Abstract
Purpose
We report a novel analytical method, named intercohort co-analysis or Ican, which aids in the discovery of genes with predictive value for the progression or outcome of diseases from small-size cohorts. We tested this premise in Ewing's sarcoma (ES), a highly metastatic cancer of bone and soft tissues that lacks validated molecular metastasis and prognostic indicators.
Experimental Design
To uncover genes significantly expressed in ES patient subsets, we first determined a non-arbitrary gene expression significance cutoff based on expression levels in validated expressing and non-expressing tissues. We next searched for genes that were consistently significantly expressed in several ES cohort and cell line datasets. Significantly expressed genes were independently validated by quantitative RT-PCR in an additional ES cohort.
Results
Analysis of ES cohorts revealed marked intercohort gene expression variability. After filtering out the intercohort variability, CXCR4 and CXCR7 were found to be consistently associated with specific ES subsets. Pairwise analyses showed CXCR4 to correlate with ES metastases, and CXCR4 and CXCR7 to patient survival, but not with several other clinicopathological variables.
Conclusion
Ican is a powerful novel method to identifying genes consistently associated with particular disease states in cancers for which large cohorts are not available, currently the case of most cancers. We report for the first time that high CXCR4 expression preferentially associates with metastatic ES and that of CXCR7 with poor patient survival.
Keywords: Chemokines, Chemokine receptors, Metastasis, Pediatric cancers, Rare neoplasms, Small-size cohorts
Introduction
Cancer genomics is a powerful approach to uncovering genes that may be involved in the pathogenesis of cancer as well as those that may serve to predict cancer spread or disease outcome. This has for instance been the case of several adult tumors for which large cohorts were available (1-3). Despite low incidence and major advances in diagnosis and treatment, cancers remain the leading cause of death by disease in children in developed countries (4). This is in part due to the fact that these cancers consist of a large and heterogeneous group of low-incidence neoplasms, thus hampering large cohort studies aimed at uncovering markers of disease occurrence, progression, recurrence, spread, or outcome (5-7).
In recent years, several small-size cohort childhood cancer genomic studies have been reported. We hypothesized that a co-analysis of unrelated small-size cohorts that filters out the high intercohort variability inherent to small-size cohorts may help uncover genes associated with subsets of tumors with common clinical features. We named this approach intercohort co-analysis, henceforth dubbed Ican, and tested this premise in Ewing tumor and related sarcomas of the bone (7, 8), a family of tumors that lacks a reliable molecular prognostic or metastasis indicator, and which we refer to herein as Ewing's sarcoma or ES. As an example of analysis, we used Ican to screen for chemokines and chemokine receptors significantly associated with ES subsets (Flow chart in Supplementary Figure 1). Chemokines are 8 to 10 kDa secreted chemoattractant cytokines, and chemokine receptors are seven-transmembrane G-protein coupled cell surface proteins. These receptors are defined by their ability to induce the directional migration of cells toward a cognate chemotactic cytokine gradient in a process known as chemotaxis. Chemokine receptors, therefore, support several physiological and pathological processes underlying cell motility, egress, and homing to distant sites. For the purpose of this study, we chose to survey chemokines and their receptors because they have been associated with tumor growth, metastasis, or neoangiogenesis in several adult cancers (9, 10). They can also serve as therapeutic targets to curtail tumor growth or metastasis (9, 11, 12) and may thus offer novel treatment options for ES.
We report here that Ican successfully identified CXCR4 (a.k.a. CD184, LESTR, WHIM, or SDF-1 receptor; Entrez gene ID # 7852) and CXCR7 (a.k.a. CMKOR1, GPR159, RDC1, or chemokine orphan receptor 1; Entrez gene ID # 57007), two chemokine receptors known to associate with metastasis and/or poor prognosis in other cancers (10, 13), as significantly associated with ES subsets. We show for the first time that high CXCR4 expression marks ES metastases and that high expression of both CXCR4 and CXCR7 serves as a predictor of adverse patient survival. Expression of these genes in the analyzed tumors is unlikely to stem from non-tumoral sources as we found them to be also expressed in ES-derived cell lines (EDCL). In EDCL, high CXCR4 expression was restricted to those derived from metastases, and both CXCR4 and CXCR7 expression was dependent on EWS-FLI1, an ES specific transcription factor. We present these findings as a proof-of-principle that Ican of publicly available gene expression profiles can yield genes with predictive value for the progression or outcome of low-incidence diseases.
Materials and Methods
Ewing's Sarcoma-derived Cell Lines (EDCL)
EDCL culture and transfection conditions have been described (14). Cell lines derived from ES metastases (M-EDCL) consisted of SK-N-MC, SMB, STA-ET-2.2, STA-ET-3, STA-ET-7.3, STA-ET-8.1, TC252, VH64, WE-68-M1 and WE-68-M2, and cell lines derived from localized ES (L-EDCL) consisted of A673, RD-ES, RM82, STA-ET-2.1, STA-ET-4, STA-ET-6, STA-ET-7.1, STA-ET-7.2, STA-ET-11 and WE-68. Additional details on these EDCL were previously reported (15).
EWS-FLI1-dependent Gene Regulation Assessed by RNA Interference
Knockdown studies were carried out using EWS-FLI1 type I- or type II-specific small-hairpin RNA (shRNA) in EDCL, and compared to non-targeting shRNA-transfected cells as previously reported (14).
Gene Expression Profiling of EDCL
Gene expression profiles of cell lines derived from localized ES or from ES metastases were surveyed by Affymetrix HG-U133A arrays (Affymetrix, Inc., Santa Clara, CA) as previously described (14), and analyses performed in R statistical environment using Bioconductor packages (see below).
Bioinformatic Analyses and Procedures
Ican of Gene Expression Datasets
Cancer genomic datasets may be retrieved from public gene expression profile repositories such as GEO1 (16), or ArrayExpress2 (17). These may contain both normalized and raw gene expression values, but usually are accompanied by little if any biographical or clinical data. For the purpose of this study, we considered those obtained on Affymetrix GeneChip® human U133A arrays which consist of more than 22,000 probesets, covering about 14,500 human genes. As outlined below, we analyzed only probesets that passed strict selection criteria and run several quality control checks prior to the analyses. Finally, to further increase dataset good quality odds, we limited our analyses to peer-reviewed, published datasets (Supplementary Figure 1).
ES cohort, EDCL and normal tissue (NT) CEL file data (accession numbers E-MEXP-353, E-MEXP-1142, GSE7007, GSE12102, GSE14543, GSE7007, GSE1133) were extracted from GEO or ArrayExpress. Similarly to the vast majority of gene expression datasets currently deposited in these databases, all four ES cohorts tested lacked biographical or clinical data. ES cohorts were normalized using the gcrma algorithm (18). Intensity distributions from all samples were normalized using quantile normalization and the Bioconductor package limma (19). Using the Bioconductor package panp, probesets expressed at levels comparable to the probability density distribution of probes with no EST hits were deemed negative. Affymetrix probesets are known to have widely varying hybridization characteristics resulting in non-uniform thresholds of expression across probesets that can be labeled expressed versus non-expressed (20). Thus, after the initial normalization steps, we empirically searched for a threshold of gene expression for each probeset that differentiated known expressing from non-expressing tissues. We found that, in our datasets, the median of the probe density distribution across NTs separated known expressing from non-expressing tissues (9, 21). This was determined following systematic testing of cutoffs in in silico analyses of gene expression in tissues demonstrated either to express or not biologically relevant levels of genes (52 genes in 75 different tissues and in 2 biological replicates; (22)). To reduce the probability of spurious data points, only expression values exceeding median expression in NT in at least two biological replicates were deemed significant. Whenever multiple probesets probing the same gene satisfied the aforementioned criteria, we chose those whose expression was most variable across the spectrum of ES and NT. This was carried out to maximize the likelihood to pick probesets that best differentiated between expressing and non-expressing ES (23). Based on these criteria, we selected 34 out of 40 chemokines and 18 out of 22 known chemokine receptors for analysis.
Differential Gene Expression Analysis of M-EDCL and L-EDCL
Differentially expressed genes between cell lines established from localized ES or from ES metastases were determined using a moderated two-sample t-test in the R package limma (17). To this end, three L-EDCL and three M-EDCL were treated as biological replicates for either condition (i.e. localized ES or ES metastases). Thus, for each gene, average expression values of replicates were determined and the significance of the difference between the means of the two groups was determined using the moderated t-test in the limma package. p values were corrected for multiple testing using the Benjamini-Hochberg correction method.
Gene Set Enrichment Analysis (GSEA)
GSEA is a bioinformatic method to assess whether genes with known biological/molecular functions are concomitantly up- or down-regulated in a certain gene expression dataset (24). The Molecular Signatures Database (MSigDB) hosted by the Broad Institute currently provides the most comprehensive collection of gene sets with annotated/known functions. The collection of Gene Ontology3 terms available from the Broad Institute offers the advantage of manual curation thereby enhancing the information content of these gene-sets (a detailed description of the curation process may be found at4). The Gene Ontology (GO) gene-set collection C5 of the MSigDB was downloaded from the Broad Institute and loaded into R statistical environment. Gene set enrichment analysis was used to calculate whether certain GO gene sets, and therefore functional gene categories, were differentially regulated between cell lines established from localized ES or from ES metastases. To this end, LogFD = Log2 x̄(M-EDCL) - Log2 x̄(L-EDCL) was calculated and used as input for the PGSEA Bioconductor package (25). GO gene sets larger than 200 genes were omitted from the analysis because they contained largely non-specific terms. All C5 MSigDB gene sets, gene IDs, and p values from the tests are listed in Supplementary Tables 1 and 2.
Quantitative Real Time RT-PCR (qRT-PCR)
Analysis of EDCL by qRT-PCR was as described earlier (14). Archived fresh-frozen ES samples were obtained from the tumor bank at the Children's Hospital Los Angeles or from St. Anna Children's Hospital in Vienna. All specimens were obtained in compliance with HIPAA regulations and with IRB ethics committee approval. Total RNA was extracted from fresh-frozen tumor sections using Qiagen columns (Qiagen, Valencia, CA) and first strand complementary DNA (cDNA) was synthesized from 250 ng RNA using the iScript Reverse Transcriptase kit (Bio-Rad, Hercules, CA). qRT-PCR was performed using validated proprietary TaqMan Gene Expression Assays (Applied Biosystems) for CXCR4 (Hs00607978_s1), CXCR7 (Hs00664172_s1), GAPDH (Hs99999905_m1) and ACTB (Hs99999903_m1) on an Applied Biosystems 7900HT system. Expression of CXCR4 or CXCR7 was normalized relative to the average expression of ACTB and GAPDH in the same sample using the formula % expression = 2-ΔCt × 100.
Statistical Analyses of Clinical Data
Correlation of gene expression to clinical variables in ES was tested using nonparametric two-tailed unpaired Mann-Whitney U-tests (significant if p < 0.05), or One-way Anova and Bonferroni's correction for multiple comparison testing (significant if p < 0.01), as indicated. Patient survival significance was tested using Mantel-Cox Log-rank (significant if p < 0.05).
Results
High intercohort variability hampers gene-disease association studies of low-incidence cancers
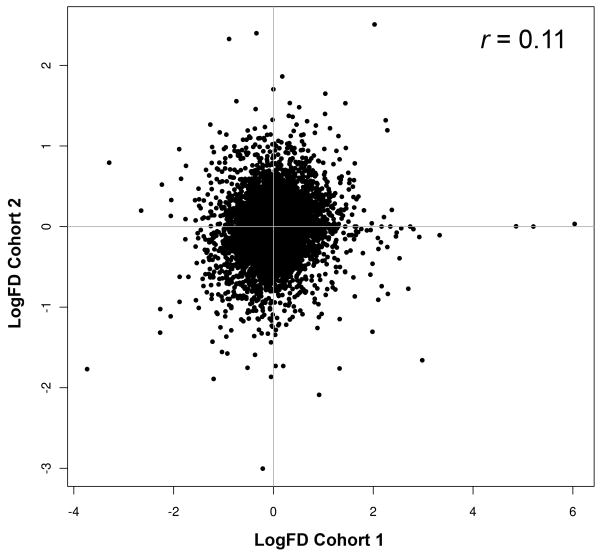
Discovery of gene-disease associations that can serve as markers of certain disease states, say of good or poor disease outcome, is greatly facilitated by the availability of large cohorts and conversely severely compromised by the small-size of cohorts in low-incidence diseases (5, 26). To illustrate this problem, there currently are two reported small-size cohort studies of gene expression signatures associated with ES metastases (27, 28). We surveyed the two signatures by calculating LogFD, the Log2 fold difference between the mean expression of genes in metastatic and localized ES, and compared the extent of convergence between the two signatures. As shown in Figure 1, there was little overlap of differentially expressed genes between the two studies (10,047 genes analyzed; Pearson correlation coefficient r = 0.111; 95% confidence interval CI 0.092-0.130). To avoid the possibility that a correlation between the two signatures might have been underestimated due to the large size of the two datasets considered, we also restricted the analysis to the expression profiles of 426 genes corresponding to the gene population subsets preferentially expressed in metastatic or localized ES in one cohort (LogFD ≥ 1 or LogFD ≤ -1, respectively; (28)), and compared them to those in the second cohort. Again, little correlation was observed between the two cohorts (426 genes analyzed; r = 0.117; 95% CI 0.023-0.209). These data exemplify the high intercohort variability manifest in a genome-wide analysis of two small-size cohorts.
Fig. 1. Metastatic versus Localized ES Differential Gene Expression Intercohort Variability.
LogFD values of two ES cohorts were plotted against each other. Positive LogFD values on the x and y abscissas denote genes expressed more highly in metastatic than in localized ES, whereas negative LogFD values point to the converse. Genes with -1 < LogFD < 1 were considered not differentially expressed. LogFD = x̄{Log2(M-ES) - Log2(L-ES)}; r: Pearson correlation coefficient.
Chemokine and chemokine receptor gene expression profiles in ES
To solve the problem posed by intercohort variability, we tested Ican on several publicly available gene expression profiles. In Ican, gene expression levels in biologically relevant normal tissues are used as a benchmark of gene expression significance in test samples in each independent cohort, and a subtractive overlay of cohort-restricted associations is carried out to uncover genes consistently associated with sample-subsets in all cohorts. To test feasibility, we retrieved gene expression profiles derived from four independent ES cohorts gathered at ES reference centers at the UK, France, Germany, and Italy (27-30). We also included gene expression profiles of six ES-derived cell lines (EDCL) and those of 75 different normal tissues (NT) (14, 22), and researched the expression of chemokine ligands and receptors in these samples.
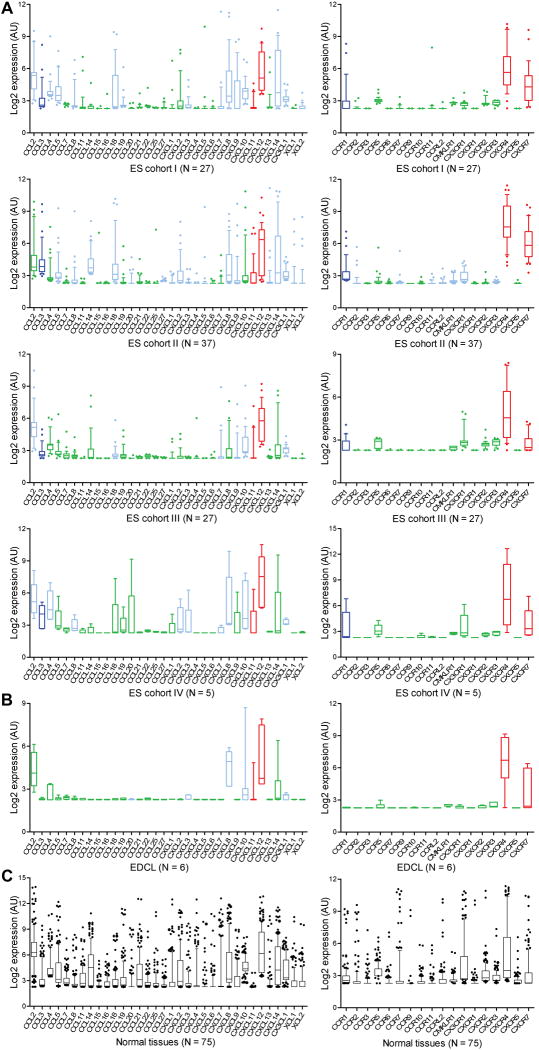
Gene expression significance is usually inferred from gene expression profiles by comparing the ranking of gene probesets to an arbitrarily chosen gene expression cutoff. The subjectiveness of this approach has resulted in cutoff values varying widely from one study to another, which in addition, do not define biological significance of gene expression. To rationally assess biological significance as determined by gene expression profiling, we first analyzed gene expression in NT. Tissues known to express biologically relevant levels of chemokines or chemokine receptors (9, 21) displayed expression values above the median expression in NT (Supplementary Tables 3, 4). We, therefore, selected the median expression in NT as a cutoff for biological gene expression significance. We next reasoned that expression of genes that mark a specific fraction of a cohort must be restricted to that fraction. To uncover chemokines and chemokine receptors that may serve as markers of ES metastases, therefore, we searched for those that are significantly expressed in a quarter of ES, approximately the fraction of patients that presents with metastases at diagnosis (31). As shown in Figure 2A-B (left panels), several chemokines were expressed in the top ES/EDCL quartile. The number and nature of significantly expressed chemokines varied amongst the four cohorts, revealing both convergent and divergent chemokine profiles, underscoring again intercohort variability. Of note, CXCL11 and CXCL12 were expressed in the top quartile of all cohorts and EDCL, indicating potential biological relevance to ES metastasis. An analogous analysis of chemokine receptors revealed CXCR4 and CXCR7 expression to be significant in ES/EDCL (Fig. 2A-B, right panels). Strikingly, these genes encode receptors that mediate the effects of CXCL11 and CXCL12 uncovered in the chemokine screen. Thus, two independent Ican tests uncovered three receptor/ligand pairs (CXCR4/CXCL12, CXCR7/CXCL11 and CXCR7/CXCL12) highlighting Ican analytical power. Significant expression of CXCL11 and CXCL12 in both ESFT and EDCL denotes possible autocrine and/or paracrine mechanisms, though in tumors, expression of these chemokines may also stem from tumor stroma leading to strictly paracrine activation of CXCR4 or CXCR7. A fourth receptor/ligand pair consisting of CCR1/CCL3 was also found significant in ESFT but not in EDCL and was not further pursued in this study.
Fig. 2. Ican uncovers chemokines and chemokine receptors significantly expressed in ES subsets.
Expression levels of chemokines (left panels) and chemokine receptors (right panels) in ES cohorts (A) are shown. Genes whose expression in the top quartile of a given cohort surpassed the median expression in normal tissues (NT; panel C) were deemed significant and depicted in pale blue. Genes consistently expressed in all cohorts are shown in dark blue, and those that are also significantly expressed in ECDL (B) are labeled in red. Expression of all genes tested could readily be detected in cognate NT (C; see also Supplementary Tables 3, 4).
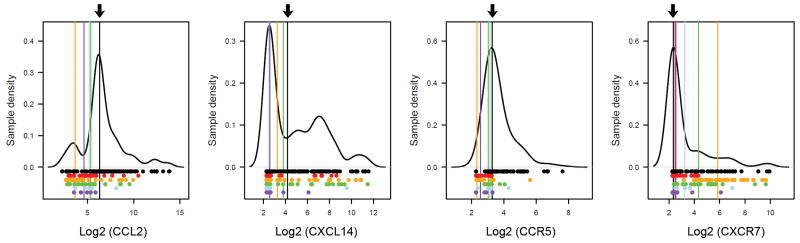
Next, to uncover chemokine ligands and receptors that may correlate to prognosis, we considered the fact that about half of all ES patients succumb to disease within five years of diagnosis (31). We thus used Ican to search for genes that associate with half of ES and which may serve as indicators of patient survival. As in the top-quartile Ican tests, this analysis revealed considerable intercohort expression variability for many genes (e.g. Fig. 3). Following Ican, only CXCR7 expression distinctly associated with half of samples in ES cohorts and EDCL. CXCR4 and CXCR7, therefore, show an association with distinct ES/EDCL subsets, and were selected for further investigation.
Fig. 3. Gene expression significance associated with half of ES.
Genes whose median expression in ES/EDCL exceeded the median expression in NT (arrows) were deemed significant. As an example, gene expression distributions of CCL2, CXCL14, CCR5 and CXCR7 are shown. The plots depict the density estimation (function “density” in R) of expression in NT. Dots represent single sample expression values in NT (black), ES cohorts (red, orange, green or light blue), or EDCL (purple). Vertical lines indicate median expression values. High intercohort variability is evident from the large spread of medians across the test cohorts.
ES-specific CXCR4 and CXCR7 expression in EDCL is EWS-FLI1-dependent
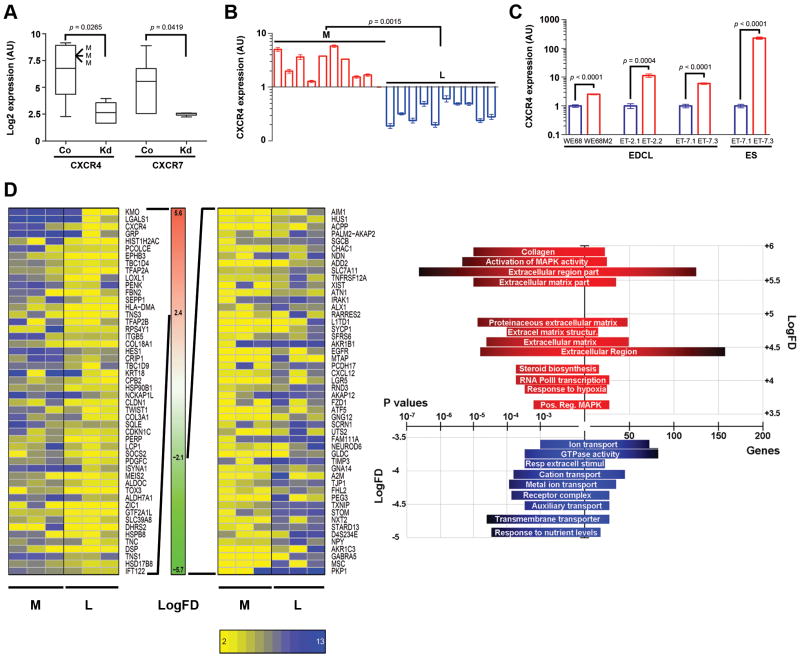
To test for ES relevance, we assayed CXCR4 and CXCR7 dependence on EWS-FLI1, an ETS (E26 Transformation-Specific) domain-containing fusion oncogene. EWS-FLI1 results from t(11;22)(q24;q12) chromosomal translocations harbored by the vast majority of ES, and is a transcription factor that imparts ES-characteristic gene expression programs causative of Ewing's sarcoma pathogenesis. We carried out shRNA-mediated EWS-FLI1 knockdown experiments in six EDCL and found expression of both CXCR4 and CXCR7 to be highly dependent on EWS-FLI1, showing that CXCR4 and CXCR7 expression is integral to EWS-FLI1-imposed, ES-specific transcriptome (Fig. 4A). No other chemokine receptor showed EWS-FLI1-dependent expression (Supplementary Fig. 2).
Fig. 4. CXCR4 expression correlates to EDCL metastatic origin.
A, EWS-FLI1 knockdown decreased CXCR4 and CXCR7 expression in six EDCL (three L-EDCL and three M-EDCL). CXCR4 was highly expressed only in M-EDCL (M). B, qRT-PCR analysis of twenty EDCL validated the preferential high-level CXCR4 expression in M-EDCL. C, qRT-PCR analysis of CXCR4 in cell lines and tumors derived from single patients further confirmed the CXCR4-metastasis association; color coding: localized in blue, metastases in red. D, Gene expression enrichment analysis yielded genes (top and bottom 50 are shown; left panel), and gene-sets (top GO-groups are shown; right panel) enriched in either M-EDCL or L-EDCL. p values and number of genes/gene-sets are shown on split x-axis. LogFD = x̄{Log2(M-EDCL) - Log2(L-EDCL)}. Co: control; Kd: knockdown.
CXCR4 expression levels are associated with metastatic phenotype in ES
As indicated in Fig. 4A, CXCR4 expression was higher in cell lines established from metastases (M-EDCL) than those derived from localized tumors (L-EDCL), suggesting a possible association with ES metastases. We extended the analysis to 20 EDCL and confirmed the higher expression of CXCR4 in M-EDCL (Fig. 4B). Because direct comparison of cell lines can be hampered by genetic background differences inherent to cells derived from different patients, we also assessed CXCR4 expression in three L-EDCL/M-EDCL pairs, each derived from a single patient (Fig. 4C). In these isogenic EDCL, CXCR4 was again expressed at higher levels in M-EDCL. For one L-EDCL/M-EDCL pair for which corresponding ES samples were available, analysis of CXCR4 expression in the original non-cultured tumors confirmed these findings, indicating that CXCR4 correlation to cells derived from ES metastases was neither due to an inter-patient genetic bias nor to a cell culture artifact (Fig. 4C). Finally, a comparison of M-EDCL and L-EDCL transcriptomes showed that CXCR4 ranked as one of the most preferentially expressed genes in M-EDCL, further validating CXCR4 expression association to ESFT metastases (Fig. 4D, left panel). A comparison of gene expression profiles in M-EDCL versus L-EDCL showed that genes (Fig. 4D, left panel) and gene groups (Fig. 4D, right panel) that can support metastasis were over-represented in M-EDCL, suggesting a functional relevance to ESFT metastasis (see also Supplementary information and Supplementary Tables 1, 2).
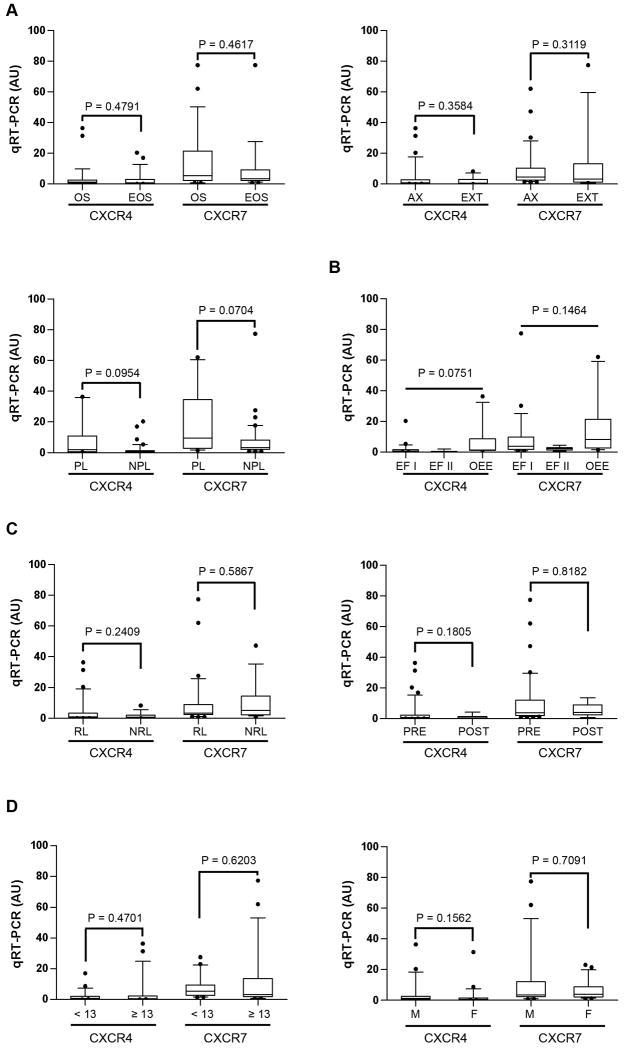
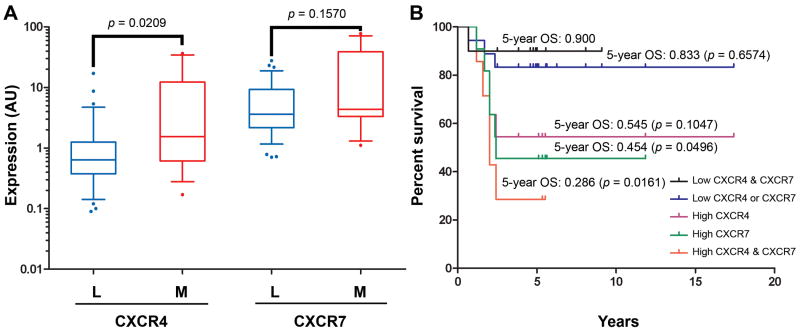
To further test the significance of CXCR4 and CXCR7 association to ES, we researched the correlation of CXCR4 or CXCR7 to several clinicopathological variables traditionally used in ES prognosis and management (32-35). As shown in Figure 5, neither gene correlated to any of the parameters tested (See Supplementary Table 5 for additional details). We next asked whether CXCR4 associated to ES metastases in an independent US cohort of 49 patients. Again, high CXCR4 (but not CXCR7) expression correlated with ES metastases (Mann-Whitney two-tailed test, p = 0.0209), validating the Ican ES/EDCL findings (Fig. 6A).
Fig. 5. CXCR4 and CXCR7 expression does not correlate to several molecular and clinical variables.
Several clinicopathological parameters were surveyed in connection with CXCR4 or CXCR7 expression. CXCR4 and CXCR7 did not correlate to: A, tumor location (OS: osseous versus EOS: extra-osseous (EOS) sites; axial (AX) or extremity (EXT) locations, and pelvis (PL) versus non-pelvic (NPL) locations); B, EWS-ETS fusions: EWS-FLI1 type I (EF I), type II (EF II), or other EWS-ETS (OEE) fusions; C, treatment status and outcome: relapsed (RL) versus non-relapsed (NRL) tumors; pre- (PRE) versus post-chemotherapy (POST); D, biographical data such as age (children less than 13-year old versus patients 13-year old or older, or gender (males: M; females: F). Pairwise univariate analyses (nonparametric two-tailed unpaired Mann-Whitney U-tests) were carried out for all analyses to assess significance (p < 0.05), except for data in (B), wherein Bonferroni's multiple comparison testing (significance if p < 0.01) and One-way Anova (p values shown) were used.
Fig. 6. CXCR4 and CXCR7 associate with metastatic and/or poor prognosis ES.
A, qRT-PCR analysis of CXCR4 and CXCR7 expression in localized (L; 36 samples) and metastatic (M; 13 samples) ES confirms the significant association of high CXCR4 (but not CXCR7) expression to ES metastases. B, Kaplan-Meier analysis of overall survival (OS) shows that high expression of either CXCR4 or CXCR7 is detrimental to patient survival. Survival of patients whose tumors highly co-expressed both chemokine receptors was most drastically curtailed. Expression was considered high if above median.
High CXCR4 and CXCR7 expression demarcates poor prognosis in ES patients
Finally, we researched the correlation of CXCR4 and CXCR7 expression to patient survival. ES patients whose tumors expressed low levels of both CXCR4 and CXCR7 had the highest 5-year overall survival (OS) rates (90%; Figure 6B). These rates were diminished for patients with either high CXCR4 (OS = 54.5%) or CXCR7 (OS = 45.4%) tumor content, and were drastically reduced for patients with high-expressing CXCR4 and CXCR7 tumors (OS = 28.6%; Log-rank test, p = 0.0161; hazard ratio 8.0). Analysis of event-free survival (EFS) rates suggested a trend wherein high CXCR4 and CXCR7 expression might negatively impacted EFS (median EFS in high CXCR4/CXCR7 = 1.5 years versus 5.4 years for patients with low CXCR4/CXCR7; Hazard ratio = 0.34; 95% CI 0.11-1.0), but this correlation did not reach statistical significance (p = 0.0586), presumably owing to the small sample size analyzed. Except for metastasis addressed above (Fig. 6A), neither CXCR4 nor CXCR7 expression levels correlated with other variables that may influence disease outcome (pairwise univariate analyses; Fig. 5A-D). CXCR4 and CXCR7 thus appear to serve as prognostic factors of metastasis and overall survival, respectively.
Discussion
Cancer is the leading cause of death by disease of children aged 1-14 years in the developed world (4). This fact underscores the urgency of uncovering markers that can help in the prognosis of tumor progression and outcome. These markers would aid in patient treatment stratification and help curtail the high mortality that afflicts subsets of cancer patients. Childhood cancers regroup a very large number of different low-incidence cancers, thus preventing large cohort studies and compromising marker discovery (5-7). Whilst several multicentre cancer consortia have formed to consolidate cohorts (5, 6), these cannot make their resources available to most non-consortium proposals without compromising critical deliverables and deadlines. These and other considerations render investigations aimed at uncovering molecular prognostic factors in childhood cancers impractical for most laboratories or even single institutions (6). We present here an analytical methodology, Ican, which enables to uncover genes that are consistently associated with patient subsets with common features from small-size cohorts.
Using Ican, we found the CCL3/CCR1, CXCL12/CXCR4, CXCL11/CXCR7 and CXCL12/CXCR7 chemokine/chemokine receptor pairs to be significantly associated with ES subsets. Relevance of these chemokine receptors to cancer is well established: CCR1 and CXCR7 contribute to growth or metastasis of breast, liver, lung, oral, and prostate cancers (9, 36, 37), whereas CXCR4 supports tumor mitogenesis, chemoresistance, or metastasis of more than twenty neoplasms of different etiologies and tissues of origin (9, 10). These considerations suggest that these chemokines and chemokine receptors may also be involved in ES. Activation of the CXCL12/CXCR4 axis, a prototypical metastasis-mediating chemokine/chemokine receptor pair, can lead to metastatic processes involving at least three steps. First, cells within the tumor (tumor cells, but also stroma cells such as tumor-associated fibroblasts and endothelial cells of the tumor vasculature) can secrete CXCL12. This leads to increased CXCR4-dependent motility, and for some CXCR4-expressing tumor cells, egress into tumor vasculature. This first step is of paracrine and/or autocrine nature. Next, CXCR4-expressing tumor cells migrate toward a CXCL12 chemotactic gradient in the blood stream to target organs producing high levels of CXCL12 (e.g. bone marrow and lungs). This second step utilizes endocrine mechanisms. Finally, CXCR4-expressing tumor cells invade CXCL12 expressing organs. The very high CXCL12 levels in these organs lead to CXCR4 internalization, decreased cell motility, and attachment of tumor cells to the site of metastasis. Paracrine mechanisms prevail at this step. These three steps are detailed along with some of the supporting published evidence in the excellent review by Burger and Kipps (10). In the subsequent analyses, we chose not to further investigate the CCL3/CCR1 pair as it was not significantly expressed in EDCL. Whilst this may be due to a lack of expression of both genes following cell culture, this may also signify that CCL3/CCR1 expression in the tumor samples stemmed from non-tumoral sources such as tumor stroma, tumor-infiltrating lymphoid cells, or tumor-associated vasculature endothelial cells, as previously reported (38).
Both CXCR4 and CXCR7 expression was highly dependent on EWS-FLI1 in EDCL. CXCR4 contains several ETS binding sites within close proximity to its transcription start site that may allow ETS-specific transcriptional regulation in normal tissues (39). CXCR4 promoter region also contains hypoxia-inducible factor 1 (HIF1α) binding sites (39). These mediate CXCR4 transcriptional activation in response to hypoxia, one of the earliest initiating events of metastasis (10). Similarly to CXCR4, we find that CXCR7 promoter proximal region contains several ETS binding sites (not shown). It is thus plausible that EWS-FLI1 activates CXCR4 and CXCR7 transcription via binding to these ETS binding sites, leading to ES-specific expression. Furthermore, we recently uncovered a two-way crosstalk between EWS-FLI1 and HIF1α that modulates ES gene signature during hypoxia (40). Here we find CXCR4 to be preferentially expressed in ES metastases as well as in cell lines derived from ES metastases. The association of CXCR4 to ES metastases, the presence of multiple ETS and HIF1α transcription factor binding sites at CXCR4 promoter, and the EWS-FLI1-HIF1α crosstalk in ES suggest that CXCR4 preferential expression in ES metastases might be the result of transcriptional activation by EWS-FLI1 and possibly HIF1α. Further research into these and other possible molecular mechanisms of CXCR4 activation in ES are warranted as they may lead to means to targeting and disrupting the CXCR4/CXCL12 axis and possibly ES metastasis.
CXCR4 association to ES metastases is also consistent with the previous finding that CXCR4 supports EDCL cellular migration in response to CXCL12 (41), an in vitro characteristic indicative of in vivo metastatic potential (42). Moreover, the bone/bone marrow and lungs are the richest sources of CXCL12 (Supplementary Table 3) and are also organs of predilection for ES metastasis (43). The strong correlation between organs that produce the most CXCL12 and those to which ES metastasize further supports the CXCR4-ES metastases association. CXCR4 correlation to ES metastases may be of clinical relevance as metastasis represents the single most significant adverse clinicopathological variable in ES patients (31). The dismal survival rates of ES patients are in part due to a lack of metastasis and prognostic indicators that would help in risk assessment and patient stratification. Our findings suggest that CXCR4 and CXCR7 may serve that purpose. Furthermore, CXCR4 and CXCR7 antagonists that can effectively reduce growth or spread of CXCR4- and CXCR7-expressing tumors in experimental models and patients have been reported (9, 12). These drugs may also be effective in treating CXCR4/CXCR7-expressing ES and merit investigation.
In the present study, we used Ican of public expression datasets to uncover gene-ES associations. Ican may be applied to any disease for which only small size cohorts are available, currently the case of most diseases. In the case of childhood cancers, several are already represented by more than 100 arrays each in GEO database alone, making Ican feasible. This approach is poised to gain further momentum as many journals now request the mandatory submission of gene expression profiles to public repositories. These policies, along with the growing number of gene expression profiling investigations, we predict, will swiftly drive upward the number of publicly available low-incidence disease expression datasets and consequently, of Ican applications.
Translational relevance.
Childhood cancers comprise a large number of low-incidence histogenetically and molecularly distinct neoplasms, allowing only for small-size cohort trials. These, however, lack the necessary statistical power for population-based genomic studies, thus hampering the discovery of markers. Using Ican, we uncovered two chemokine receptors that can serve as metastasis or prognosis indicators in Ewing's sarcoma, a highly metastatic childhood cancer. These markers may help in Ewing's sarcoma metastasis diagnosis and patient treatment stratification. Additionally, both chemokine receptors are drug targets suggesting possible novel treatment options for poor prognosis Ewing's sarcoma. Our findings in Ewing's sarcoma serve as a proof-of-principle for Ican translational applications in childhood and other low-incidence cancers.
Supplementary Material
Acknowledgments
We thank CHLA, COG, and Peter Ambros for tumor samples, Ynnez Gwye and Betty Schaub for technical assistance, Ulrike Pötschger for help with statistics, and Barbara Bennani-Baiti for suggestions throughout this work and critical reading of the manuscript.
Financial support: This work was supported by funds from the NIH SPECS grant 1U01CA114757-04 to E.R.L., the European Community FP6-project EET-pipeline grant 037260 to H.K., and the National Austrian Bank Medical Sciences Fund Grant OeNB-12765 to I.M.B-B.
Footnotes
References
- 1.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature medicine. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 3.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 5.Brown RC, Dwyer T, Kasten C, et al. Cohort profile: the International Childhood Cancer Cohort Consortium (I4C) Int J Epidemiol. 2007;36:724–30. doi: 10.1093/ije/dyl299. [DOI] [PubMed] [Google Scholar]
- 6.Carter A, Landier W, Schad A, et al. Successful coordination and execution of nontherapeutic studies in a cooperative group setting: lessons learned from Children's Oncology Group studies. Cancer Epidemiol Biomarkers Prev. 2008;17:1665–73. doi: 10.1158/1055-9965.EPI-07-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steliarova-Foucher E, Stiller C, Lacour B, Kaatsch P. International Classification of Childhood Cancer, third edition. Cancer. 2005;103:1457–67. doi: 10.1002/cncr.20910. [DOI] [PubMed] [Google Scholar]
- 8.Ushigome S, Machinami R, Sorensen PH, editors. Ewing sarcoma/primitive neuroectodermal tumour (PNET) Lyon: IARC Press; 2002. [Google Scholar]
- 9.Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–44. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 10.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–7. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Lee VC, Chevalier E, Hwang ST. Chemokine receptors as targets for cancer therapy. Curr Pharm Des. 2009;15:742–57. doi: 10.2174/138161209787582165. [DOI] [PubMed] [Google Scholar]
- 12.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23:43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- 13.Miao Z, Luker KE, Summers BC, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15735–40. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ban J, Bennani-Baiti IM, Kauer M, et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's sarcoma. Cancer research. 2008;68:7100–9. doi: 10.1158/0008-5472.CAN-07-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Valen F, Masters J. Human Cell Culture. Great Britain: Kluwer Academic Publishers; 1999. Ewing's Sarcoma Family of Tumors; pp. 55–85. [Google Scholar]
- 16.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkinson H, Kapushesky M, Kolesnikov N, et al. ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–72. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadota K, Nakai Y, Shimizu K. Ranking differentially expressed genes from Affymetrix gene expression data: methods with reproducibility, sensitivity, and specificity. Algorithms Mol Biol. 2009;4:7. doi: 10.1186/1748-7188-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 20.Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat Methods. 2007;4:911–3. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–21. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 22.Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauer M, Ban J, Kofler R, et al. A molecular function map of Ewing's sarcoma. PLoS One. 2009;4:e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitley E, Ball J. Statistics review 4: sample size calculations. Crit Care. 2002;6:335–41. doi: 10.1186/cc1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer KL, Eisenacher M, Braun Y, et al. Microarray analysis of Ewing's sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur J Cancer. 2008;44:699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Scotlandi K, Remondini D, Castellani G, et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J Clin Oncol. 2009;27:2209–16. doi: 10.1200/JCO.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- 29.Henderson SR, Guiliano D, Presneau N, et al. A molecular map of mesenchymal tumors. Genome Biol. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer cell. 2007;11:421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 31.Subbiah V, Anderson P, Lazar AJ, Burdett E, Raymond K, Ludwig JA. Ewing's sarcoma: standard and experimental treatment options. Curr Treat Options Oncol. 2009;10:126–40. doi: 10.1007/s11864-009-0104-6. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Galindo C, Liu T, Krasin MJ, et al. Analysis of prognostic factors in ewing sarcoma family of tumors: review of St. Jude Children's Research Hospital studies. Cancer. 2007;110:375–84. doi: 10.1002/cncr.22821. [DOI] [PubMed] [Google Scholar]
- 33.Leavey PJ, Mascarenhas L, Marina N, et al. Prognostic factors for patients with Ewing sarcoma (EWS) at first recurrence following multi-modality therapy: A report from the Children's Oncology Group. Pediatr Blood Cancer. 2008;51:334–8. doi: 10.1002/pbc.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacci G, Longhi A, Ferrari S, Mercuri M, Versari M, Bertoni F. Prognostic factors in non-metastatic Ewing's sarcoma tumor of bone: an analysis of 579 patients treated at a single institution with adjuvant or neoadjuvant chemotherapy between 1972 and 1998. Acta Oncol. 2006;45:469–75. doi: 10.1080/02841860500519760. [DOI] [PubMed] [Google Scholar]
- 35.Ferrari S, Bertoni F, Palmerini E, et al. Predictive factors of histologic response to primary chemotherapy in patients with Ewing sarcoma. J Pediatr Hematol Oncol. 2007;29:364–8. doi: 10.1097/MPH.0b013e3180640d08. [DOI] [PubMed] [Google Scholar]
- 36.Silva TA, Ribeiro FL, Oliveira-Neto HH, et al. Dual role of CCL3/CCR1 in oral squamous cell carcinoma: implications in tumor metastasis and local host defense. Oncol Rep. 2007;18:1107–13. [PubMed] [Google Scholar]
- 37.Yang X, Lu P, Fujii C, et al. Essential contribution of a chemokine, CCL3, and its receptor, CCR1, to hepatocellular carcinoma progression. International journal of cancer. 2006;118:1869–76. doi: 10.1002/ijc.21596. [DOI] [PubMed] [Google Scholar]
- 38.Lorusso G, Ruegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol. 2008;130:1091–103. doi: 10.1007/s00418-008-0530-8. [DOI] [PubMed] [Google Scholar]
- 39.Maroni P, Bendinelli P, Matteucci E, Desiderio MA. HGF induces CXCR4 and CXCL12-mediated tumor invasion through Ets1 and NF-kappaB. Carcinogenesis. 2007;28:267–79. doi: 10.1093/carcin/bgl129. [DOI] [PubMed] [Google Scholar]
- 40.Aryee DNT, Niedan S, Kauer M, et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances malignant properties of Ewing's sarcoma cells in-vitro. Cancer research. 2010;70:4015–23. doi: 10.1158/0008-5472.CAN-09-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chansky HA, Barahmand-Pour F, Mei Q, et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing's sarcoma cells in vitro. J Orthop Res. 2004;22:910–7. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–57. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 43.Paulussen M, Ahrens S, Burdach S, et al. Primary metastatic (stage IV) Ewing tumor: survival analysis of 171 patients from the EICESS studies. European Intergroup Cooperative Ewing Sarcoma Studies. Ann Oncol. 1998;9:275–81. doi: 10.1023/a:1008208511815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.