Abstract
The PIK3 signaling pathway has been identified as one of the most important and most frequently mutated pathways in breast cancer. Somatic mutations in the catalytic subunit of PIK3CA have been found in a significant fraction of breast carcinomas, and it has been proposed that mutant PIK3CA plays a role in tumor initiation. However, the majority of primary human tumors analyzed for genetic alterations in PIK3CA have been invasive breast carcinomas and the frequency of PIK3CA mutations in pre-invasive lesions has not been explored. To investigate this, we sequenced exons 9 and 20 of PIK3CA in pure ductal carcinoma in situ (DCIS), DCIS adjacent to invasive carcinoma, and invasive ductal breast carcinomas (IDC). In a subset of cases both in situ and invasive areas were analyzed from the same tumor. We found that the frequency of PIK3CA mutations was essentially the same (~30%) in all three histologic groups. In some cases in situ and invasive areas of the same tumor were discordant for PIK3CA status and in two cases where multiple invasive and adjacent in situ areas within the same tumor were analyzed independently, we detected intra-tumor heterogeneity for PIK3CA mutations. Our results suggest that mutation of PIK3CA is an early event in breast cancer that is more likely to play a role in breast tumor initiation than in invasive progression, although a potential role for exon 9 mutations in the progression of a subset of DCIS cases cannot be excluded.
Keywords: DCIS, PIK3CA, tumor progression, invasion, breast cancer
INTRODUCTION
The phosphatidylinositol 3 kinase (PIK3) signaling pathway is an important regulator of cell growth, metabolism, proliferation, survival, motility, and invasion (1, 2). Highlighting the importance of PIK3 signaling in human cancer, genetic alterations have been reported in several components of the pathway in various tumor types including deletion of PTEN (3), amplification of AKT1 and PIK3CA, and somatic mutations of PIK3CA (2) and AKT1 (4). In human breast cancer, mutations in PIK3CA have been reported to occur in 8–40% of tumors making it one of the most frequently mutated genes in this tumor type (5, 6). The majority of mutations have been identified in the helical domain (exon 9) and in the kinase domain (exon 20) of PIK3CA.
Expression of cancer derived PIK3CA mutants in cultured cells increases kinase activity, invasion, resistance to apoptosis, and in immortalized human mammary epithelial cells it is sufficient to induce soft agar growth and tumorigenicity suggesting that mutant PIK3CA may play a role in the initiating steps of breast tumorigenesis (5, 6). In contrast, somatic mutations in PIK3CA have been reported to occur at higher frequency in advanced stage invasive tumors suggesting a role in tumor progression (7, 8). Analysis of pre-invasive tumors including colon adenomas and Barrett’s esophagus has demonstrated a paucity of mutations in PIK3CA compared to invasive carcinomas (7, 8). Similarly the frequency of PIK3CA mutations was significantly higher in advanced gastric carcinomas compared to early stage tumors (9) and gain of PIK3CA was more frequent in high-grade dysplasias and carcinomas than in low-grade dysplasias in head and neck cancer (10). Each of these studies analyzed only a few cases of in situ tumors and did not test adjacent in situ and invasive areas from the same patient.
In human breast cancer only a few published studies on small number of samples have analyzed PIK3CA mutations in pre-invasive tumors including DCIS (9, 11–13). Campbell et al. performed SSCP (Single-Strand Conformational Polymorphism) analysis of PIK3CA exons 1–20 in 70 breast tumors including 3 DCIS and detected mutations in 28/67 invasive tumors and 0/3 DCIS (11), whereas Lee et al. analyzed exons 9 and 20 by SSCP in 93 breast tumors and identified mutations in 24/78 of invasive breast tumors and only 2/15 DCIS (9). Neither report provided information on the type of DCIS analyzed (i.e., pure DCIS or in situ areas adjacent to invasive tumor), but, they nonetheless, implicated PIK3CA in the in situ to invasive carcinoma progression. A third study analyzed 11 DCIS adjacent to IDC and found concordant mutations in all cases (12).
Based on these data we hypothesized that mutational activation of the PIK3CA pathway may play a role in the progression of in situ carcinomas to invasive disease. To test this hypothesis we analyzed the sequence of the two mutational hotspot regions of PIK3CA (exons 9 and 20) in pure DCIS, and in invasive breast carcinomas and adjacent DCIS regions.
MATERIALS AND METHODS
Tissue specimens
Fresh, frozen, or formalin fixed and paraffin embedded (FFPE) tumor specimens were obtained for the pilot cohort from Harvard affiliated Hospitals (Boston, MA), Duke University (Durham, NC), University Hospital Zagreb (Zagreb, Croatia), Baylor College of Medicine (Houston, TX), and the National Disease Research Interchange (Philadelphia, PA); the larger cohort’s specimens were obtained from Bundang Hospital of Seoul National University College of Medicine and Samsung Medical Center (Seoul, South Korea). All human tissue was collected without patient identifiers using protocols approved by the Institutional Review Boards.
Manual and Laser Capture Microdissection (LCM)
Six-micron thick paraffin sections were deparaffinized in xylene and rehydrated in graded alcohols. For manual dissection slides were stained with hematoxylin and eosin, and tumor cell-enriched areas collected with a razor blade. For LCM, slides were stained with 0.5% methyl green for 30 seconds, rinsed in distilled water and left to air dry. Stained slides were microdissected with an IR laser enabled microscope (Arcturus Engineering, Mountain View, CA) in the Dana-Farber Cancer Institute Pathology Core facility. A 30-micron laser spot size was used to isolate a minimum of 2,000 selected cells onto CapSure LCM caps (Molecular Devices, Sunnyvale, CA).
DNA extraction, mutation and statistical analyses
DNA preparation and mutation analyses were performed essentially as described (14), detailed procedures are available from the authors upon request. Statistical analysis used Fisher’s exact test and exact power calculations for it, as well as logistic regression (with likelihood ratio tests) and exact binomial confidence intervals (CI); all p values are two-sided and no correction for multiple comparisons was done. Each case with matched IDC and DCIS lesions analyzed for mutations could only be used in a logistic regression (or a Fisher test) once (to avoid violating the assumption of independence), so analyses were done twice (once using the DCIS mutation status and categorizing them as DCIS adjacent to IDC and once using the IDC mutation status).
RESULTS AND DISCUSSION
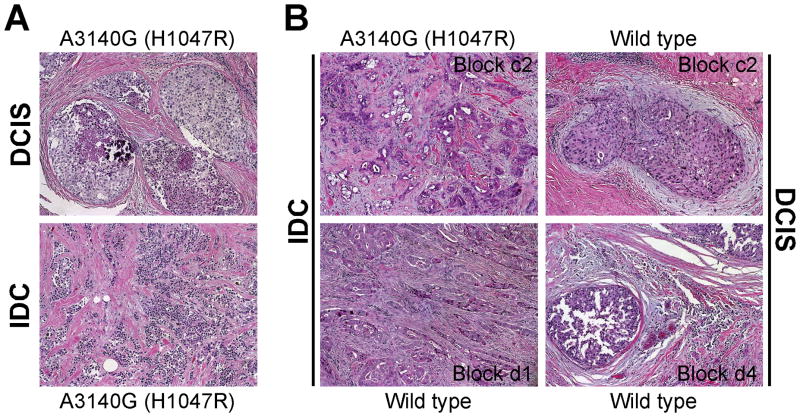
We determined the frequency of PIK3CA mutations in pure DCIS, DCIS adjacent to invasive cancer, and in invasive ductal breast carcinomas (IDC). All samples were microdissected to ensure the proper identification and selection of in situ and invasive areas (Fig. 1). Due to the limited amount of DNA recovered, our mutational analysis was limited to two previously characterized mutational hot spots: exons 9 and 20.
Figure 1. Examples of DCIS and invasive tumors used for microdissection.
Hematoxylin& eosin stained sections of in situ and invasive areas selected for microdissection from two independent cases in which DCIS and IDC had the same mutation (A) or demonstrated intra-tumor heterogeneity (B).
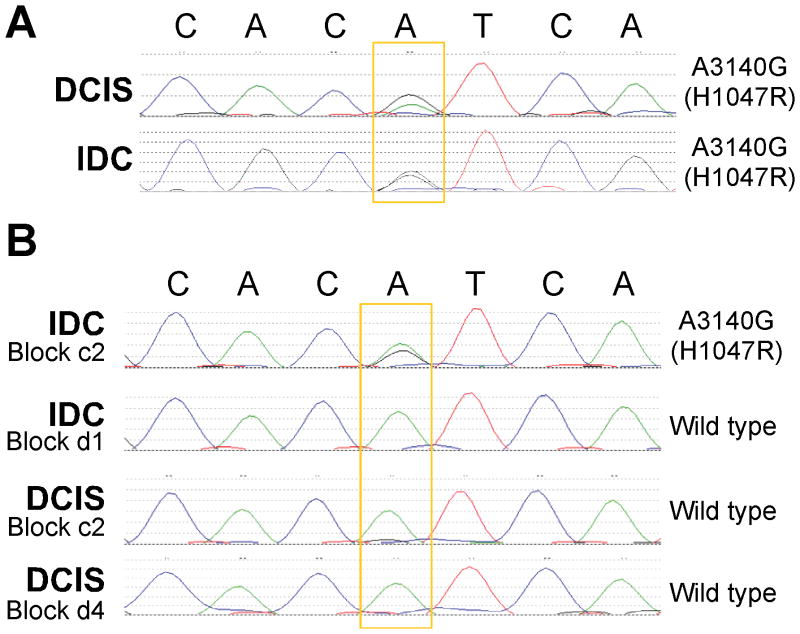
First, we analyzed mutations using Sanger sequencing in a smaller cohort: specimens from 83 patients, including 31 patients with matched DCIS and IDC lesions; two of the cases had several IDC and DCIS areas isolated for analysis (Supplementary Table S1). PIK3CA mutations were found in only 5% of pure DCIS (95% CI 1–16%) but in about 16% of DCIS adjacent to IDC and 9% of IDC. Of the 29 matched cases with a single DCIS and a single IDC lesion evaluated, only two (7%) had discordant results (95% CI of 1 to 25%); when both IDC and adjacent DCIS were mutant they had identical mutations confirming their clonal relatedness (Fig. 2A). In the two cases with multiple DCIS and IDC regions analyzed, in one case the IDC was wild type but two out of three DCIS areas had a mutation whereas in the other case one invasive area had a mutation and the remaining invasive and DCIS areas were all wild type (Fig. 2B).
Figure 2. Examples of PIK3CA Sanger sequencing results.
A, Concordant somatic PIK3CA mutation (A3140G) in IDC and adjacent DCIS. B, Intra-tumor heterogeneity for PIK3CA mutation. One of the invasive areas of this tumor had exon 20 A3140G (H1047R) mutation whereas other invasive and adjacent DCIS were wild type.
Because the frequency of PIK3CA mutations in the initial cohort was low and the 95% CIs were so large, we expanded our study to 374 Korean patients (including 48 patients with matched DCIS and IDC lesions). In addition, due to the intratumor heterogeneity of PIK3CA mutations we observed in our pilot study, we analyzed the most frequent exon 9 (E542K) and exon 20 (H1047R and H1047L) mutations by mass spectrometry (14) (Supplementary Figure S1), which is more sensitive for mutation detection than Sanger sequencing (14, 15). Using this approach, the frequency of any PIK3CA mutation was between 28% and 31% in each histologic subgroup and not significantly different (Table 1 and Supplementary Table S2). The higher frequency of mutations detected by mass spectrometry is unlikely to be due to ethnic or other differences between the pilot and extended cohorts, but highlights the advantage of using this method for the testing of known mutations in cancer genes that may be present only in a subset of cancer cells (14, 15).
Table 1.
Summary of PIK3CA mutations by histology.
| PIK3CA mutation | Number of cases with specific mutations ** | ||||
|---|---|---|---|---|---|
| E542K | H1047L | H1047R | |||
| Pure DCIS | 61/202 (30%) | 5 | 5 | 52 | |
| DCIS adj. to IDC (not matched) | 15/49 (31%) | 2 | 0 | 14 | |
| Matched | DCIS | 14/48 (29%) | 2 | 0 | 13 |
| IDC* | 14/45 (31%) | 0 | 0 | 14 | |
| IDC alone | 21/75 (28%) | 4 | 0 | 17 | |
| DCIS adj. to IDC (matched or not) | 29/97 (30%) | 4 | 0 | 27 | |
| IDC (matched or not) | 35/120 (29%) | 4 | 0 | 31 | |
| P-value of difference between histologic groups (DCIS used for matched cases) | 0.94 | ||||
| P-value of difference between histologic groups (IDC used for matched cases) | 0.97 | ||||
Three cases had matched DCIS and IDC specimens but mutation analysis for IDC lesion was inconclusive.
Three cases (1 pure DCIS and 2 DCIS adjacent to IDC) had more than 1 mutation.
To determine if the sample size used has enough power to detect significant differences in the three histology groups, we performed power calculations. We found that when the matched cases were analyzed as DCIS, there was 80% power to detect as significantly different true mutation frequencies of 38% and 22% in pure DCIS and DCIS adjacent to IDC and there was 80% power to detect true mutation frequencies of 41% and 20% in DCIS adjacent to IDC and IDC. Of the 45 matched cases, 15 (33%; 95% CI 20–49%) had discordant results, 8 with a mutation only in IDC and 7 with a mutation only in DCIS (Supplementary Table S3). These results indicate that divergence for PIK3CA mutations occur in a significant fraction of breast tumors during in situ to invasive progression. Furthermore, the equal frequency of the two possible discordant patterns (i.e., mutant DCIS and wild type IDC and vice versa) implies the lack of selective advantage of cells with mutant PIK3CA during invasive progression.
We also explored associations between any PIK3CA mutation and age or pathologic variables (when histologic group was forced into the model), as well as interactions between histologic group and other variables (Supplementary Table S4). Neither histologic group nor any other variable was significantly associated with frequency of any mutation (or specific H1047R, H1047L, E542K mutations). When matched cases were analyzed as DCIS, the only significant interaction was between mutant p53 (based on positivity by immunohistochemistry) and pure DCIS (p=0.03); mutant p53 was associated with a greater percentage of patients having a PIK3CA mutation among pure DCIS cases (Supplementary Table S5). When matched cases were analyzed as IDC, the p53 interaction with DCIS remained significant (p=0.01), and the interaction of grade and pure DCIS was also significant (p=0.02); high grade associated with a lower percentage of patients having any PIK3CA mutation in pure DCIS (Supplementary Table S5). The interaction of Her2 and histology was close to significant (p=0.07) with the greatest mutation frequency in tumors with an imunohistochemical staining score of 3+ among DCIS adjacent to IDC (Supplementary Table S5). Models for H1047R mutations were similar to models of any mutation Supplementary Table S6) and there were too few H1047L mutations to analyze separately. However, the H1047L mutation was only seen in pure DCIS, but the frequencies of the mutation do not differ significantly between histologic groups (Supplementary Table S4, p=0.21 and 0.17 depending on whether you counted the matched cases as DCIS or IDC). There were too few E542K mutations to estimate interactions. However, this mutation was not detected among patients diagnosed with pure DCIS who were older than 46 (p=0.03) or had high-grade tumors (p=0.02) (Supplementary Table S4), implying that E542K mutation may increase the risk of DCIS progression in these subsets. Correlating with our results, previous studies have found exon 9 mutations to be more frequent in IDCs of older women (16) and that they are associated with shorter disease-free and overall survival (17).
In summary, this is the first study to demonstrate that PIK3CA mutation is a relatively early event in breast tumorigenesis preceding invasion, since the frequency of PIK3CA mutations is the same in pure DCIS as in DCIS adjacent to IDC and in IDC. Furthermore, we also demonstrated that invasive and in situ areas of the same tumor have identical PIK3CA sequence in the majority of cases although clonal divergence and intra-tumor heterogeneity can also be observed in a significant fraction of tumors. Our results support the hypothesis that mutational activation of the PIK3CA pathway may play a role in breast tumor initiation and it is not likely to be involved in promoting invasive progression. However, the association between exon 9 mutations with the risk of DCIS progression merits further investigation.
Supplementary Material
Acknowledgments
Grant support: Funded by the National Cancer Institute Specialized Program in Research Excellence in Breast Cancer at Dana-Farber/Harvard Cancer Center (CA89393) grant awarded to AM, DI, RG, and KP.
We thank Drs. Myles Brown and Ian Krop for their critical reading of the manuscript.
LIST OF ABBREVIATIONS
- DCIS
ductal carcinoma in situ
- IDC
invasive ductal carcinoma
- HER2
human epidermal growth factor receptor 2
- PIK3CA
phosphatidylinositol 3-kinase catalytic alpha
- LCM
laser capture microdissection
References
- 1.Richardson CJ, Schalm SS, Blenis J. PI3-kinase and TOR: PIKTORing cell growth. Semin Cell Dev Biol. 2004;15:147–59. doi: 10.1016/j.semcdb.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 2.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 3.Saal LH, Gruvberger-Saal SK, Persson C, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–7. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–44. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 5.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 6.Paradiso A, Mangia A, Azzariti A, Tommasi S. Phosphatidylinositol 3-kinase in breast cancer: where from here? Clin Cancer Res. 2007;13:5988–90. doi: 10.1158/1078-0432.CCR-07-1106. [DOI] [PubMed] [Google Scholar]
- 7.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 8.Phillips WA, Russell SE, Ciavarella ML, et al. Mutation analysis of PIK3CA and PIK3CB in esophageal cancer and Barrett’s esophagus. Int J Cancer. 2006;118:2644–6. doi: 10.1002/ijc.21706. [DOI] [PubMed] [Google Scholar]
- 9.Lee JW, Soung YH, Kim SY, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2004 doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 10.Woenckhaus J, Steger K, Werner E, et al. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;198:335–42. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- 11.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 12.Dunlap J, Le C, Shukla A, et al. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat. 2010;120:409–18. doi: 10.1007/s10549-009-0406-1. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Zhu R, Wang L, et al. PIK3CA mutations mostly begin to develop in ductal carcinoma of the breast. Exp Mol Pathol. 2010;88:150–5. doi: 10.1016/j.yexmp.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 14.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fumagalli D, Gavin PG, Taniyama Y, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 17.Barbareschi M, Buttitta F, Felicioni L, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–9. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.