Abstract
Purpose
Elevated levels of human telomerase (hTERT) mRNA in tumors is a marker for poorer survival in patients with stage I non-small cell lung cancer (NSCLC). A functional variant of MNS16A-Short tandem repeats in hTERT (S allele) is associated with higher expression levels of hTERT mRNA compared with the MNS16A-long (L) allele. However, it is unknown whether or not the hTERT MNS16A variant genotype predicts survival of NSCLC patients.
Experimental Design
The hTERT genotypes of 808 patients with NSCLC were determined by direct PCR with genomic DNA. Overall median survival times were estimated by the life-table method, and the log-rank test was used to test for homogeneity of the survival curves. Both univariate and multivariate Cox proportional hazards models were used to assess the associations between survival time and the hTERT genotype as well as other known risk factors.
Results
The hTERT variant genotype was not associated with overall survival among the 808 patients. However, among 221 patients with stage I or II NSCLC, the S allele was associated with shorter survival time (P = 0.027, by Log-Rank test). The adjusted hazard ratios (HR) were 1.30 (95% CI = 0.79–2.14, P = 0.310) for the SL-genotype and 2.34 (95% CI = 1.20–4.56, P = 0.012) for the SS-genotype compared with the LL-genotype (P = 0.021 for trend test). These findings were not evident in 587 patients with stage III or IV NSCLC.
Conclusion
The functional MNS16A-SS genotype may be a marker for poorer survival in early stage NSCLC.
Keywords: NSCLC, VNTR, hTERT, Genotype, Prognosis
Introduction
Lung cancer remains the leading cause of cancer-related death for men and women in the United States (1). Non-small cell lung cancer (NSCLC) accounts for more than 80% of lung cancers. Although innovative surgical procedures, novel treatment and effective clinical management have somehow improved the survival (2, 3), few validated biomarkers can predict response to treatment and survival (4). Individual patients vary widely in their response to therapy, which could attribute in part to genetic differences in the patients.
Recent genome-wide studies of lung cancer have identified a lung cancer susceptibility TERT-CLPTM1L locus on chromosome 5p15.33 (5, 6), particularly for lung adenocarcinoma (7, 8), and this locus has also been reported to predict DNA adduct formation (9). Telomeres, the structure capping the distal ends of chromosomes, function to prevent chromosome degradation, end-to-end fusions, rearrangements, and chromosome loss (10). Genomic instability characteristic of shortened telomeres or telomere dysfunction is associated with senescence and cellular crisis in humans (11). Although the telomere functions to maintain the potential of cellular immortalization, stabilization of telomeres by telomerase more likely leads to immortalization (12). Telomerase is a holoenzyme, and its catalytic subunit (TERT) is the core component responsible for its enzymatic activity. In humans, telomerase activity is undetectable in most of normal cells, although peripheral and cord blood (or bone marrow) leukocytes do exhibit telomerase activity (13, 14). However, telomerase reactivation has been reported as one of the most common events in almost all types of human cancers (15).
In a previously published study, we demonstrated that hTERT mRNA levels in tumors were an independent predictor of survival in patients with stage I NSCLC after surgical resection (16). Subsequently, we identified a functional variable number of tandem repeats (VNTR), a variant named MNS16A, in the downstream region of the hTERT gene locus (5p15.33), which has four different alleles that were classified as either short (S) or long (L) alleles on the basis of their functionality (16). Further experiments showed that the MNS16A VNTR was associated with initiation of an antisense RNA transcript (unpublished data) whose function remains uncertain. However, the MNS16A-S allele was associated with higher levels of the MNS16A-antisense RNA transcripts, compared with the MNS16A-L allele, and the level of this antisense RNA transcript parallels that of hTERT mRNA in all telomerase-positive NSCLC cell lines and primary lung cancer tissues (16). Based on these observations, we hypothesized that the MNS16A-S allele, compared with the L allele, is associated with poorer survival in NSCLC patients. In this study, we evaluated the association between the hTERT genotype (based on the S and L alleles defined by MNS16A VNTR) and survival in NSCLC patients with different TNM staging tumors.
Materials and Methods
Study Population
Patients were recruited for an ongoing hospital–based case–control epidemiologic study of lung cancer from The University of Texas M. D. Anderson Cancer Center, Houston, Texas during the period between July 1995 and September 2003 (17). A total of 1056 patients newly diagnosed with histopathologically confirmed NSCLC were available for analysis. There were no age, stage, or histology restrictions but all patients were untreated. Blood samples were obtained from each subject after written informed consent and completion of a standardized personal interview before the therapy was instituted. The research protocol was approved by the institutional review board of M. D. Anderson Cancer Center.
Patient clinical follow-up information was obtained from chart review and from the M. D. Anderson institutional database. Only non-Hispanic white patients who were residents of Texas at the time of diagnosis were included because there were too few patients of other ethnicities for meaningful statistical analysis. As a result, a total of 808 NSCLC patients were included for this survival analysis. Patients were followed from the date of diagnose to the date of last follow-up or date of death through September 2004. The patients were followed up to 10 years for the longest. Survival in months was calculated as the time between the diagnosis date and the last contact date (or date of death).
DNA Extraction and Genotyping of MNS16A
For each blood sample, a leukocyte cell pellet was obtained from the buffy-coat layer by centrifugation of 1 ml of whole blood. The genomic DNA was extracted by using the Qiagen DNA blood mini kit (Qiagen, Valencia, CA) according to the manufacture's instruction. The DNA purity was evaluated by electrophoresis on a 1% agarose gel and the concentration was determined by UV spectrophotometry.
We genotyped the MNS16A VNTR by PCR using the primer set as previously reported (18). The sequence of forward primer was 5′-AGGATTCTGATCTCTGAAGGGTG-3′, and the reverse primer 5′-TCTGCCTGAGGAAGGACGTATG-3′. A 10 μl of PCR reaction mixture was assembled with 40-ng genomic DNA, 2.5 pmol of each primer, 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, pH 8.3), 1.5 mM MgCl2, 0.1 mM each dNTP, and 1 U Taq polymerase (Sigma-Aldrich Biotechnology, St. Louis, MO). The PCR reaction was performed with a PTC-200 DNA Engine (Peltier Thermal Cycler, MJ Research Inc, Watertown, MA). The amplification procedure consisted of an initial denaturing step with 5 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C, 45 seconds at 60°C, and 1 minute at 72°C as well as a final extension step with 10 minutes at 72°C. All PCR products were visualized on a 2% agarose gel containing 0.25 μg/ml of ethidium bromide.
Among the cases genotyped for the MNS16A, 4 samples (0.4%) were excluded because the genotyping results were not consistent on repeated assay. About 14% of the samples were randomly repeated with 100% concordance of results.
Statistical analysis
We grouped the patients by age at diagnosis (<45, 45–59 and ≥ 60 years old), common histological type (adenocarcinoma, squamous cell carcinoma and large cell carcinoma), and clinical stage (TNM stage I or II, stage IIIa or IIIb and stage IV). Subjects who had smoked more than 100 cigarettes in their lifetime were defined as “ever smokers” and they were further divided into “former smokers,” who had quit smoking at least 1 year prior to diagnosis, and the rest were “current smokers”. The χ2-test was performed to test for the difference in the distributions of hTERT genotypes defined by the MNS16A alleles as well as the demographic variables and clinicopathological features. Overall median survival times were estimated by the Kaplan-Meier method and the log-rank test was used to test for homogeneity of the survival curves. Both univariate and multivariate Cox proportional hazards models were used to assess the association between survival time and risk factors including age at diagnosis, sex, pack-years smoked, histopathological types, treatment and the hTERT MNS16A/VNTR genotype. The hazard ratio (HR) was calculated with adjustment for these covariates in the same model. All P values were determined by two-sided tests. All statistical analysis was performed with Statistical Analysis System software (Version 8e; SAS Institute Inc., Cary, NC).
Results
In the 808 eligible NSCLC patients, there were 432 (53.5%) males and 376 (46.5%) females with ages ranging between 33 and 85 years old (mean diagnosis age 61.5 ± 10.0 for males and 60.3 ± 10.2 for females). All patients were self-reported non-Hispanic whites. There were 139 (17.2%) never-smokers and 669 (82.8%) ever smokers including 348 (43.1%) former smokers and 321 (39.7%) current smokers. The majority of patients were diagnosed with adenocarcinoma (57.4%), and the remaining diagnoses were 204 (25.2%) squamous cell carcinoma, 32 (4.0%) large cell carcinoma, and 108 (13.4%) unclassified NSCLC tumors. Overall, these 808 NSCLC patients consisted of 27.3% (221) clinical stage I (156) or II (65), 39.0% (315) stage III, and 33.7% (272) stage IV.
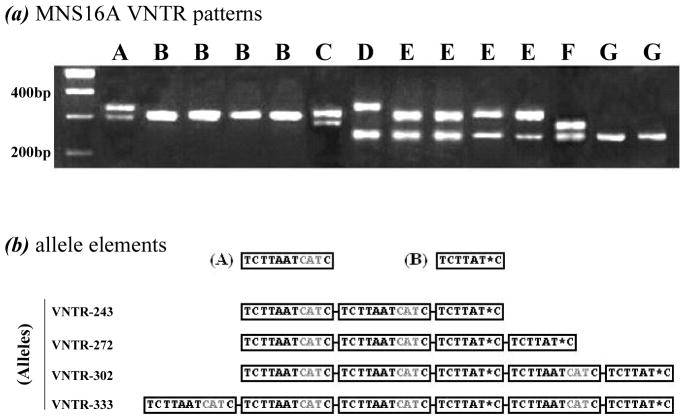
Genotyping results of the MNS16A VNTR showed seven (A to G) distinct patterns in this study population (Figure 1a) with a frequency distribution of A (2.0%), B (42.1%), C (1.7%), D (0.7%), E (42.6%), F (1.5%), and G (9.4%) based on the combinations of the four alleles named as MNS16A VNTR-243, VNTR-272, VNTR-302, and VNTR-333 (18). The schematic sequence difference of these four alleles is shown in Figure 1b. These alleles consist of two basic elements (i.e., A and B) with or without an “AATC” insertion, and we classified the allele VNTR-243 and VNTR-272 as the S-allele, and VNTR-302 and VNTR-333 as the L-allele on the basis of their functional relevance (16). Therefore, the hTERT genotypes of the MNS16A VNTR were defined as either SS, SL or LL genotypes.
Fig. 1. Genotyping pattern of hTERT genotypes defined by MNS16A VNTR.
(a) MNS16A VNTR patterns detected by PCR; (b) Sequence comparison of four alleles VNTR-243, VNTR-272, VNTR-302, and VNTR-333.
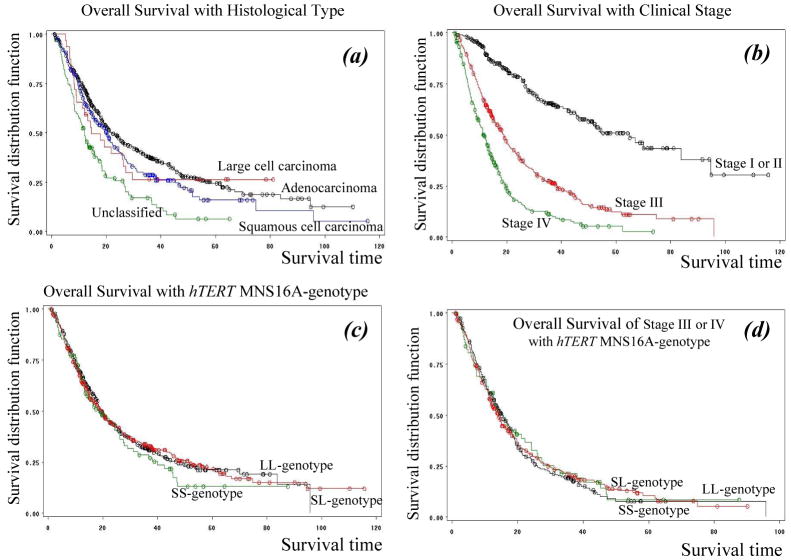
Although there were no overall statistically significant differences in distribution of the hTERT MNS16A genotypes by age, sex, smoking status, tumor histology or clinical stage of 808 patients with NSCLC (data not shown), the SS genotype was present in 5.8% of never smokers, 11.1% of former smokers and 12.8% of current smokers (P = 0.038 for trend test). The overall survival of the 808 NSCLC patients grouped by histopathological types or stages is graphed in Fig. 2a and 2b. As expected, tumor stage remains the best predictor for overall survival of NSCLC patients. The median survival times were 64.9 months for stage I or II NSCLC, 18.9 for stage III, and 11.6 for stage IV (P < 0.001, by Log-Rank test). There were no survival differences by the hTERT MNS16A genotype overall (Fig. 2c).
Fig. 2. Survival analysis of 808 NSCLC patients stratified by different variable groups.
(a) Histopathological types: red line-large cell, black line-adenocarcinoma, blue-squamous and green-unclassified; (b) Clinical stages; (c) hTERT MNS16A-genotypes: black line-LL, red-line SL and green line-SS; and (d) Stage III or IV (advanced stages) stratified by hTERT MNS16A-genotypes: red line-SL, green line-LL and black line-SS.
Because we previously found an association between tumor hTERT mRNA expression and poorer survival in stage I NSCLC patients (19), we focused the 221 stage I or II patients who were grouped by surgery alone, or in combination with radiation or chemotherapy. The majority of these stage I or II patients (132 cases) were diagnosed with adenocarcinoma and the remaining (non-adenocarcinoma) were squamous cell carcinoma (75 cases), large cell carcinoma (6 cases) and unclassified (8 cases).
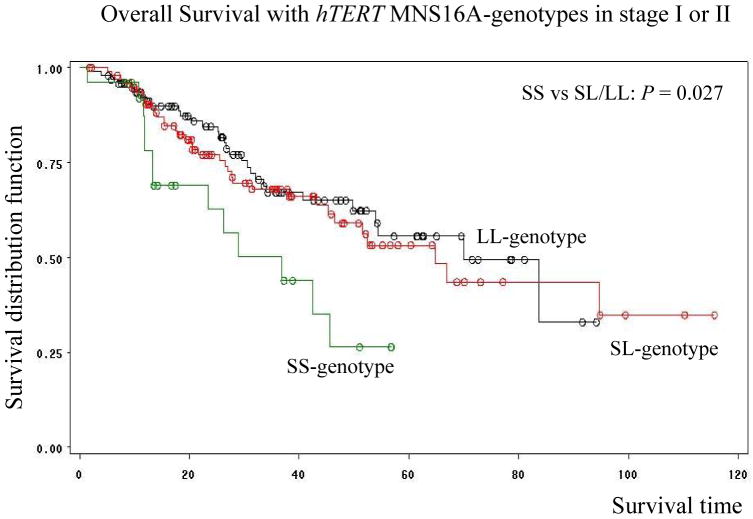
There was no association by genotypes in 587 stage III and IV patients (Fig. 2d). However, the median survival time (MST) in patients with the hTERT MNS16A-S variant (SL and SS) genotypes was statistically shorter than that (MST; P = 0.027 by Log-Rank test) in patients with the hTERT MNS16A-LL genotype in these 221 stage I or II NSCLC patients (Fig. 3). Among these 221 cases, there were 143 (64.7%) patients still alive and 78 dead (35.3%) at the last follow-up. The median survival rates for these patients were significantly different by hTERT MNS16A-genotypes: 36.8 months for the SS-genotype, 64.9 months for the SL-genotype, and 70.0 for the LL-genotype (Fig. 3).
Fig. 3. Survival analysis of 221 stage I or II NSCLC patients stratified by hTERT MNS16A-genotypes: black line-LL, red-line SL and green line-SS.
We then used the univariate and multivariate Cox proportional hazard models to evaluate hazard ratios (HRs) associated with the hTERT MNS16A genotypes with adjustment for age at diagnosis, sex, smoking status, tumor histology, and treatment. Among all known variables shown in Table 1, the only significant predictor was non-adenocarcinoma compared to adenocarcinoma (P = 0.044). However, compared with the LL genotype, the hTERT MNS16A-S variant genotypes were also associated with increased hazard ratios in an allele-dose response manner (HR = 1.30, 95% CI = 0.79–2.14 for the SL genotype and HR = 2.34, 95% CI = 1.20–4.56 for the SS genotype). The trend test for increased hazard ratios with the increasing numbers of S-allele was statistically significant (P = 0.021).
Table 1.
Univariate and Multivariate analysis of 221 stage I or II NSCLC patients with Cox proportional hazard model for the death relative risk associated with clinical variables
| Variable | No. | HR (95% CI) | P* | Adjusted HR (95% CI) ‡ | P* |
|---|---|---|---|---|---|
| Age at diagnosis (years old) | |||||
| <45 | 9 | Ref. | 1 | ||
| 45-59 | 57 | 1.13 (0.26–4.96) | 0.87 | 0.88 (0.20–3.93) | 0.87 |
| 360 | 155 | 2.10 (0.51–8.61) | 0.3 | 1.48 (0.35–6.27) | 0.6 |
| Sex | |||||
| Male | 102 | Ref. | |||
| Female | 119 | 0.63 (0.40–0.99) | 0.04 | 0.76 (0.48–1.20) | 0.24 |
| Smoking status | |||||
| Never smoker | 20 | Ref. | 1 | ||
| Former smoker | 118 | 1.13 (0.48–2.67) | 0.79 | 0.83 (0.35–2.01) | 0.68 |
| Current smoker | 83 | 1.47 (0.62–3.52) | 0.39 | 1.12 (0.46–2.72) | 0.81 |
| Tumor histology | |||||
| Adenocarcinoma | 132 | Ref. | 1 | ||
| Non-adenocarcinoma | 89 | 1.89 (1.21–2.96) | 0.01 | 1.61 (1.01–2.57) | 0.04 |
| Treatments | |||||
| Surgery + Radiation | 18 | Ref. | 1 | ||
| Others | 203 | 0.88 (0.40–1.91) | 0.74 | 0.77 (0.35–1.69) | 0.52 |
| Surgery + Chemotherapy | 27 | Ref. | 1 | ||
| Others | 194 | 1.01 (0.49–2.12) | 0.97 | 1.01 (0.48–2.11) | 0.98 |
| Surgery + Combination † | 37 | Ref. | 1 | ||
| Other | 184 | 0.86 (0.47–1.56) | 0.61 | 0.82 (0.45–1.50) | 0.52 |
| hTERT MNS16A-genotypes | |||||
| LL-genotype | 96 | Ref. | 1 | ||
| SL-genotype | 99 | 1.12 (0.69–1.83) | 0.65 | 1.30 (0.79–2.14) | 0.31 |
| SS-genotype | 26 | 2.35 (1.22–4.53) | 0.01 | 2.34 (1.20–4.56) | 0.01 |
| PTrend = 0.021 |
Two-sided c2-test.
Combination of radiation and chemotherapy.
Adjusted for sex, smoking status, histology and treatment, where appropriate.
Discussion
In the present survival analysis, we demonstrated that the hTERT MNS16A SS genotype was associated with a significantly shorter survival time in early stage (I or II) NSCLC patients, compared with the LL genotype as assessed in the multivariate Cox hazard model. In a previous study, we had shown that the hTERT MNS16A-S allele was correlated with elevated hTERT mRNA expression levels compared with the L allele and that the elevated hTERT mRNA expression levels in tumors were associated with poorer survival in patients with stage I NSCLC (16). Therefore, our present findings are consistent with previously observed association between increased hTERT expression in tumors and poorer survival in stage I NSCLC patients but was not evident in patients with advanced diseases (19). It is known that hTERT mRNA overexpression is correlated with reactivated telomerase in the lungs of cigarette smokers (18) and that the shorter telomeres are associated with increased risks for human cancers (20). However, our results provide a possible genetic mechanism for the previously observed association between increased hTERT expression in tumors and poorer survival in stage I NSCLC patients (19). Our findings are biologically plausible, because both telomere and telomerase activity have implications in tumorigenesis.
The hTERT gene is located on chromosome 5p15.33 and has 16 exons (21, 22) that can be further divided into four exon-clusters according to the distribution of many minisatellites and microsatellites (16). All seven reported major conserved motifs of the telomerase subunit (23) are located in the second cluster (T, 1, 2 and A motifs) and third cluster (B, C, D and E motifs). Except for a 430-bp EcoR V–BamH I fragment from the hTERT B–E motif, we found that a clear antisense RNA expression signal was always accompanied with hTERT mRNA expression signal detected by in situ hybridization with single-strand specific riboprobes made from several regions of hTERT cDNA (unpublished data). Further exploration of this phenomenon eventually led to identification of the first antisense RNA transcript in the hTERT gene locus (unpublished data). To date, approximately 85% of all cancer tissues tested are reported to be telomerase positive (12). Similarly, hTERT expression is also detectable in approximately 85% of NSCLC tissues, and the remaining small telomerase-negative minority maintain their telomeres by an alternative lengthening of telomeres (ALT) mechanism (24). Studies have shown that after surgical treatment, the levels of hTERT mRNA levels in serum were significantly reduced (25), further suggesting the origin of hTERT mRNA levels was in tumors (26).
Because higher expression levels of MNS16A-antisense RNA transcript were found to be associated with the MNS16A-S allele (16), we tested the hypothesis in this study that the MNS16A-S allele is associated with poorer prognosis in NSCLC patients, especially in early stage disease. Indeed, we found significantly shorter median survival time for the 221 patients with stages I or II NSCLC, who were carriers of the hTERT MNS16A SS variant genotype, whereas we did not observe any effect of the hTERT MNS16A/VNTR genotypes on survival in patients with stages III and IV NSCLC. However, these data need to be replicated by larger studies.
In general, the length of telomeres and telomerase activity do not have the same implications in cancer etiology. One of the underlying mechanisms for age-related cancer risk is an age-related telomere shortening that limits the replicative lifespan of primary human cells, which might play a role in both aging and cancer etiology (27). Telomeres may also indirectly affect DNA repair mediated by the binding of Ku80, a critical protein for nonhomologous DNA double-strand break repair and site-specific recombination of V(D)J gene segments (28). Such interference with repair activity may increase genetic instability due to unrepaired DNA damage, suggesting that telomere dysfunction may be one of the most important molecular mechanisms responsible for genetic instability and thus may play an important role during tumorigenesis (29, 30). Short telomeres were found to be associated with increased risks for human bladder, head and neck, lung, and renal cell cancers in an epidemiological case–control studies (20). Interestingly, it is reported that there is a significant correlation between telomere length and hTERT mRNA expression level in cancer tissues and adjacent mucosa samples and that telomeres were significantly shorter in colorectal carcinoma tissue than that in adjacent mucosa (10).
Telomerase is inactivated in almost all normal somatic epithelial cells in humans and telomerase activation leads to cell immortalization and tumorigenesis (15, 31, 32). We have previously reported that hTERT mRNA expression was detectable in about 64% of bronchial biopsy specimens (170 of 266) obtained from chronic smokers without lung cancer, suggesting that hTERT reactivation could be a smoking-induced event, one of the precursors in tumorigenesis (18), although the exact underlying mechanism needs to be further explored.
In summary, on the basis of our early observations that 1) the hTERT mRNA expression was an independent prognostic marker for stage I NSCLC patients after surgery resection treatment; 2) an antisense RNA transcript expression level was well correlated with hTERT mRNA expression level; 3) a genetic variant VNTR MNS16A is responsible for initiation of this antisense RNA transcript; and 4) the MNS16A-S allele was associated with higher expression level of MNS16A-antisense RNA transcript, we further evaluated the prognostic value of the hTERT genotypes defined by MNS16A/VNTR in the TNM staging subgroups of NSCLC patients. We concluded that the hTERT MNS16A-SS genotype was associated with poorer prognosis in early stage NSCLC patients, and therefore the hTERT genetic variation, i.e., the hTERT MNS16A genotype, may serve as a potential prognostic biomarker of early stage NSCLC. Future large studies with clinically homogenous subsets of patients, which would allow further stratification analysis, such as by adenocarcinoma and non-adenocarcinoma, are needed to confirm our findings. The functional relevance of the MNS16A/VNTR as well as the therapeutic potential in pharmacogenomic studies by targeting hTERT also warrants further investigations.
Translational Relevance.
This study has provided some evidence that the hTERT MNS16A variant genotype may be a marker for survival of stages I and II NSCLC patients. This finding is consistent with previous reports that elevated levels of human telomerase (hTERT) mRNA in tumors is an independent marker for poorer survival in patients with stage I NSCLC and that this MNS16A-Short tandem repeats in hTERT (S allele) is associated with higher in vitro expression levels of hTERT mRNA compared with the MNS16A-long (L) allele. This finding has some clinical implication, given TERT has been identified in the genome-wide association studies of lung cancer as a susceptibility locus, particularly for lung adenocarcinoma.
Acknowledgments
We thank Susan Honn for assistance in recruiting the subjects, Zhensheng Liu, Hongbing Shen, John I. Calderon, Jianzhong He and Kejin Xu for their laboratory assistance, Li-e Wang and Qiong Dong for maintaining the database, Dr. Qiuling Shi and Shengying Fang for statistical analysis discussion, and Rachel Williams for scientific editing.
Grant Support: This study was in part supported by National Institutes of Health grants CA 86390 (to M. R. S.), R01 ES 11740 and CA 131274 (to Q. W.) and Cancer Center Core Grant CA P30 16672 (to M. D. Anderson Cancer Center).
The Abbreviations used are
- NSCLC
non-small cell lung cancer
- SCC
squamous cell carcinoma
- ADC
adenocarcinoma
- hTERT
human telomerase
- VNTR
variable number of tandem repeats
- HR
hazard ratio
- CI
confidence interval
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Pearson FG. Lung cancer. The past twenty-five years. Chest. 1986;89:200S–5S. doi: 10.1378/chest.89.4.200s. [DOI] [PubMed] [Google Scholar]
- 3.Pearson FG. Non-small cell lung cancer: role of surgery for stages I-III. Chest. 1999;116:500S–3S. doi: 10.1378/chest.116.suppl_3.500s. [DOI] [PubMed] [Google Scholar]
- 4.Scagliotti GV, Novello S. Adjuvant therapy in completely resected non-small-cell lung cancer. Curr Oncol Rep. 2003;5:318–25. doi: 10.1007/s11912-003-0074-y. [DOI] [PubMed] [Google Scholar]
- 5.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–7. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin G, Xu L, Shu Y, et al. Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis. 2009;30:987–90. doi: 10.1093/carcin/bgp090. [DOI] [PubMed] [Google Scholar]
- 8.Kohno T, Kunitoh H, Shimada Y, et al. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis. doi: 10.1093/carcin/bgq003. [DOI] [PubMed] [Google Scholar]
- 9.Zienolddiny S, Skaug V, Landvik NE, et al. The TERT-CLPTM1L lung cancer susceptibility variant associates with higher DNA adduct formation in the lung. Carcinogenesis. 2009;30:1368–71. doi: 10.1093/carcin/bgp131. [DOI] [PubMed] [Google Scholar]
- 10.Greider CW. Chromosome first aid. Cell. 1991;67:645–7. doi: 10.1016/0092-8674(91)90058-7. [DOI] [PubMed] [Google Scholar]
- 11.Wright WE, Shay JW. The two-stage mechanism controlling cellular senescence and immortalization. Exp Gerontol. 1992;27:383–9. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- 12.Counter CM, Gupta J, Harley CB, Leber B, Bacchetti S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood. 1995;85:2315–20. [PubMed] [Google Scholar]
- 13.Counter CM, Hahn WC, Wei W, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci U S A. 1998;95:14723–8. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–9. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–91. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Soria JC, Chang YS, Lee HY, Wei Q, Mao L. Association of a functional tandem repeats in the downstream of human telomerase gene and lung cancer. Oncogene. 2003;22:7123–9. doi: 10.1038/sj.onc.1206852. [DOI] [PubMed] [Google Scholar]
- 17.Wei Q, Cheng L, Amos CI, et al. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst. 2000;92:1764–72. doi: 10.1093/jnci/92.21.1764. [DOI] [PubMed] [Google Scholar]
- 18.Soria JC, Moon C, Wang L, et al. Effects of N-(4-hydroxyphenyl)retinamide on hTERT expression in the bronchial epithelium of cigarette smokers. J Natl Cancer Inst. 2001;93:1257–63. doi: 10.1093/jnci/93.16.1257. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Soria JC, Kemp BL, Liu DD, Mao L, Khuri FR. hTERT expression is a prognostic factor of survival in patients with stage I non-small cell lung cancer. Clin Cancer Res. 2002;8:2883–9. [PubMed] [Google Scholar]
- 20.Wu X, Amos CI, Zhu Y, et al. Telomere dysfunction: a potential cancer predisposition factor. J Natl Cancer Inst. 2003;95:1211–8. doi: 10.1093/jnci/djg011. [DOI] [PubMed] [Google Scholar]
- 21.Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 22.Bryce LA, Morrison N, Hoare SF, Muir S, Keith WN. Mapping of the gene for the human telomerase reverse transcriptase, hTERT, to chromosome 5p15.33 by fluorescence in situ hybridization. Neoplasia. 2000;2:197–201. doi: 10.1038/sj.neo.7900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura TM, Morin GB, Chapman KB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–9. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 24.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–4. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 25.Miura N, Nakamura H, Sato R, et al. Clinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor marker for lung cancer. Cancer Sci. 2006;97:1366–1373. doi: 10.1111/j.1349-7006.2006.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu CQ, Cutz JC, Liu N, et al. Amplification of telomerase (hTERT) gene is a poor prognostic marker in non-small-cell lung cancer. Br J Cancer. 2006;94:1452–9. doi: 10.1038/sj.bjc.6603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–8. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 28.Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci U S A. 1999;96:12454–8. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudolph KL, Chang S, Lee HW, et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–12. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 30.Artandi SE, Chang S, Lee SL, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–5. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 31.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–52. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 32.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]