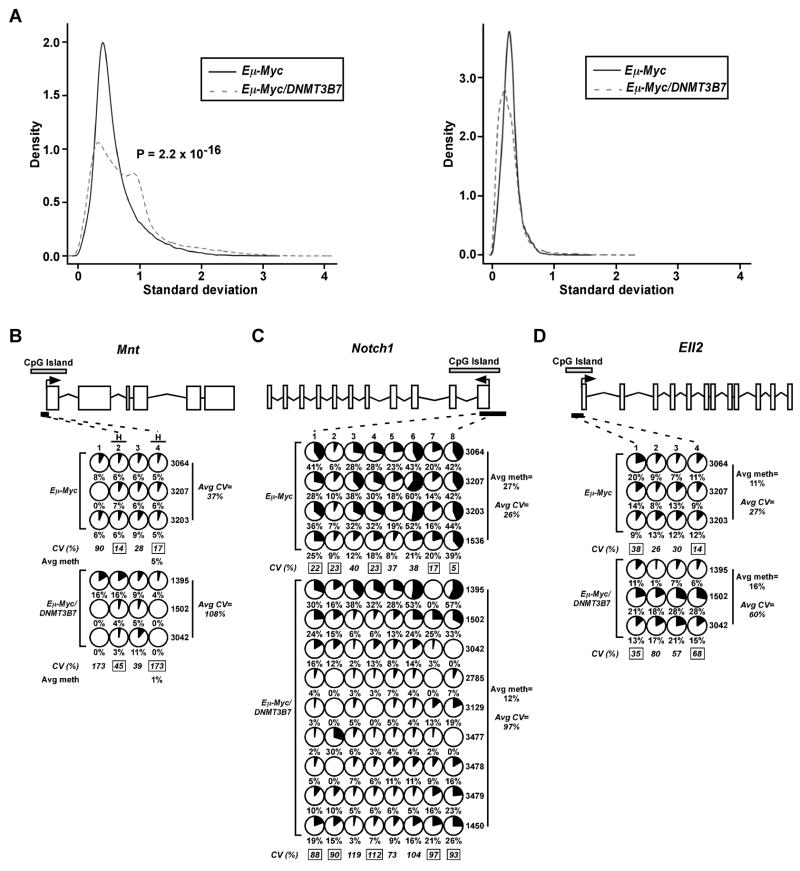
In addition to uncovering consistent changes in DNA methylation, the HELP assay revealed significant heterogeneity in the DNA methylation profiles of particular CpG dinucleotides within individual Eμ-Myc/DNMT3B7 tumors as measured by an increase in the gene methylation standard deviation across these tumors, whereas the Eμ-Myc lymphomas had much more consistent DNA methylation patterns (Fig. 5A). This variability in DNA methylation among the Eμ-Myc/DNMT3B7 tumors was observed for individual genes (e.g. Mnt and Supt4h2) identified by the HELP assay and validated using bisulfite sequencing, in which the variability was quantified by calculating the coefficient of variance (CV) (Fig. 5B and Supplementary Fig. S9). For example, in Notch1 (Figure 5B), at the CpG at position 8 in Eμ-Myc tumors, the amount of methylation varies between 39–44% in each tumor, with a CV of 5. However, in Eμ-Myc/DNMT3B7 tumors, methylation of this CpG dinucleotide varies from 0–57%, with a significantly higher CV of 93, reflecting the generalized deregulation in DNA methylation patterns observed in these tumors.
Figure 5.
Increased variability in DNA methylation of particular CpG dinucleotides within gene promoters is observed in mediastinal lymphomas of Eμ-Myc/DNMT3B7 mice. A, Left, Standard deviation of the signal density obtained from the HpaII channel of the HELP assay performed on Eμ-Myc lymphomas (solid line) and Eμ-Myc/DNMT3B7 lymphomas (dashed line). Right, Standard deviation of the signal density obtained from the MspI (control) channel of the HELP assay performed on Eμ-Myc lymphomas and Eμ-Myc/DNMT3B7 lymphomas. B–D, Overall DNA hypo- or hypermethylation is mediated by individual CpG dinucleotides to varying degrees, as measured by bisulfite sequencing. Methylation changes in the CpG island of B, Mnt, C, Notch1, and D, Ell2. Schematic diagrams of each gene and the amount of methylation at CpG dinucleotides are as described in the legend of Figure 4. In panel B, CpG positions with an “H” over the number indicate the location of a HpaII site. Average percent methylation for each genotype is indicated to the right, except in panel B, where it is shown under the significant HpaII site. The average coefficient of variance (CV) is given as a percentage underneath each CpG position. CV values with a black box around the number denote P≤0.05 for Eμ-Myc versus Eμ-Myc/DNMT3B7 lymphomas at specific CpGs. The average CV across all positions for each genotype is indicated to the right.
This high degree of variability in DNA methylation levels was seen in the CpG islands/promoters of genes identified through gene expression profiling as well. Specifically, Notch1, Foxp4, and Sox4 showed increased expression levels (Supplementary Table S8) in double transgenic tumors and were generally hypomethylated. Each showed a high degree of variability in which specific CpG dinucleotides displayed hypomethylation as well as in the degree of DNA methylation at those positions (Fig. 5C and Supplementary Fig. S9D-E). We also observed DNA methylation changes in the “CpG island shore” (3) of Notch1 (Supplementary Fig. S9F). Additionally, other genes, such as Ell2, which were underexpressed (Supplementary Table S8), showed an increase in DNA methylation of their CpG islands/promoters, again with variability in the DNA methylation profile (Fig. 5D). This observation argues in favor of a generalized perturbation of DNA methylation in the Eμ-Myc/DNMT3B7 lymphomas.
Discussion
Human tumors are characterized by a re-distribution of DNA methylation, enabling “DNA methylation signatures” to be described for many cancers (30–32). Our work tests the hypothesis that a truncated DNMT3B isoform commonly found in human cancer cells, DNMT3B7, can alter DNA methylation. Our findings of disrupted mammalian development and accelerated lymphomagenesis with increased chromosomal instability and perturbed DNA methylation patterns supports the hypothesis that truncated DNMT3B proteins can disrupt DNA methylation and alter tumorigenesis in vivo.
The precise embryonic defects that we observe in two independent lines of transgenic mice, craniofacial abnormalities, ventricular septal defects, and lymphopenia, are reminiscent of those seen in Dnmt3b-deficient animals. Mice lacking Dnmt3b die prior to E14.5, with sub-aortic ventricular septal defects identical to those seen in DNMT3B7 animals (14). In addition, mice expressing ICF Syndrome-associated mutations within the catalytic domain survive birth, but display low body weight, craniofacial malformations, and T cell apoptosis (15). Furthermore, human ICF patients have documented developmental abnormalities, including ventricular or atrial septal defects, cleft palate, and immunodeficiency, with decreased levels of IgA, IgM, and IgG2 (12). Notably, three of the described DNMT3B mutations in patients with ICF Syndrome are premature termination mutations within the 5′ end of the gene, like those in cancer cells: Q42Term, R54Term, and Q204Term. DNMT3B7 consists of the first 360 amino acids of the full-length protein and may possess a biologic activity similar to the ICF-associated isoforms.
Given the potential of DNMT3B7 to disrupt DNMT function, one possible outcome of expression of a truncated DNMT3B isoform was the development of lymphoma or another hematopoietic tumor. Interestingly, mice transplanted with hematopoietic progenitors deficient in Lsh, a chromatin remodeling protein known to bind Dnmt3b, develop erythroleukemia (33), the same type of leukemia that developed spontaneously within our colony. However, the fact that DNMT3B7 transgenic mice do not themselves have a strong predisposition to tumor formation argues that the expression of a truncated DNMT3B protein does not provide a major oncogenic stimulus.
In contrast, when placed in the context of c-Myc, the effects of DNMT3B7 expression are profound, with alterations of many tumor features, including tumor spectrum, kinetics, cytogenetic abnormalities, and DNA methylation distribution. The Eμ-Myc tumors showed only chromosomal gains, as shown previously (34). In contrast, the Eμ-Myc/DNMT3B7 mediastinal tumors demonstrated complex cytogenetic abnormalities involving multiple chromosomes, including polyploidy, translocations, insertions, and chromosomal losses in addition to gains, . similar to the types of chromosomal changes seen in human tumors. Although DNMT3B7 expression leads to chromosomal instability and DNA hypomethylation in some contexts (Supplementary Fig. S4), current technology makes it difficult to determine the specific regions or loci contributing to increased global DNA methylation in Eμ-Myc/DNMT3B7 tumors.
Our results suggest that the introduction of DNMT3B7 promotes large-scale disarray in DNA methylation changes in Eμ-Myc/DNMT3B7 mediastinal tumors that, in turn, induce changes in gene expression that promote mediastinal lymphomagenesis. DNA methylation alterations included genes with hypermethylation within their promoters as well as others that became hypomethylated. For some genes, we observed a high degree of individual variability in the DNA methylation changes of specific CpG nucleotides, but with an overall consistent change seen across a CpG island (e.g. hypomethylation within Mnt and Notch1 promoters and hypermethylation within the Ell2 promoter), a phenomenon remarkably similar to what is observed in individual human primary tumors (29, 30).
Several of the gene expression changes that we observed were similar to what is seen in other tumors. Sox4 is overexpressed in endometrial tumors due to hypermethylation of miR-129-2, which normally suppresses SOX4 (35). Notch1, although not altered by DNA methylation in human tumors, is overexpressed in hypomethylated thymic tumors that develop in Dnmt1 hypomorphic mice (36). Additionally, Mnt forms heterodimers with Myc that lead to transcriptional repression of some genes in many cancers (37). Examining the effects of DNMT3B7 within other tumor models will test whether these DNMT3B7-induced effects are specific to Myc itself (38), or are common to tumors that form under the direction of other oncogenes.
The co-expression of a truncated DNMT-like protein along with the full-length enzymes, as we have modeled for cancer cells, is a well-established paradigm in other systems. DNMT3L, a protein highly homologous to the DNMTs, is known to bind to the intact DNMT3A and DNMT3B enzymes and regulate their function (39–41). Therefore, the interaction of DNMT3B7 with DNMT3B suggests that DNMT3B7 influences the activity of intact DNMT3B.
Aberrant DNMT3B transcription is one example of a widespread phenomenon of alternative splicing seen in human tumors (6). Investigation of splicing signatures in ovarian and breast tumors showed that they are characteristic for tumors and can be used to distinguish normal tissues from cancer (42, 43). DNMT3B was among the few genes that could be used to distinguish normal tissue from ovarian or breast tumors, providing further evidence that aberrant DNMT3B transcription is a common and defining event in tumorigenesis.
Our results show that DNMT3B7, a truncated DNMT3B isoform found in tumors, disrupts murine embryonic development and accelerates lymphomagenesis with dramatic changes in DNA methylation patterns. This system represents the first in vivo model for the redistribution of DNA methylation seen in human tumors and argues that the presence of truncated DNMT3B proteins within human tumors play an important role in the observed changes in DNA methylation that characterize virtually all human tumors.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI R01 grant CA129831, American Cancer Society grant 08-02, the Kimmel Scholars Program (to LAG), and LLS SCOR 7410-07 (to MML). We thank Sonja Volker, Gopal Patel, Jesús Expósito-Céspedes, Cara Rosenbaum, Bohao Liu, Ken Cohen, Linda Degenstein, Dirk Kuck, Frank Lyko, Adam Karpf, Alexander Schilling, Albert Shaw, and Scott Lowe for their contributions to this work.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 5.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 6.Fackenthal JD, Godley LA. Aberrant RNA splicing and its functional consequences in cancer cells. Dis Model Mech. 2008;1:37–42. doi: 10.1242/dmm.000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostler KR, Davis EM, Payne SL, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Wang J, Sun S, et al. A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int J Oncol. 2006;29:201–207. [PubMed] [Google Scholar]
- 9.Wang J, Walsh G, Liu DD, et al. Expression of Delta DNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 2006;66:8361–8366. doi: 10.1158/0008-5472.CAN-06-2031. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Bhutani M, Pathak AK, et al. Delta DNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res. 2007;67:10647–10652. doi: 10.1158/0008-5472.CAN-07-1337. [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishnan S, Van Emburgh BO, Shan J, et al. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol Cancer Res. 2009;7:1622–1634. doi: 10.1158/1541-7786.MCR-09-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagleitner MM, Lankester A, Maraschio P, et al. Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome) J Med Genet. 2008;45:93–99. doi: 10.1136/jmg.2007.053397. [DOI] [PubMed] [Google Scholar]
- 13.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 14.Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 15.Ueda Y, Okano M, Williams C, et al. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- 16.Corcoran LM, Tawfilis S, Barlow LJ. Generation of B lymphoma cell lines from knockout mice by transformation in vivo with an Emu-myc transgene. J Immunol Methods. 1999;228:131–138. doi: 10.1016/s0022-1759(99)00094-0. [DOI] [PubMed] [Google Scholar]
- 17.Le Beau MM, Bitts S, Davis EM, et al. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice parallel human acute promyelocytic leukemia. Blood. 2002;99:2985–2991. doi: 10.1182/blood.v99.8.2985. [DOI] [PubMed] [Google Scholar]
- 18.Song L, James SR, Kazim L, et al. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- 19.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Team RCD R. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 21.http://www.ncbi.nlm.nih.gov/geo/
- 22.Kelly TL, Li E, Trasler JM. 5-aza-2′-deoxycytidine induces alterations in murine spermatogenesis and pregnancy outcome. J Androl. 2003;24:822–830. doi: 10.1002/j.1939-4640.2003.tb03133.x. [DOI] [PubMed] [Google Scholar]
- 23.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 24.Broske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 25.Frith CH, McConnell RF, Johnson AN. Erythroleukemia in a mouse. Lab Anim Sci. 1990;40:418–419. [PubMed] [Google Scholar]
- 26.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 27.Harris AW, Pinkert CA, Crawford M, et al. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa ME, Melnick A, Greally JM. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- 29.Figueroa ME, Skrabanek L, Li Y, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figueroa ME, Lugthart S, Li Y, et al. DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer Cell. 2010 doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stumpel DJ, Schneider P, van Roon EH, et al. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood. 2009;114:5490–5498. doi: 10.1182/blood-2009-06-227660. [DOI] [PubMed] [Google Scholar]
- 32.Goto Y, Shinjo K, Kondo Y, et al. Epigenetic Profiles Distinguish Malignant Pleural Mesothelioma from Lung Adenocarcinoma. Cancer Res. 2009;69:9073–9082. doi: 10.1158/0008-5472.CAN-09-1595. [DOI] [PubMed] [Google Scholar]
- 33.Fan T, Schmidtmann A, Xi S, et al. DNA hypomethylation caused by Lsh deletion promotes erythroleukemia development. Epigenetics. 2008;3:134–142. doi: 10.4161/epi.3.3.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helmrich A, Lee S, O’Brien P, et al. Recurrent chromosomal aberrations in INK4a/ARF defective primary lymphomas predict drug responses in vivo. Oncogene. 2005;24:4174–4182. doi: 10.1038/sj.onc.1208600. [DOI] [PubMed] [Google Scholar]
- 35.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129–2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard G, Eiges R, Gaudet F, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 37.Dang CV, O’Donnell KA, Zeller KI, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Brenner C, Deplus R, Didelot C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. Embo J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata K, Okano M, Lei H, et al. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 40.Chen ZX, Mann JR, Hsieh CL, et al. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 41.Jia D, Jurkowska RZ, Zhang X, et al. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klinck R, Bramard A, Inkel L, et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68:657–663. doi: 10.1158/0008-5472.CAN-07-2580. [DOI] [PubMed] [Google Scholar]
- 43.Venables JP, Klinck R, Bramard A, et al. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.