Abstract
The EB-eye is a fast and efficient search engine that provides easy and uniform access to the biological data resources hosted at the EMBL-EBI. Currently, users can access information from more than 62 distinct datasets covering some 400 million entries. The data resources represented in the EB-eye include: nucleotide and protein sequences at both the genomic and proteomic levels, structures ranging from chemicals to macro-molecular complexes, gene-expression experiments, binary level molecular interactions as well as reaction maps and pathway models, functional classifications, biological ontologies, and comprehensive literature libraries covering the biomedical sciences and related intellectual property. The EB-eye can be accessed over the web or programmatically using a SOAP Web Services interface. This allows its search and retrieval capabilities to be exploited in workflows and analytical pipe-lines. The EB-eye is a novel alternative to existing biological search and retrieval engines. In this article we describe in detail how to exploit its powerful capabilities.
Keywords: text search, biological databases, integration, interoperability, web services, Apache Lucene
INTRODUCTION
Searching for accurate and functionally related biological concepts through stacks of journals and articles is time consuming. Furthermore, establishing the relationships between genes, transcripts, proteins, expression, and molecular structures using the web is often an error-prone process. Scientists need to use different web resources, which have different search engines that are syntactically and semantically incompatible: results are returned in heterogeneous formats, making deriving a coherent view of the biological meaning of these data cumbersome.
The availability of new tools and libraries for the development of search engines and web portals, allows us to build a system that enables interoperability between distinct data resources and channel these through a single hub. Scientists can now quickly search and identify biological entities, relationships and simply navigate to expert primary resources.
We present here a high-performance, full-feature text search engine that finds and displays biological entities and their associations (i.e. the relationship between genomic sequences, transcripts, proteins and their function, molecular structures, gene expression profiles, protein–protein interactions, pathways and published scientific and patent literature), in much the same way as scientists do searches in a library. This web-based search engine is called the ‘EB-eye’ and is built on the free Open Source Apache Lucene JavaTM library [1].
What is the EB-eye?
EB-eye is a catalogue of biological entities, similar to a library catalogue that describes publications, and contains enough information to allow for efficient searching. Unlike indexing warehouses such as Entrez [2], SRS [3] and MRS [4], which provide complete access to the data and allow searching over fields with specialist value and which are difficult to search without prior knowledge of their contents, EB-eye focuses on indexing selected textual content, which are the most meaningful while searching biological data (e.g. database names and database identifiers, gene names and synonyms, protein names, chemistry identifiers, reaction equations, authors, titles, various types of descriptions and importantly, cross-references that link entries between distinct databases). This excludes data which requires specialist searching, such as sequences, structure coordinates, expression profiles and ambiguous data, such as numeric counts, for which there exists search tools associated with the primary resources. EB-eye improves the user’s experience by providing a search engine that presents consistent result pages and navigation across all the data resources maintained by EMBL-EBI.
EB-eye is not limited to specific data formats. As well as indexing database dumps, such as flat-files from EMBL-Bank [5] and UniProt [6], database specific XML, web content (e.g. HTML and XHTML), EB-eye uses a custom XML dump format, which has been designed for data resources without an export format. These dumps focus on the essential content required to describe a biological concept in the database, and are produced by the data providers.
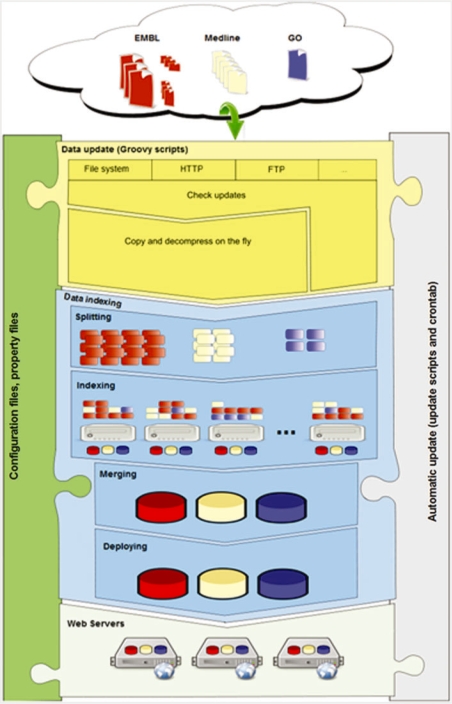
The EB-eye is composed of a set of modules, each designed to carry out a distinct task (Figure 1). In the following we will describe the web interface, the back-end data-management and indexing system, and finally, the SOAP Web Services interface that is used to integrate and/or embed its functionality into analytical pipe-lines, external applications and web portals.
Figure 1:
Modules available in the overall architecture for the search engine back-end.
THE EB-Eye’s WEB INTERFACE
Like internet search engines, the EB-eye has adopted established design principles to provide a simple, coherent and intuitive interface for querying and presentation of results. Of particular importance is achieving good performance when obtaining results for queries against large biological datasets while maintaining the number of operations that separate the user from results within the constraints described by the ‘Three-Click-Rule’ [7].
Global search input box
At the top of every web page of the EMBL-EBI portal (http://www.ebi.ac.uk) there is a text search box. This is the main entry point to the EB-eye search engine. Search terms such as entry identifiers, gene names, article titles, biological or chemical nomenclature terms, or a set of keywords can be used. By default all available data resources are searched and the results are initially presented in the summary overview.
Summary overview
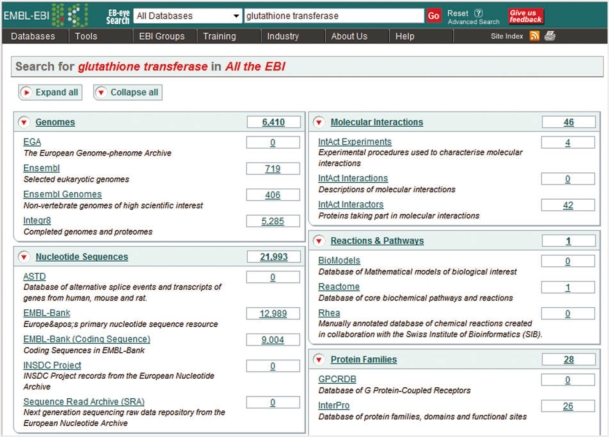
Data within the EB-eye are organised into categories representing biological knowledge domains. Each domain is composed of a hierarchy of sub-domains that focus on related data. For example, the ‘Small molecules’ knowledge domain comprises ChEBI [8]: a dictionary of small chemical compounds of biological interest; Ligands [9]: a dictionary of small chemical components and RESID [10]: a comprehensive collection of annotations and structures for residue modifications. Similarly, the ‘Nucleotide Sequences’ domain, contains the Alternative Splicing and Transcript Diversity database (ASTD) [11]; EMBL Nucleotide Sequence Archive (EMBL-Bank) and the EMBL Coding Sequences database. EMBL-Bank is further broken down into specific sub-domains related to the internal structure of the database (e.g. EMBL-Bank (Release) and EMBL-Bank (Updates)). Following the strategy of initially performing a broad search and then narrowing down the scope if necessary, the results are initially presented in a summary page showing the number of hits in each domain (Figure 2). Clicking the domain name or the number of hits takes the user to a domain-specific results page containing a list of entries found by the query.
Figure 2:
Summary overview page for the results of a search (expanded mode).
Domain-specific results page
For a domain which does not contain sub-domains, an overview of each entry found in the domain is presented in pages of 15 entries. For a domain containing sub-domains the most relevant three entries found in each sub-domain are shown with a ‘more’ link to navigate to all the results found in the sub-domain.
An overview is shown for each entry. This typically contains the primary identifier of the entry, which is displayed first and is hyperlinked to the primary resource, and a descriptive title. Additional annotation may be displayed, which commonly includes secondary database identifiers and classification, dates, authors, alternative names, etc. For example, for an entry in UniProtKB, the identifier, accession numbers, gene names with synonyms and the descriptions are displayed. In contrast, for a literature database (e.g. MEDLINE [1]) a citation style summary is presented for each entry.
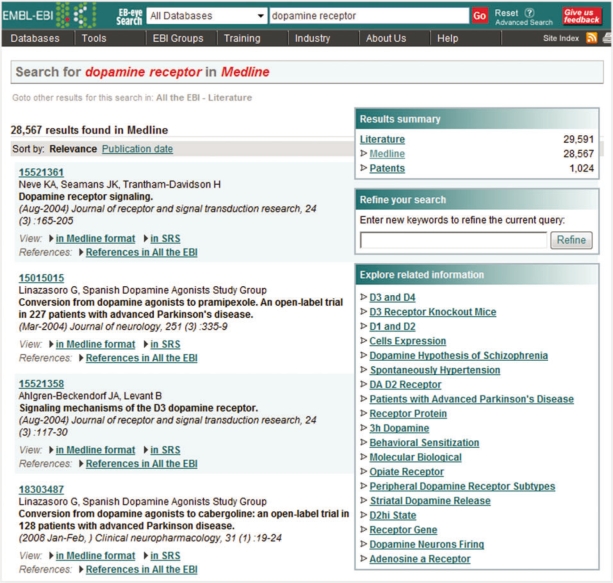
On the right of the domain specific results page there are three boxes: ‘Results summary’ containing a count of the results found within the domain hierarchy; ‘Refine your search’, allowing query refinement through adding additional terms to the query; and ‘Explore related information’ (Figure 3) which displays query refinement suggestions that are dynamically generated from the query results using techniques provided by Carrot2 [12], a search results clustering engine. For example, querying MEDLINE for ‘dopamine receptor’ yields a list of related terms including: ‘Hypothesis of Schizophrenia’, ‘Patients with Parkinson's; Disease’, ‘Depression Component’ and ‘Dopamine Receptors and Hypertension’.
Figure 3:
The ‘Explore related information’ box in the domain specific results page displays the terms related to a given search query using Carrot2 clustering techniques.
Views
An entry may have ‘Views’ that provide access to other formats and portals. For example, a nucleotide entry in EMBL-Bank has views which show the entry in EMBL flat-file format, in SRS and the entry’s history using the Sequence Version Archive. Likewise, an entry in PDBe [13] can be viewed in PDBSum [14], PDB format and in SRS.
Cross-references
The data indexed by the EB-eye contains cross-references. These are displayed as hyperlinks in the ‘References’ section. These allow navigation between related entries, helping the user build a coherent overview of the biological entities described. For example, from protein sequences (in UniProtKB) discovering associated coding genes (in EMBL-CDS [5]), protein families (in InterPro [15]), literature (in MEDLINE), organism taxonomy (in the NCBI Taxonomy), etc.
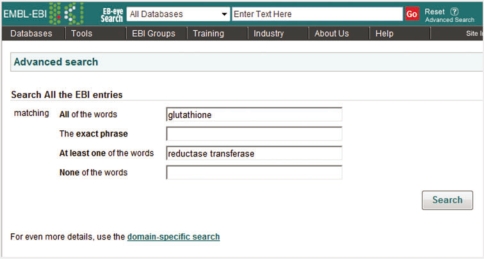
The advanced search
To help the user combine terms for querying across all the data resources, the advanced search has four text input fields, which address fields for querying ‘All the words’, ‘The exact phrase’, ‘At least one of the words’ and ‘None of the words’. It provides easy access to the boolean query operators (i.e. AND, OR and NOT) needed to build complex queries (Figure 4). For example: searching for ‘insulin’ yields overlapping results that contain both ‘insulin’ and ‘insulin-like’. To overcome this ambiguity the user can type ‘insulin’ into the ‘All words matching’ box and ‘insulin-like’ into the ‘None of the words’ box to perform a search which excludes entries containing the term ‘insulin-like’ from the results.
Figure 4:
The Advanced Search page for a query allows for the use of boolean operators to write complex queries.
For a ‘domain-specific search’ a tree of domains and sub-domains is shown, from which a single domain can be selected. If a data source (i.e. a leaf of the tree), rather than a collection of data sources is selected, the search can be further constrained to specific fields and cross-references. Multiple fields and/or cross-references can be selected, and the query form will be updated to include specific options for these fields and cross-references.
The EB-eye’s query syntax
The EB-eye uses the Apache Lucene query syntax [16], which is similar to that used by Google and other major internet search engines. Table 1 describes the major syntactical elements supported by EB-eye. A more detailed description can be found in the EB-eye help page at: http://www.ebi.ac.uk/inc/help/search_help.html.
Table 1:
Main syntactical elements of the Lucene library used in the EB-eye.
| EB-eye Lucene syntax token | Meaning | Usage | Example |
|---|---|---|---|
| AND ( + ) | In addition to | term1 AND term2 | glutathione AND transferase |
| term1 + term2 | glutathione + transferase | ||
| OR | Equivalence | term1 OR term2 | human OR ‘homo sapiens’ |
| NOT (-) | Exclusion | term1 NOT term2 | coding NOT fragment |
| term1 –term2 | coding -fragment | ||
| * | Wild card | partialterm1* | gluta* = (glutathione, glutamate, glutamic… ) |
| ? | Replacement | Str?ng | str?ng = (string, strong) |
| “” | Quoted text | “complete sentence” | “molecular evolution” |
| () | Grouping | (term1 AND term2) OR term3 | (reducatase OR transferase) AND glutathione |
| Field: | Field searching | fieldname:term | description:dopamine |
Unlike the aforementioned search engines, multiple search terms are combined with a ‘AND’, which is analogous to the behaviour in Entrez, SRS and MRS. Thus a query containing ‘glutathione transferase’ is treated as ‘glutathione AND transferase’ and will find only those entries containing both terms. The default sort order of results is based on the proximity of the terms in the entries, thus entries where the phrase ‘glutathione transferase’ occurs, will appear first in the list of results.
MRS, Entrez and SRS provide similar capabilities in their query languages (see some examples in Table 2), however the results obtained differ. These differences are related to the data being searched and the nature of the query systems. The syntactical differences between these systems highlight an issue, characteristic to all biological search engines. Although the syntax of each search engine is internally consistent, the names of the fields indexed in these systems are different. EB-eye implements aliasing for fields to common names with equivalent meaning. For example, common field names such as ‘id’, ‘accession’, ‘name’ and ‘description’, which are used across many of the databases in the system to describe fields that have semantically equivalent meaning.
Table 2:
Examples of equivalent ways of expressing search queries in EB-eye, MRS, SRS and Entrez.
| Search Engine | Query |
|---|---|
| EB-eye | (reductase OR transferase) AND glutathione |
| MRS | (reductase OR transferase) AND glutathione |
| SRS | [UNIPROT-all:(reductase|transferase)&glutathione] |
| Entrez | (reductase OR transferase) AND glutathione |
| EB-eye | gene_primary_name:(gst) |
| MRS | gn:gst |
| SRS | [UNIPROT-genename:gst] |
| Entrez | gst[Gene Name] |
| EB-eye | gene_primary_name:(gst*) |
| MRS | gn:gst* |
| SRS | [UNIPROT-genename:gst*] |
| Entrez | gst*[Gene Name] |
UPDATING AND INDEXING DATABASES
Currently, the EB-eye provides access to more than 200 million entries from 56 data sources (http://www.ebi.ac.uk/ebisearch/statistics.ebi). Keeping the system up to date requires a system that automatically updates and re-indexes data on a daily basis. This comprises two modules (Figure 1), controlled by a set of configuration files:
(i) When data updates are detected, the data are downloaded and optionally decompressed into an off-line directory. For each data resource, a Groovy [17] script is used to configure this process. This addresses the complexity of fetching data, which are accessible using diverse protocols (e.g. http, ftp, fasp (http://www.asperasoft.com), ssh, rsh or simple file system copy). These scripts also handle any additional post-processing required.
(ii) Generating indices from the new data by parsing the data to extract the relevant information. An initial stage examines the data and determines a strategy for performing the parsing in parallel. The indexing task is then farmed out to a number of machines, each machine creating a partial index. Once all the tasks are completed the partial indexes are merged to form the final index.
To ensure the consistency of the index the number of entries in the data resource is checked against the number of entries recorded by the index. In addition, the cross-references are checked against the know cross-references for the data resource. Any discrepancies in the number of entries or cross-references prevent deployment of the index into the on-line environment, and are logged as errors for investigation. If no errors were encountered the completed indexes are deployed and made available to the public.
EB-eye WEB SERVICES
Scientists require biological data searches in desktop tools and analytical pipe-lines. Many analytical tools in bioinformatics also require the ability to perform searches to obtain the required data for performing and enriching their analysis. EB-eye provides a web service interface to address these requirements. This web service exposes the functionality available in the web interface allowing EB-eye to be integrated into other systems. Detailed documentation including example clients is available from http://www.ebi.ac.uk/Tools/webservices/services/eb-eye, where the reader can find descriptions of the input required for each method as well as of output structures returned.
The web service uses the, widely supported, Simple Object Access Protocol (SOAP) [18] standard, coupled with a Web Services Description Language (WSDL) [19] interface description document. Clients programs can access the service without the need to develop custom code. Web services technologies are platform and programming language neutral, thus EB-eye can be incorporated into existing applications as well as those specifically developed to exploit the EB-eye's; features.
Methods
The methods provided by the web service can be grouped into three broad categories:
Meta-data: information about the EB-eye and the data domains available.
Search and Retrieval: performing searches and retrieving data from the results.
Navigation: using cross-reference information to navigate between related entries and domains.
The following sections provide an overview of the methods in each of these categories.
Meta-data
To build a user interface it is necessary to be able to obtain information about the search system, such as the data resources available, the fields available for each resource, which fields can be searched and which fields can be retrieved. The web service provides a set of methods to access the meta-data describing the search domains:
listDomains(): list of search domains available. These domains correspond to the leaves of the hierarchy of domains.
listFields(): list fields for a domain from which data can be retrieved.
getDomainsReferencedInDomain(): list of EB-eye domains referenced by a domain.
getDomainsReferencedInEntry(): list of EB-eye domains referenced by a specific entry in a domain.
listAdditionalReferenceFields(): list of fields referencing data resources which are not available in EB-eye.
getDomainsHierarchy(): get the complete tree of domains.
listFieldsInformation(): detailed information about the fields for a domain, this describes both searchable and retrievable fields.
Search and Retrieval
Fundamental features of a search engine are performing searches, retrieving summary data for the results and obtaining pointers to the complete data. The following methods cater for different types of search:
getNumberOfResults(): get the number of entries which are found by a query.
getDetailedNumberOfResults(): for a search covering multiple domains get the number of entries found in each of the domains, either as a tree covering the relevant section of the domains hierarchy or a flattened list.
getResultsIds() and getAllResultsIds(): get the identifiers of the entries matching a query.
getResults(): retrieve data from specified fields for the entries matching a query.
getEntry() and getEntries(): retrieve data from specified fields for an entry or a set of entries.
Cross-references navigation
Navigation in the EB-eye allows scientists to explore relationships within and between diverse biological knowledge domains. Methods are provided to navigate the cross-references given a specific entry identifier or a search result as the starting point:
getEntryFieldUrls() and getEntriesFieldUrls(): get the URLs associated with the specified fields. Used to obtain references to the main site for the data resource.
getReferencedEntries(): for a query get the identifiers of entries in a referenced domain.
getReferencedEntriesSet() and getReferencedEntriesFlatSet(): for a query get data from specified fields in the referenced domain.
Using EB-eye Web Services in pipe-lines and workflows
Workflow design tools such as Taverna [20], Triana [21] and KNIME [22] can use the web service WSDL to create the components required to combine the EB-eye with other services in order to create complex workflows. As well as purpose built workflow engines, like the aforementioned, scripting environments such as the UNIX shells (e.g. Bourne shell, C-shell, etc.), or the Microsoft Windows scripting environments (e.g. batch, VBscript, Jscript or PowerShell) can be used with the example clients written in .NET [23], Java [24] or Perl [25] to create similar pipelines.
One example of such a workflow, which could provide a foundation for an annotation process, is using the EB-eye to obtain consolidated identifier mappings from the results of a BLAST [26] search against the UniProtKB. Using the EMBL-EBI's; tools web services [27] a workflow can be constructed which performs a BLAST search using the WSWUBlast (http://www.ebi.ac.uk/Tools/webservices/services/wublast) web service against the UniProtKB database. The BLAST hit identifier list from the sequence search can be used as query terms in an EB-eye search against the UniProt Archive [6] (UniParc). The resulting UniParc entry identifiers are used to retrieve the complete entry using the WSDBfetch web service (http://www.ebi.ac.uk/Tools/webservices/services/dbfetch). Cross-references to RefSeq [28], Ensembl [29], PDB [30] and EMBL-CDS are extracted from these entries to characterise the protein sequence space actually covered by the initial BLAST search. Example implementations of this workflow using Taverna and Bourne shell are available from http://www.ebi.ac.uk/Tools/webservices/.
It is noteworthy, that the services mentioned above as well as more than 1100 other life-sciences relevant web services can be found in the BioCatalogue project portal (http://www.biocatalogue.org) and its content is indexed in the EB-eye.
Integration and embedding
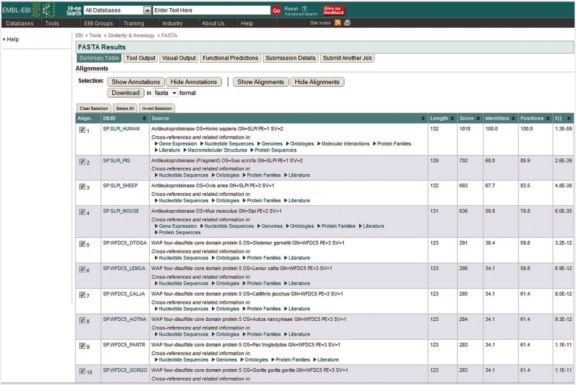
Third-parties can use the EB-eye web services to provide fast full text searching capabilities across their data in their own portal. For example, Ensembl Genomes (http://www.ensemblgenomes.org), which is built on the, Perl-based, Ensembl framework, delegates its text searches to EB-eye and the results obtained are mapped to the entries within the database and presented to the user. As well as the integration of search capabilities the web service can also be used to provide access to the data network, for example in the EBI Sequence Similarity Search Services (http://www.ebi.ac.uk/Tools/sss) the web service is used to obtain details of the domains referenced by each hit in the search result (Figure 5), this provides additional context to the sequence search allowing the scientist to determine which hits provide the most relevant information for the type of search they are performing.
Figure 5:
Results of a Smith-Waterman search against UniProtKB/SwissProt showing the cross-reference information obtained for each hit using the EB-eye web service.
CONCLUSIONS AND FUTURE DIRECTIONS
Finding information about biological entities is a cumbersome and error prone process. Unlike systems such as Entrez, SRS and MRS, which provide both search and data retrieval capabilities, EB-eye focuses solely on the search and cross-reference navigation aspect of the data integration process. By providing access to navigate to the primary data source, where data is up-to-date, well maintained, and displayed in the way expected by the specialists, the EB-eye can integrate a larger range of data sources for an equivalent resource cost. Not to be confused with an integration platform, the EB-eye enables interoperability between resources and allows the user to cross-navigate between heterogeneous knowledge domains in a fast and consistent manner. EB-eye aims to always give the user comprehensive, reproducible and easy to interpret results.
Plans for future work include the capability to search with ranges in numerical fields such as dates, molecular weights and sequence length. In the context of web services, REST-styled interfaces are also on the agenda. Novel types of data, including image metadata and raw experimental data are also being considered for inclusion. Improving the accuracy and integrity of the cross-references network and displaying third party links (i.e. non-EBI resources) in the web interface is high on the list of priorities.
Key points.
The EB-eye is a novel approach for searching biological information. It was designed to index key concepts in the data that are relevant to scientists in no particular scientific knowledge domain (e.g. information about a gene may be found in different data resources, ranging from protein and nucleotide sequences, gene expression databases, 3D structures, literature, etc).
Using the EB-eye is very easy and intuitive. The web interface has been designed to use navigation metaphors most users should be familiar with (e.g. Google).
The EB-eye search engine is a fast and scalable solution for finding core biological entities as well as related knowledge about these.
The EB-eye makes extensive use of the cross-references that exist between heterogeneous data resources to help the user identify biological concepts and maintain uniform navigation within the system.
The EB-eye is an interoperability platform. Integrating the EBI-eye functionality into other web sites, using web services, allows third-parties to use it to build complex pipe-lines, workflows and provide their users with links and annotations from other databases.
FUNDING
European Union (contract number 021902 as part of the FELICS Research Infrastructure; contract number LHSG-CT-2004-12092 as part of the EMBRACE project; and contract number IST-2001-32688 as part of the ORIEL Project), the Wellcome Trust; the European Patent Office; the National Institutes of Health (as part of the UniProt project, grant 1 U01 HG02712-01); and core funding from the European Molecular Biology Laboratory (EMBL).
Biographies
Franck Valentin is a senior software engineer with M.Sc. in Computer Science from the University of Rennes, France. He is a specialist in software architecture design, programming patterns and frameworks.
Silvano Squizzato is a senior software developer with M.Sc. from the University of Padua, Italy. He specialises in the development and implementation of Web Services technologies and programmatic interface design and testing.
Mickael Goujon is a senior Java software engineer with M.Sc. in Computer Science from the University of Bordeaux, France. He specialises in Software architecture, web development and new technologies.
Hamish McWilliam is a senior software developer with M.Sc. in Biological Computation from the University of York in the United Kingdom. He specialises in data-warehousing, data-management and bioinformatics tools integration.
Juri Paern is a senior software engineer with a Diplom degree from the University of Marburg, Germany. His main work focuses on data-mining, machine-learning and drug-design.
Rodrigo Lopez is Head of the External Service Group at EMBL-EBI. He has Cand. Scient. degree in Molecular Toxicology from the University of Oslo, Norway.
References
- Apache Lucene. [(20 October 2009, date last accessed)]. http://lucene.apache.org/java/docs/index.html. [Google Scholar]
- Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold T, Ulyanov A, Argos P. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–28. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- Hekkelman ML, Vriend G. MRS: a fast and compact retrieval system for biological data. Nucleic Acids Res. 2005;33:W766–9. doi: 10.1093/nar/gki422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane G, Akhtar R, Bonfield J, et al. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37:D19–25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Universal Protein Resource (UniProt) 2009. The UniProt Consortium. Nucleic Acids Res. 2009;37:D169–74. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldman J. Taking Your Talent to the Web: Making the Transition from Graphic Design to Web Design. New Riders; 2001. p. 448 pp. [Google Scholar]
- Degtyarenko K, de Matos P, Ennis M, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–50. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JM, Cho DH. PDB-Ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures. Nucleic Acids Res. 2005;33:D238–41. doi: 10.1093/nar/gki059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4(6):1527–33. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- Koscielny G, Le Texier V, Gopalakrishnan C, et al. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009;93(3):213–20. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Carrot2. [(20 October 2009, date last accessed)]. http://project.carrot2.org. [Google Scholar]
- PDBe: [(21 October 2009, last accessed)]. http://www.ebi.ac.uk/pdbe. [Google Scholar]
- Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–9. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apache Lucene. [(20 October 2009, date last accessed)]. Query Parser Syntax. http://lucene.apache.org/java/2_9_0/queryparsersyntax.html. [Google Scholar]
- [(20 October 2009, date last accessed)]. Groovy, an agile dynamic language for the Java platform. http://groovy.codehaus.org. [Google Scholar]
- Simple Object Access Protocol (SOAP) [(20 October 2009, date last accessed)]. http://www.w3c.org/TR/soap. [Google Scholar]
- Web Services Description Language (WSDL) [(20 October 2009, date last accessed)]. http://www.w3c.org/TR/wsdl. [Google Scholar]
- Hull D, Wolstencroft K, Stevens R, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–32. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, Shields M, Wang I, Harrison A. The Triana Workflow Environment: architecture and applications. In: Taylor I, Deelman E, Gannon D, Shields M, editors. Workflows for e-Science. New York: Secaucus, NJ, USA: Springer; 2007. pp. 320–39. [Google Scholar]
- KNIME. [(20 October 2009, date last accessed)]. http://www.knime.org. [Google Scholar]
- NET framework. [(20 October 2009, date last accessed)]. http://www.microsoft.com/NET. [Google Scholar]
- Java. [(20 October 2009, date last accessed)]. http://java.sun.com. [Google Scholar]
- Perl language. [(20 October 2009, date last accessed)]. http://www.perl.com. [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- McWilliam H, Valentin F, Goujon M, et al. Web services at the European Bioinformatics Institute-2009. Nucleic Acids Res. 2009;37(Suppl 2):W6–10. doi: 10.1093/nar/gkp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]