Abstract
Objective
We examined the association between genetic variants in the apelin system and blood pressure (BP) responses to low- and high-sodium interventions in the GenSalt Study.
Methods
A 7-day low-sodium intervention (51.3 mmol sodium/day) followed by a 7-day high-sodium intervention (307.8 mmol sodium/day) was conducted among 1,906 participants from 637 Han Chinese families. BP measurements were obtained at baseline and following each intervention using a random-zero sphygmomanometer. Twenty-three single nucleotide polymorphisms (SNPs), including both tag and functional SNPs, were selected from three candidate genes (APLN, APLNR, and ACE2). Single marker and haplotype analyses were conducted using the FBAT program. The false discovery rate method was used to correct for multiple testing.
Results
SNPs rs2282623 and rs746886 of the APLNR gene were significantly associated with diastolic (DBP) (both P=0.002) and mean arterial pressure (MAP) (P=0.001 and 0.005, respectively) responses to low-sodium intervention. Six SNPs of the ACE2 gene were significantly associated with systolic (SBP), DBP or MAP responses to low-sodium intervention. Three of them, rs1514283, rs1514282, and rs4646176, were also significantly associated with MAP response to high-sodium (all P≤0.006). Haplotype analysis indicated the A-T-T haplotype of APLNR SNPs rs721608-rs2282623-rs746886 was associated with decreased DBP and MAP responses to low-sodium intervention (P=0.001 and 0.003, respectively), while G-C-C was associated with increased SBP and MAP responses to high-sodium intervention (P=0.004 and 0.01, respectively).
Conclusions
This large family-based study indicates that genetic variants in the APLNR and ACE2 genes are significantly associated with BP responses to dietary sodium intervention.
Keywords: ACE2, apelin, apelin receptor, blood pressure, polymorphism, dietary sodium, salt sensitivity
Introduction
Blood pressure (BP) response to changes in dietary sodium intake varies considerably between individuals, a phenomenon known as salt-sensitivity [1, 2]. Salt-sensitivity of BP has been associated with increased risk of hypertension, cardiovascular disease (CVD) and premature death [3, 4]. Previous studies suggested that genetic factors might play an important role in the salt-sensitivity of BP [5–8]. Family studies also documented a moderately high heritability of salt-sensitivity [5, 6]. Examining the genetic determinants of salt-sensitivity will enhance our understanding of its mechanism and thus facilitate the development of targeted prevention and treatment strategies for reducing hypertension and related CVD burden.
The apelin system, including apelin and its receptor APJ, is a peptidic signaling pathway which is implicated in the regulation of cardiovascular function and fluid homeostasis [9]. APJ belongs to a family of seven-transmembrane domain receptors and is coupled to inhibitory G-proteins. Apelin is synthesized as a 77 amino acid pre-propeptide that can be cleaved into fragments of different sizes that activate APJ [10,11]. Although physiological experiments in animal models have yielded conflicting results about the peripheral role of apelin in regulating vascular tone, acting as vasodilator or vasoconstrictor, a recent study showed that exogenous apelin caused nitric oxide-dependent arterial vasodilation in vivo in man [12–16]. In addition, there is accumulated evidence suggesting the central effect of apelin system in neural control of cardiovascular function. Most recent studies indicate that overexpression of the apelin gene in the rostral ventrolateral medulla results in chronic BP elevation and cardiac hypertrophy in normotensive rats [17] and APJ knockout mice do not show decreased urine volume and urine osmolality during water deprivation, suggesting an anti-diuretic effect of apelin in vivo [18]. Apelin peptides are specific substrates of angiotensin-converting enzyme 2 (ACE2), an enzyme which acts as an important regulator of the renin-angiotensin-aldosterone system (RAAS) [19,20]. Single nucleotide polymorphisms (SNPs) of the apelin and APJ genes have recently been reported to be associated with hypertension [21]. The aim of this study was to examine the association between genetic variants in the apelin system (apelin and APJ) and ACE2 and salt-sensitivity measured by BP responses to changes in dietary sodium intake among 637 Chinese families.
Methods
Study Participants
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) study was conducted in rural areas of northern China from October 2003 to July 2005. All study participants were of Han Chinese ethnicity. The GenSalt sampling design targeted families at high risk for developing hypertension, who might be more sensitive to sodium intervention [22]. Probands with a mean systolic BP (SBP) between 130–160 mmHg and/or diastolic BP (DBP) between 85–100 mmHg were identified by community-based BP screening among all 18–60 years in the study villages. Both 2-generation (probands, their parents and siblings) and 3-generation (additionally including the probands’ spouse and offspring) families were recruited for the study. Probands, along with their siblings, spouses, and offspring participated in the dietary intervention. Individuals who had stage-2 hypertension, secondary hypertension, a history of clinical CVD, diabetes, or chronic kidney disease (including microproteinuria), use of antihypertensive medications, or were pregnant, heavy alcohol drinkers or currently on a low-sodium diet were excluded from the dietary intervention. Among the 1,906 participants eligible for dietary intervention, 1,871 (98.2%) and 1,860 (97.6%) completed the low and high dietary sodium interventions, respectively, and were included in the current analysis. Institutional Review Boards at all participating institutes approved the study. Written informed consents for the baseline observation and for the intervention program were obtained from each participant.
Dietary Sodium Intervention
After a 3-day baseline observation, the study participants received a 7-day low-salt diet (3 grams of sodium chloride or 51.3 mmol of sodium per day) followed by a 7-day high-salt diet (18 grams of sodium chloride or 307.8 mmol of sodium per day). During the period of sodium intervention, dietary potassium intake remained unchanged. Total energy intake was varied according to each participant’s baseline energy intake. All foods were cooked without salt, and pre-packaged salt was added to the individual study participant’s meal when it was served by the study staff. To ensure study participants’ compliance to the intervention program, they were required to have their breakfast, lunch, and dinner at the study kitchen under supervision of the study staff during the entire study period. The study participants were instructed to avoid consuming any foods that were not provided by the study. In addition, three timed urinary specimens (one 24–hour and two overnight) were collected at baseline and in each phase of intervention (days 5, 6, and 7) to monitor participants’ compliance to dietary sodium intervention. The timed overnight urinary excretions of sodium and potassium were converted to 24-hour values based on a formula developed from data in this study. The results from the 24-hour urinary excretions of sodium showed excellent compliance with the study diet: the mean (standard deviation) 24-hour urinary excretions of sodium and potassium were 242.4 (66.7) mmol and 36.9 (9.6) mmol at baseline, 47.5 (16.0) and 31.4 (7.7) during the low-sodium intervention, and 244.3 (37.7) and 35.7 (7.5) during the high-sodium intervention, respectively.
Phenotype Measurement
A standard questionnaire was administered by trained staff at the baseline examination just preceding dietary salt intervention. Information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors were collected. Body weight, height, and waist circumference were measured twice in light indoor clothing without shoes during the baseline examination. Body mass index (BMI) was calculated as kilograms per meters squared (kg/m2). Three BP measurements were obtained each morning of the 3-day baseline observation, and on days 5, 6 and 7 of each intervention period by the trained and certified observers using a random-zero sphygmomanometer according to a standard protocol [23]. BP was measured with the participant in the sitting position after 5 minutes of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes prior to their measurement. All BP observers were blinded to the dietary intervention.
SNP Selection and Genotyping
Both the apelin coding gene (APLN) and ACE2 are located on the X chromosome and the APJ coding gene (APLNR) on chromosome 11. TagSNPs from these three genes were selected based on empirical patterns of linkage disequilibrium (LD) structure in the Chinese Han of Beijing (CHB) HapMap sample using Tagger software. We also included SNPs which were previously reported to be associated with BP or hypertension. SNPs were genotyped using SNPlex assays (Applied Biosystems, Foster City, CA, USA) based on oligonucleotide ligation assay for capillary electrophoresis on ABI 3700 DNA Analyzers (Applied Biosystems, Foster City, CA, USA). To provide better coverage of the candidate genes, we included additional SNPs genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA, USA), whose minor allele frequencies were no less than 0.01. Finally, a total of 23 SNPs (14 SNPlex SNPs and 9 Affymetrix SNPs) were analyzed in this study (Table 1).
Table 1.
Characteristics of all SNPs analyzed in this study
| Gene | Chr | SNP | Physical Location |
Region in Gene |
Allelesa | MAF | P-Valuesb |
|---|---|---|---|---|---|---|---|
| APLN | X | rs3116754 | 128603256 | 3’flanking | A:T | 0.337 | 0.1649 |
| rs17267847 | 128604715 | 3’flanking | C:T | 0.209 | 0.6554 | ||
| rs3131264 | 128605307 | 3’flanking | G:T | 0.454 | 0.8702 | ||
| rs3115759 | 128609197 | 3’UTR | A:G | 0.333 | 0.0768 | ||
| rs909657 | 128611248 | intron1 | G:A | 0.332 | 0.4509 | ||
| rs2235306 | 128611779 | intron1 | T:C | 0.408 | 0.0652 | ||
| rs3761581 | 128617402 | 5’flanking | T:G | 0.314 | 0.0186 | ||
| APLNR | 11 | rs721608 | 56754499 | 3’flanking | A:G | 0.149 | 0.9602 |
| rs2282623 | 56758402 | 3’UTR | C:T | 0.411 | 0.4338 | ||
| rs746886 | 56758687 | 3’UTR | C:T | 0.411 | 0.3791 | ||
| rs10501367 | 56762163 | 5’flanking | C:T | 0.226 | 0.8741 | ||
| rs7119375 | 56762540 | 5’flanking | G:A | 0.223 | 0.8671 | ||
| rs948844 | 56766174 | 5’flanking | T:G | 0.259 | 0.5169 | ||
| ACE2 | X | rs1514283 | 15492668 | intron16 | T:C | 0.034 | 0.757 |
| rs1514282 | 15492684 | intron16 | T:C | 0.034 | 0.757 | ||
| rs2074192 | 15492711 | intron16 | C:T | 0.417 | 0.308 | ||
| rs714205 | 15493825 | intron16 | G:C | 0.451 | 0.2775 | ||
| rs4646176 | 15497425 | intron14 | C:G | 0.035 | 0.0891 | ||
| rs4646174 | 15498192 | intron14 | C:G | 0.037 | 0.0961 | ||
| rs879922 | 15500728 | intron11 | C:G | 0.036 | 0.0687 | ||
| rs4646155 | 15507430 | intron8 | G:A | 0.033 | 0.0406 | ||
| rs4646140 | 15515773 | intron6 | G:A | 0.029 | 0.0879 | ||
| rs2285666 | 15520269 | intron3 | T:C | 0.446 | 0.8699 |
Chr, chromosome; SNP, single nucleotide polymorphism; MAF, minor allele frequency.
Alleles are presented as major : minor allele.
P values of Hardy-Weinberg Equilibrium test.
Statistical Analysis
BP levels at baseline and during intervention were calculated as the mean of 9 measurements from 3 clinical visits during the 3-day baseline observation or on days 5, 6, and 7 of each intervention period. Mean arterial pressure (MAP) was calculated as (SBP-DBP)/3 + DBP. The percent changes in BP from baseline to low-sodium and low to high-sodium interventions were used to measure BP responses to changes in dietary sodium intake. Percent changes in BP were adjusted for the effects of age and examination room temperature separately within sex-field center groups. In summary, each measure was regressed on the covariates in a stepwise manner, and only significant terms (0.05 level) were retained. The residual variance was also examined by regressing the squared residual from the first regression on the same covariates (stepwise) and retaining significant terms. The final adjusted phenotype was computed as the residual from the first regression, divided by the square root of the predicted score from the second regression. A final standardization step was taken to ensure a mean of 0 and an SD of 1. These adjusted and standardized scores were used as the phenotype variables in the association analyses.
The Mendelian consistency of the SNP genotype data was assessed by PLINK and PedCheck [24,25]. We used Haploview software (version 4.0, http://www.broad.mit.edu/mpg/haploview) to test Hardy-Weinberg Equilibrium (HWE) for each SNP and estimate the extent of pairwise LD between SNPs [26]. The solid spine LD method, as implemented in Haploview software, was used to define LD blocks.
We used the Family Based Association Test (FBAT) program (version 2.0.2, http://www.biostat.harvard.edu/~fbat/default.html) to test the association of single marker and haplotypes with adjusted phenotypes. FBAT performs family-based tests of association that are efficient and robust to population admixture, phenotype distribution and ascertainment based on phenotype. Three genetic models (additive, dominant, and recessive) were tested. To assess the effect of genetic variants on the trait value, a univariate FBAT test was performed for each allele and haplotype. This test provides a Z-statistic with its corresponding p-value. In our study, a positive Z-statistic for an allele or a haplotype indicates a decreased response to low-sodium intervention and an increased response to high-sodium intervention. The false discovery rate (FDR) method was used to correct for multiple testing [27].
Results
Table 2 shows the baseline characteristics and BP responses to the low- and high-sodium intervention among family members. The probands had higher mean baseline levels of BMI, SBP, and DBP than their siblings, spouses, and offspring whereas the parents had the highest baseline BP levels among all of the groups. Overall, BP levels decreased from baseline to low-sodium intervention but increased from low-sodium to high-sodium intervention. For both low- and high-sodium interventions, the probands had greater percentage changes in MAP than their siblings, spouses and offspring.
Table 2.
Characteristics at baseline and BP responses to sodium intervention for all study participants
| Probands | Siblings | Spouses | Offspring | Parents | |
|---|---|---|---|---|---|
| No. of subjects | 676 | 956 | 69 | 205 | 1236 |
| Age, yrs | 41.0 ± 8.3 | 39.6 ± 7.7 | 49.1 ± 6.7 | 23.5 ± 6.5 | 67.6 ± 8.4 |
| Male, % | 60.4 | 51.1 | 33.3 | 44.4 | 48.6 |
| Body mass index, kg/m2 | 24.2 ± 3.3 | 23.1 ± 2.8 | 23.4 ± 3.7 | 21.5 ± 3.3 | 22.8 ± 3.4 |
| BP at baseline, mm Hg | |||||
| Systolic | 128.0 ± 11.4 | 111.6 ± 11.5 | 112.6 ± 14.9 | 106.6 ± 10.3 | 136.6 ± 23.9 |
| Diastolic | 80.3 ± 9.0 | 71.0 ± 8.9 | 72.6 ± 10.0 | 65.3 ± 9.0 | 75.0 ± 11.7 |
| Mean arterial pressure | 96.7 ± 8.5 | 84.5 ± 9.2 | 85.9 ± 11.2 | 79.1 ± 8.7 | 95.6 ± 14.3 |
| BP response to low salt, % | |||||
| Systolic | −6.1 ± 6.0 | −3.7 ± 5.0 | −4.4 ± 5.8 | −2.0 ± 4.5 | – |
| Diastolic | −5.1 ± 7.2 | −2.4 ± 7.4 | −3.4 ± 6.8 | −2.4 ± 8.1 | – |
| Mean arterial pressure | −5.6 ± 5.8 | −3.0 ± 5.5 | −3.8 ± 5.8 | −2.3 ± 5.5 | – |
| BP response to high salt, % | |||||
| Systolic | 4.9 ± 5.6 | 4.4 ± 5.5 | 5.4 ± 5.5 | 2.5 ± 4.2 | – |
| Diastolic | 3.6 ± 8.1 | 2.8 ± 7.7 | 2.9 ± 6.5 | 1.7 ± 8.5 | – |
| Mean arterial pressure | 4.1 ± 6.2 | 3.4 ± 6.0 | 3.9 ± 5.4 | 2.0 ± 5.3 | – |
Continuous variables are indicated as mean ± standard deviation. BP, blood pressure.
Table 3 shows all SNPs which were significantly associated with BP response to low-sodium or high-sodium intervention after adjustment for multiple testing (FDR <0.05). SNPs rs2282623 and rs746886 of APLNR were significantly associated with DBP (both P =0.002) and MAP (P =0.001 and 0.005, respectively) responses to low-sodium intervention. Compared to their major alleles, the minor alleles were associated with decreased BP response during the low-sodium intervention. Six SNPs of the ACE2 gene were also significantly associated with BP responses to low-sodium intervention. Among them, SNPs rs2074192, rs714205 and rs2285666 were associated with SBP response to low-sodium intervention (P =0.0007, 0.003 and 0.003, respectively). The other three SNPs were significantly associated with MAP response to both low-sodium and high-sodium interventions. Their minor alleles predicted greater MAP response to changes in dietary sodium intake. We did not find any association between APLN SNPs and BP responses.
Table 3.
Summary of individual SNPs significantly associated with BP response to low- and high-sodium interventions
| Gene | SNP | Allele | SBP Response | DBP Response | MAP Response | |||
|---|---|---|---|---|---|---|---|---|
| Z | P-value | Z | P-value | Z | P-value | |||
| Low-sodium Intervention | ||||||||
| APLNR | rs2282623 | T | 1.851 | 0.06 | 3.100 | 0.002a* | 3.275 | 0.001a* |
| rs746886 | T | 0.993 | 0.32 | 3.165 | 0.002a* | 2.786 | 0.005a* | |
| ACE2 | rs1514283 | C | 0.031 | 0.98 | −2.787 | 0.005b* | −2.863 | 0.004b* |
| rs1514282 | C | 0.031 | 0.98 | −2.787 | 0.005b* | −2.863 | 0.004b* | |
| rs2074192 | T | 3.376/3.066 | 0.0007c*/0.002a* | 2.682 | 0.007b* | 2.723/2.903 | 0.006a*/0.004b* | |
| rs714205 | C | 2.990 | 0.003c* | 1.272 | 0.20 | 1.999 | 0.045 | |
| rs4646176 | G | −0.004 | 1 | −2.787 | 0.005b* | −2.863 | 0.004b* | |
| rs2285666 | C | 2.923 | 0.003c* | 1.886 | 0.06 | 2.409 | 0.02 | |
| High-sodium Intervention | ||||||||
| ACE2 | rs1514283 | C | −0.851 | 0.40 | 0.734 | 0.46 | 2.738 | 0.006b* |
| rs1514282 | C | −0.851 | 0.40 | 0.734 | 0.46 | 2.738 | 0.006b* | |
| rs4646176 | G | −0.890 | 0.37 | 0.614 | 0.54 | 2.738 | 0.006b* | |
For associations that were not significant under any model, Z and P values for an additive model are listed. All genetic models are based on the minor allele of each SNP. SNP, single nucleotide polymorphism; BP, blood pressure; SBP, systolic blood pressure; DBP, diastolic blood pressure. Z indicates test statistic for FBAT.
dominant model;
recessive model;
additive model.
P-values are significant after correction for multiple testing (FDR <0.05).
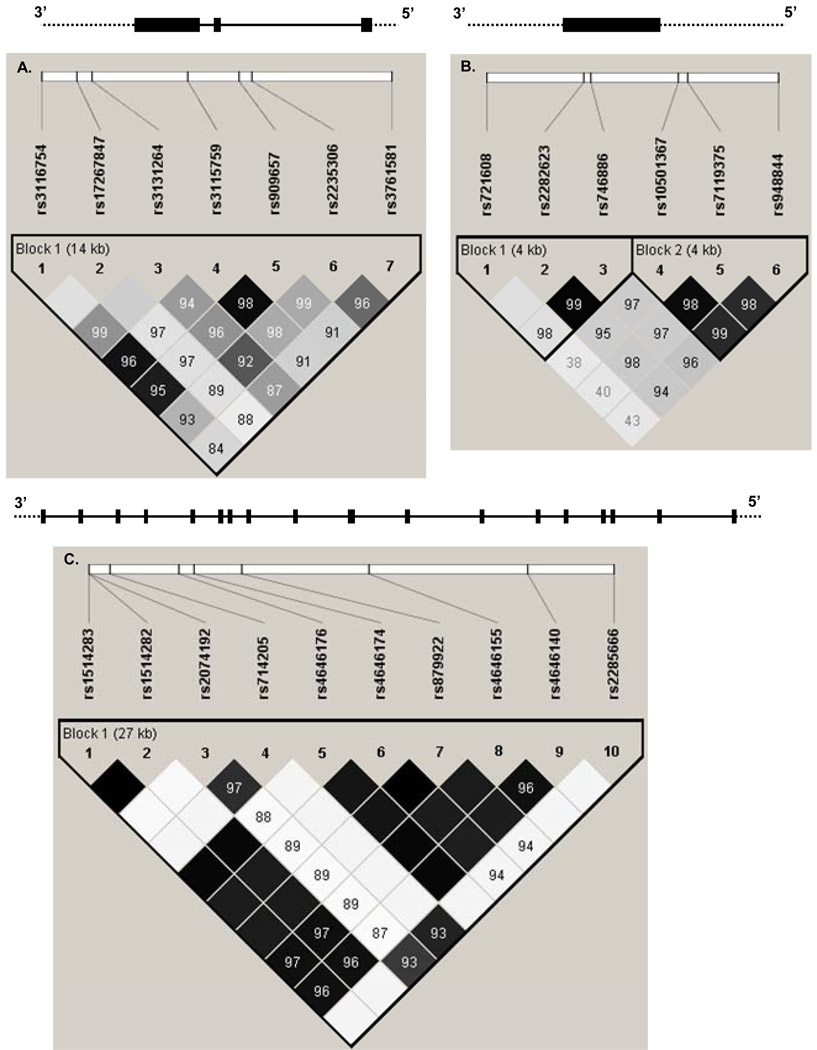
We also performed haplotype analysis based on LD blocks of the three genes defined by Haploview (Figure 1). For the APLNR gene, there were three major haplotypes (≥0.05) in block1 which were formed by rs721628, rs2282623, and rs746886. Of the three haplotypes, haplotype ATT was significantly associated with DBP and MAP responses to low-sodium intervention (P =0.001 and 0.003, respectively) (Table 4). Another haplotype of this block (GCC) was significantly associated with SBP and MAP responses to high-sodium intervention (P =0.004 and 0.01, respectively). For the ACE2 gene, only one block was defined, within which two haplotypes (TTTCCCCGGC and TTCGCCCGGT) were significantly associated with SBP response to low-sodium intervention. Haplotype TTTCCCCGGC was associated with reduced SBP response to low-sodium, whereas haplotype TTCGCCCGGT was associated with increased SBP response. These haplotypes only differed at three loci, rs2074192, rs714205, and rs2285666, which also showed significant associations with SBP response to low-sodium in the single marker analysis.
Figure 1.
The gene structures of APLN (A), APLNR (B), and ACE2 (C) and the position of single nucleotide polymorphisms. The black boxes in each gene are exons. Solid lines show introns and dashed lines show flanking regions. Pairwise linkage disequilibrium (LD) coefficients D’×100 are shown in each cell (D’ values of 1.0 are not shown). The r2 color scheme of Haploview was applied (r2 =0 shown in white, 0 < r2 <1 shown in shades of grey, and r2 =1 shown in black).
Table 4.
Summary of haplotypes significantly associated with BP response to low- and high-sodium interventions
| Gene | Haplotype | Frequency | SBP Response | DBP Response | MAP Response | |||
|---|---|---|---|---|---|---|---|---|
| Z | P-value | Z | P-value | Z | P-value | |||
| Low-sodium Intervention | ||||||||
| APLNR | ATTa | 0.38 | 1.183 | 0.24 | 2.607/3.241 | 0.009b*/0.001c* | 2.924 | 0.003c* |
| ACE2 | TTCGCCCGGTd | 0.52 | −2.153 | 0.03b* | −0.929 | 0.35 | −1.391 | 0.17 |
| TTTCCCCGGCd | 0.40 | 2.395/2.365 | 0.02b*/0.02c* | 0.558 | 0.58 | 1.206 | 0.23 | |
| High-sodium Intervention | ||||||||
| APLNR | GCCa | 0.14 | 2.483/2.868 | 0.01b*/0.004c* | 1.887 | 0.06 | 2.519/2.437 | 0.01b*/0.01c* |
Z indicates test statistic for FBAT. For association not significant under any model, Z and P values for additive model are listed. BP, blood pressure; SBP, systolic blood pressure; DBP, diastolic blood pressure.
rs721608-rs2282623-rs746886.
additive model.
dominant model.
rs1514283-rs1514282-rs2074192-rs714205-rs4646176-rs4646174-rs879922-rs4646155-rs4646140-rs2285666.
P-values are significant after correction for multiple testing (FDR <0.05).
Discussion
This large family-based association study identified multiple SNPs in the apelin receptor and ACE2 genes associated with BP responses to changes in dietary sodium intake. These results indicated that the apelin system and ACE2 genes might be mechanistically involved in BP salt-sensitivity and their genetic variants might contribute to the variation of this complex phenotype.
It is well-known that BP is a complex phenotype and influenced by multiple genetic and environmental factors and their interactions. Although recent large-scale genome-wide association studies have identified some genetic variants highly significantly associated with BP, they only explained a very small proportion of the total variation of BP [28, 29]. Failure to measure gene and environment interactions may result in the inability to fully detect genetic contribution to BP variability in those studies. The dietary sodium intervention in our study provides a good opportunity to identify genes related to salt-sensitive hypertension.
Based on its physiological function in the cardiovascular system, we selected candidate genes in the apelin system and examined their association with BP responses to changes in dietary sodium intake. This novel pathway has broad effects in the cardiovascular system and is emerging as an important mediator of cardiovascular homeostasis [30]. Recently, the apelin system has been implicated in BP regulation and reported to be associated with hypertension [14, 21].
In this study, we observed that two SNPs in the 3’ UTR region of APLNR, rs2282623 and rs746886, were significantly associated with BP response to low-sodium intervention. The haplotype analysis of the block including these two SNPs further confirmed their association with BP response to salt interventions. To speculate on the possible function of these significant SNPs, we analyzed them using the web tool SNPseek, a database of putatively functional human SNPs (http://snp.wustl.edu/cgi-bin/SNPseek/index.cgi). SNPseek shows that both of them are in regulatory potential regions where a marked resemblance of typical regulatory elements exists. This annotation of SNPseek is integrated from a regulatory potential score, which is computed from alignments of human DNA with that of six other species and provided by UCSC Genome browser. It is based on the principle that regions conserved across long evolutionary timescales are likely to be functionally significant [31]. Thus, it is possible that these two SNPs of APLNR influence BP response to salt intake through their impact on gene expression of APLNR. On the other hand, we still cannot exclude the possibility that these two SNPs are in strong LD with the true functional SNP which was not analyzed in our study. These results highlight a potential region for targeted resequencing in the future.
We found multiple intronic SNPs of the ACE2 gene associated with BP responses to changes in salt intake. Of them, SNP rs2285666, also known as 8790G/A, has been reported to be associated with hypertension among those with metabolic syndrome [32]. SNPseek analysis shows SNP rs2285666 is located in a splice site of the ACE2 gene and may alter splicing patterns. We could not find functional support for the association of the other SNPs, which are also not in strong LD with any other SNPs based on the HapMap data. However, there is accumulating evidence that intronic polymorphisms may cause complex disorders [33–35]. In order to fully explain these associations, further dissection of the regions surrounding these intronic SNPs and functional studies are warranted.
Large sample size, homogenous population, and excellent compliance to the dietary intervention highlight the strengths of the study. A possible concern of this study is the definition of BP response to low-sodium intervention. We took the BP difference between low-sodium intervention and baseline observation as the BP response to low-sodium intervention. Since the baseline dietary salt intake was not controlled, it might influence the study power and validity. However, all study participants were living in a homogenous environment with high dietary salt intake. Through measuring urine sodium excretion, we found that the baseline dietary sodium intake was much higher than that during low-sodium intervention and even as high as that during the high-sodium intervention. This might reduce the variation of BP response to low-sodium caused by the inter-individual variation in baseline sodium intake. Furthermore, some associations observed for BP response to low-sodium intervention were replicated by BP response to high-sodium intervention in our study.
In summary, we reported here for the first time significant associations between genetic variants of the APLNR and ACE2 genes and BP responses to changes in dietary sodium intake. These findings may contribute to a better understanding of the genetic mechanisms underlying BP regulation and may have potential clinical and public health implications. Replication of these results in other populations and further functional studies to identify causal variants are critically important.
Acknowledgements
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland.
Appendix
GenSalt Collaborative Research Group
Tulane University Health Sciences Center, New Orleans, USA: Jiang He (PI), Lydia A. Bazzano, Chung-Shiuan Chen, Jing Chen, Tanika N. Kelly, L. Lee Hamm, Paul Muntner, Kristi Reynolds, Paul K. Whelton, Wenjie Yang, and Qi Zhao.
Washington University School of Medicine, St. Louis, USA: DC Rao (PI), Matthew Brown, Charles Gu, Treva Rice, Karen Schwander, and Shiping Wang.
Chinese Academy of Medical Sciences, Beijing, China: Dongfeng Gu (PI), Jie Cao, Jichun Chen, Xiufang Duan, Jianfeng Huang, Jinghan Huang, Jianxin Li, Depei Liu, Donghua Liu, Enchun Pan, Yang Wei, and Xiqui Wu. Shandong Academy of Medical Sciences, Shandong, China: Fanghong Lu (PI), Shikuan Jin, Qingjie Meng, Fan Wu, and Yingxin Zhao; Shandong Center for Diseases Control and Prevention, Shandong, China: Jixiang Ma (PI), Weika Li, and Jiyu Zhang; Zhengzhou University: Dongsheng Hu (PI), Yaxin Ding, Hongwei Wen, Meixi Zhang, and Weidong Zhang; Xinle Traditional Chinese Medicine Hospital, Hebei, China: Xu Ji (PI), Rongyan Li, Haijun Zu; Nanjing University of Medical Sciences, Jiangsu, China: Cailiang Yao (PI), Yongchao Li, Chong Shen, and Jiayi Zhou; Xi’an Jiaotong University, Shanxi, China: Jianjun Mu (PI), Enrang Chen, Qinzhou Huang, and Man Wang.
Chinese National Human Genome Center at Beijing: Zhi-Jian Yao (PI), Shufeng Chen, Dongfeng Gu, Hongfan Li, Laiyuan Wang, Penghua Zhang.
University of Texas Health Sciences Center at Houston: James E. Hixson (PI) and Lawrence C. Shimmin.
National Heart, Lung, and Blood Institute: Cashell E. Jaquish
Footnotes
Disclosure: The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland.
Conflicts of interest: None.
References
- 1.Weinberger MH. Salt sensitive human hypertension. Endocr Res. 1991;17:43–51. doi: 10.1080/07435809109027188. [DOI] [PubMed] [Google Scholar]
- 2.Luft FC, Weinberger MH. Heterogeneous responses to changes in dietary salt intake: the salt-sensitivity paradigm. Am J Clin Nutr. 1997;65:612S–617S. doi: 10.1093/ajcn/65.2.612S. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, et al. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet. 1997;350:1734–1737. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 4.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37:429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 5.Miller JZ, Weinberger MH, Christian JC, Daugherty SA. Familial resemblance in the blood pressure response to sodium restriction. Am J Epidemiol. 1987;126:822–830. doi: 10.1093/oxfordjournals.aje.a114719. [DOI] [PubMed] [Google Scholar]
- 6.Svetkey LP, McKeown SP, Wilson AF. Heritability of salt sensitivity in black Americans. Hypertension. 1996;28:854–858. doi: 10.1161/01.hyp.28.5.854. [DOI] [PubMed] [Google Scholar]
- 7.Svetkey LP, Chen YT, McKeown SP, Preis L, Wilson AF. Preliminary evidence of linkage of salt sensitivity in black Americans at the beta 2-adrenergic receptor locus. Hypertension. 1997;29:918–922. doi: 10.1161/01.hyp.29.4.918. [DOI] [PubMed] [Google Scholar]
- 8.Beeks E, Kessels AG, Kroon AA, van der Klauw MM, de Leeuw PW. Genetic predisposition to salt-sensitivity: a systematic review. J Hypertens. 2004;22:1243–1249. doi: 10.1097/01.hjh.0000125443.28861.0d. [DOI] [PubMed] [Google Scholar]
- 9.Reaux A, De Mota N, Skultetyova I, Lenkei Z, El Messari S, Gallatz K, et al. Physiological role of a novel neuropeptide, apelin, and its receptor in the rat brain. J Neurochem. 2001;77:1085–1096. doi: 10.1046/j.1471-4159.2001.00320.x. [DOI] [PubMed] [Google Scholar]
- 10.Masari B, Knibiehler B, Audigier Y. Apelin signalling: a promising pathway from cloning to pharmacology. Cell Signal. 2005;17:415–426. doi: 10.1016/j.cellsig.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Masari B, Morin N, Pedebernarde L, Knibiehler B, Audigier Y. The apelin receptor is coupled to Gi1 or Gi2 protein and is differentially desensitized by apelin fragments. J Biol Chem. 2006;281:18317–18326. doi: 10.1074/jbc.M600606200. [DOI] [PubMed] [Google Scholar]
- 12.Cheng X, Cheng XS, Pang CC. Venous dilator effect of apelin, an endogenous peptide ligand for the orphan APJ receptor, in conscious rats. Eur J Pharmacol. 2003;470:171–175. doi: 10.1016/s0014-2999(03)01821-1. [DOI] [PubMed] [Google Scholar]
- 13.Ishida J, Hashimoto T, Hashimoto Y, Nishiwaki S, Iguchi T, Harada S, et al. Regulatory roles for APJ, a seven-transmembrane receptor related to angiotensin-type 1 receptor in blood pressure in vivo. J Biol Chem. 2004;279:26274–26279. doi: 10.1074/jbc.M404149200. [DOI] [PubMed] [Google Scholar]
- 14.Kagiyama S, Fukuhara M, Matsumura K, Lin Y, Fujii K, Iida M. Central and peripheral cardiovascular actions of apelin in conscious rats. Regul Pept. 2005;125:55–59. doi: 10.1016/j.regpep.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 15.Charles CJ, Rademaker MT, Richards AM. Apelin-13 induces a biphasic haemodynamic response and hormonal activation in normal conscious sheep. J Endocrinol. 2006;189:701–710. doi: 10.1677/joe.1.06804. [DOI] [PubMed] [Google Scholar]
- 16.Japp AG, Cruden NL, Amer DA, Li VK, Goudie EB, Johnston NR, et al. Vascular effects of apelin in vivo in man. J Am Coll Cardiol. 2008;52:908–913. doi: 10.1016/j.jacc.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Yao F, Raizada MK, O'Rourke ST, Sun C. Apelin gene transfer into the rostral ventrolateral medulla induces chronic blood pressure elevation in normotensive rats. Circ Res. 2009;104:1421–1428. doi: 10.1161/CIRCRESAHA.108.192302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts EM, Newson MJ, Pope GR, Landgraf R, Lolait SJ, O'Carroll AM. Abnormal fluid homeostasis in apelin receptor knockout mice. J Endocrinol. 2009;202:453–462. doi: 10.1677/JOE-09-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 20.Burrell LM, Johnston CI, Tikellis C, Cooper ME. ACE2, a new regulator of the renin-angiotensin system. Trends Endocrinol Metab. 2004;15:166–169. doi: 10.1016/j.tem.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li WW, Niu WQ, Zhang Y, Wu S, Gao PJ, Zhu DL. Family-based analysis of apelin and AGTRL1 gene polymorphisms with hypertension in Han Chinese. J Hypertens. 2009;27:1194–1201. doi: 10.1097/HJH.0b013e32832a3eb1. [DOI] [PubMed] [Google Scholar]
- 22.GenSalt Collaborative Research Group. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88:2460–2470. doi: 10.1161/01.cir.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical powerful approach to multiple testing. J. Roy. Statist. Soc. Ser.B. 1995;57:289–300. [Google Scholar]
- 28.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009 doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinz MJ, Skepper JN, Davenport AP. Immunocytochemical localisation of the apelin receptor, APJ, to human cardiomyocytes, vascular smooth muscle and endothelial cells. Regul Pept. 2005;126:233–240. doi: 10.1016/j.regpep.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong J, Yan Z, Liu D, Ni Y, Zhao Z, Zhu S, et al. Association of angiotensin-converting enzyme 2 gene A/G polymorphism and elevated blood pressure in Chinese patients with metabolic syndrome. J Lab Clin Med. 2006;147:91–95. doi: 10.1016/j.lab.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 34.Uitterlinden AG, Burger H, Huang Q, Yue F, McGuigan FE, Grant SF, et al. Relation of alleles of the collagen type Ialpha1 gene to bone density and the risk of osteoporotic fractures in postmenopausal women. N Engl J Med. 1998;338:1016–1021. doi: 10.1056/NEJM199804093381502. [DOI] [PubMed] [Google Scholar]
- 35.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003;35:341–348. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]