Abstract
Rat models have been used to investigate physiological and pathophysiological mechanisms for decades. With the availability of the rat genome and other online resources, tools to identify rat models that mimic human disease are an important step in translational research. Despite the large number of papers published each year using rat models, integrating this information remains a problem. Resources for the rat genome are continuing to grow rapidly, while resources providing access to rat phenotype data are just emerging. An overview of rat models of disease, tools to characterize strain by phenotype and genotype, and steps being taken to integrate rat physiological data is presented in this article. Integrating functional and physiological data with the rat genome will build a solid research platform to facilitate innovative studies to unravel the mechanisms resulting in disease.
Keywords: phenotype, physiological genomics, database, rat strains, disease models, genome
INTRODUCTION
Rat models for disease
Rat models have been used for many decades to study physiological processes and disease mechanisms. Rat strains were developed primarily by physiologists by selectively breeding for traits with specific biomedical interest [1]. The rat is well characterized in the areas including cardiovascular, pulmonary, pharmacology, immunology, cancer, toxicology, nutrition, behavior, aging, and transplantation [2, 3]. Because of its size, rats can be successfully used for a wide variety of experiments, involving surgical manipulation, drug or chemical administration, as well as behavioral and physiological measurements in multiple states. Economically, rats can be bred, maintained and studied at relatively low cost due to the ease of breeding and short generation time. For these reasons, rat models have been become a primary model to study basic physiological mechanisms and pathophysiological changes leading to disease.
Rat strains are available in a variety of strain types; thus, providing researchers with many options when designing experiments. The laboratory rat strains, derived from the Rattus norvegicus species, include outbred stocks of fairly heterogeneous strains of animals and inbred strains, including consomic (chromosomal substitution), congenic (chromosomal region substitution), recombinant inbred (rat strains derived from an F2 population), transgenic and mutant strains. To study human disease mechanisms, rat models that mimic human disease traits have been developed by administering drugs or chemicals to pharmacologically induce disease, by surgically altering organ function, by identifying spontaneous mutations that result in altered structure or function, by using molecular techniques or mutagens to produce genetic modifications, and through breeding strategies that move a gene, segment of chromosome, or whole chromosome from one rat strain to another [4]. Although the development of knock-out rats has been slower than in mouse, several new techniques to create mutant rats have recently been reported. These technologies are very recent, so the number of mutant rat models is expected to increase dramatically in the near future.
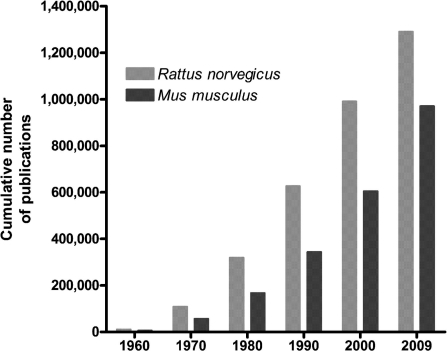
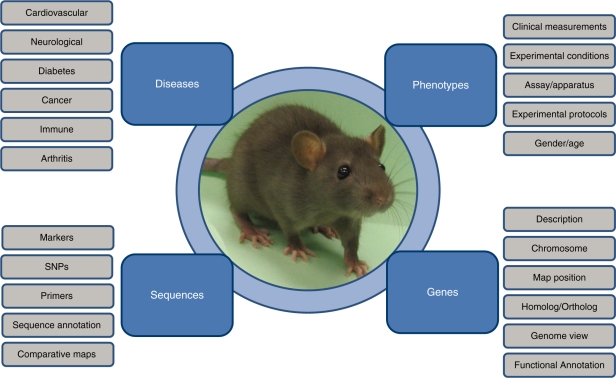
The use of rat models to study physiological mechanisms began in the 19th century [2] with a focus on diseases increasing in the late 20th century as spontaneous models of disease were identified. As larger mammalian models became cost prohibitive to use or difficult to acquire, the use of rat models to investigate physiological mechanisms began to increase. Despite the large number of available mouse strains and the common view that mouse is the most widely used model system, a PubMed review using genus species names demonstrates that rat has the most publications in biomedical literature after humans (Table 1). In fact, the number of rat publications is continuing to grow and mouse is not outpacing rat (Figure 1). As methods to manipulate the rat genome become more efficient and more effective, the gap between rat and mouse publications using genetic models is expected to close. The laboratory rat, Rattus norvegicus, can be used to study many diseases through the development of rat models that display multiple disease traits or through experiments focusing on specific physiological traits, or phenotypes, genes, and sequence (Figure 2).
Table 1:
Number of publications by species generated by querying PubMed using genus species names
| Organism | Number of publications |
|---|---|
| Homo sapiens | 10 864 554 |
| Rattus norvegicus | 1 290 588 |
| Mus musculus | 970 971 |
| Oryctolagus cuniculus (rabbit) | 305 935 |
| Saccharomyces cerevisiae | 86 523 |
| Drosophila melanogaster | 32 990 |
| Danio rerio (zebrafish) | 12 128 |
Figure 1:
Cumulative number of publications available in PubMed for Rattus norvegicus (light gray bars) and Mus musculus (dark gray bars) per decade.
Figure 2:
Laboratory rat models are used as disease models, to investigate the physiological and pathophysiological mechanisms leading to traits or phenotypes, to identify genes involved in disease, and sequence variants that can result in disease or increased disease risk.
Rat strains for physiological research
Rat models have been used to study physiological mechanisms for most major organ systems, including cardiovascular, pulmonary, neurological, renal, gastrointestinal, muscular and skeletal, immune, and reproductive function. In addition to normal physiological function, rat models are the primary model used for drug testing in biotechnology and pharmaceutical research [5]. To study human disease, distinct rat models have been developed by selecting for phenotypes that resemble human disease traits. The development and progression of complex diseases is a result of both genetic and environmental conditions. Rat models for disease that control for genetic and environmental variability provide ideal models to study disease mechanism and develop preventive strategies and treatment for disease. An ideal rat model for disease, as described by Koch and Britton [6], would be ‘mechanistically’ correct, polygenic, display clinical traits, and be influenced by environmental factors. Whether a researcher is focused on normal (non-diseased) physiological function or changes in mechanism that result in altered function, a wide variety of rat models are available.
A variety of methods have been used to generate rat models for disease. Commonly used strains, such as outbred Sprague-Dawley rats, are frequently used to capture the heterogeneous nature of the human population in studies not focused the genetic impact on disease. Outbred strains typically breed well and are robust in size, thus making many experimental protocols more efficient and reproducible. Outbred strains are regularly used for safety or toxicological testing of drug candidates [7]. Inbred strains have the advantage of being isogenic following brother–sister mating for 20 generations or backcrossing for 10 generations, allowing studies to include genetic variability in the experimental design. Many inbred strains were developed by selectively breeding rats with distinct phenotypes that mimic disease traits to fix the genes related to that trait. These rat models were initially developed for physiological and pharmacological studies [8], but have become repeatedly used to identify gene(s) related to disease traits. By utilizing two inbred strains with distinguishable phenotypes, linkage analysis can be used to identify genes underlying traits by developing segregating crosses of inbred rats (strains in which a particular allele or mutation is maintained in heterozygous state). This is a powerful method to map quantitative traits to the genome, but requires large populations of rat.
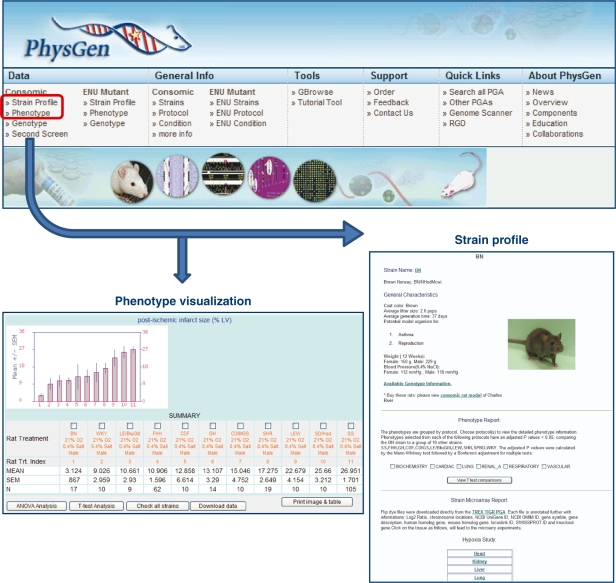
Inbred rat strains have been used to create many other valuable rat models. Recombinant inbred strains are produced by outcrossing two inbred strains, followed by inbreeding for at least 20 generations to create a group of inbred strains with a mosaic of the parental genomes [7]. Each recombinant inbred strain has a unique, fixed genome through combination of the original parental genomes. Although each recombinant inbred strain can have unique phenotypes, the limitation with these strains is the number of strains in the set which impacts the resolution in identifying small genomic regions associated with disease traits. Consomic (chromosomal substitution) and congenic strains have been developed using a breeding strategy to introgress an entire chromosome (consomic) or a portion of a chromosome (congenic) from a control inbred strain into a susceptible inbred strain. Physiological studies can be used to validate the functional importance of the genomic region and associate phenotypes to the genome [9]. The PhysGen Program for Genomic Applications (PGA) at the Medical College of Wisconsin (http://pga.mcw.edu) developed two inbred consomic panels on two genetic backgrounds to enable traits to be mapped to chromosomes [9]. Because all of the consomic strains have a fixed genetic background, ample numbers of genetically identical rats can be used to generate sufficient statistical power to detect linkage. The PhysGen PGA provides several modes to view and analyze the phenotype data, including visualization and statistical analysis of individual phenotype results across multiple strains or by accessing the strain profiles which summarize both general and phenotype data for individual strains (Figure 3). Congenic strains have the additional power of narrowing the list of possible candidate genes and prioritize genes for further study. This strategy has been successfully used to separate salt-sensitivity and hypertension quantitative trail loci (QTLs) on chromosome 18 in the spontaneously hypertensive rat (SHR) [10]. Another research team developed congenic strains to further dissect four blood pressure QTLs on chromosomes 2, 4 and 16 in the SHR. Results from this study demonstrated that the four regions influence blood pressure differently under different diet conditions and provided evidence for the influence of genetic background on the expression of the mapped QTLs [11]. The list of complex traits being positionally cloned through the development of congenic strains combined with other genomic tools has been growing rapidly. Some examples of the successes in identification of genes underlying complex traits include Ncf1 regulating severity of arthritis [12, 13], Cyp11b1 [14] and Adamts16 [15] both involved in blood pressure regulation, as well as other traits involved in heart failure, left ventricular mass, mammary cancer, and diabetes [3].
Figure 3:
The PhysGen PGA website provides access to strain and phenotype data for more than 70 strains. Users can query by phenotype or strain. Phenotype queries (lower left) return data for all strains for a single phenotype. Strain profile data for a particular strain across all phenotypes (lower right).
Spontaneous mutations in inbred strains provided a natural path to study the normal physiological pathways and pathophysiological changes that occur in disease [16]. However, genetic manipulation using a variety of transgenic techniques has resulted in many strains with altered expression of specific genes resulting in disease phenotypes [8]. Recent advances in direct manipulation of the rat genome have established that embryonic stem cells and inducible pluripotent stems can be used to knock out genes in the rat [17–20]. By using engineered zinc- finger nucleases, disruption of a targeted locus is feasible and efficient, thus providing the mechanism to create targeted gene models of disease in the rat [21]. These state-of-the-art technologies will provide excellent models to study the functional effect of genes identified as candidates for human disease [22]. Despite the novel nature of these techniques, it can be anticipated that the number of targeted mutations in rats will rise dramatically in coming years.
Investigators interested in obtaining rat models of disease to study physiological or pathophysiological function can have difficulty if the strain has not been preserved or maintained at a commercial rodent facility. To address this need, several centralized facilities were established to acquire, maintain, and preserve valuable rat strains for the scientific community. The Rat Resource and Research Center (RRRC, http://www.nrrrc.missouri.edu/), through the National Center for Research Resources, was established in 2001 to provide the scientific community with a centralized resource for archiving and distributing rat models. In Japan, the National BioResource Project (NBRP, http://www.anim.med.kyoto-u.ac.jp/nbr/) was started in 2002 to facilitate the collection, preservation and provision of rat models [7]. Although these large repositories provide access to many rat models, many valuable strains are maintained by individual investigators or research teams at academic institutions. The Rat Genome Database (RGD, http://rgd.mcw.edu) curates strain information and presents general and detailed information about each strain registered. References used to capture the strain information contain contact information for the investigators who developed each strain, providing interested users a starting point to locate relevant strains (Table 2).
Table 2:
Overview of available tools for physiological genomics using rat models.
| Tool | URL | Description |
|---|---|---|
| PhysGen PGA | http://pga.mcw.edu | This program developed unique rat models for complex disease and provides extensive phenotyping results for each strain. |
| National BioResource Project for the Rat in Japan (NBRP) | http://www.anim.med.kyoto-u.ac.jp/nbr/ | A Japanese rat repository that collects, preserves, and supplies rat strains with associated phenotype and genetic profiles. |
| RGD | http://rgd.mcw.edu | RGD serves the research community by providing access to genomic, genetic, pathway, phenotype, and strain information. |
| RGD Phenotypes & Models Portal | http://rgd.mcw.edu/wg/physiology | A portal within RGD that provide an entryway to physiological data and disease model information. |
| RGD Strain Search | http://rgd.mcw.edu/rgdweb/search/strains.html | An RGD query function that returns all strains with associated key words or traits. |
| RRRC | http://www.nrrrc.missouri.edu/ | An NIH funded repository for distributing inbred, hybrid and mutant rat strains to investigators. |
| NCBI Rat Genome Resources | http://www.ncbi.nlm.nih.gov/genome/guide/rat/ | An NCBI site developed to provide an overview of available rat data to benefit researchers using rat models of disease. |
| LONI Rat Atlas Image Database | http://www.loni.ucla.edu/Atlases/Atlas_Detail.jsp?atlas_id=1 | A three dimensional computerized map of rat brain anatomy developed by the Laboratory of Neuro Imaging. |
| Knock-out Rat Consortium (KORC) | http://www.knockoutrat.org/ | The KORC’s goal is to create single gene disruptions (knockouts) in every gene in the rat genome. |
| Rat Community Forum | http://gray.hmgc.mcw.edu/mailman/listinfo/rat-forum | The Rat Community Forum (RCF) is a moderated online forum, for use by members of the scientific community to ask questions or post information related to the rat. |
Identifying and tracking specific strains used within each study can be challenging if the authors do not include full strain information. Rat strain nomenclature guidelines have been established and approved by the Rat Genome Nomenclature Committee to provide documentation on the rules for naming strains. Detailed guidelines are available on the RGD website (http://rgd.mcw.edu/nomen/rules-for-nomen.shtml) and assistance can be provided by the RGD curation staff. New strains should be registered through RGD to assign a Laboratory Registration Code identifing the particular institute, laboratory, or investigator that produced the rat strain. This information can be used to locate strains of interest for new studies. In addition, the strain nomenclature includes information on the type of strain (inbred, hybrid, recominbant inbred, congenic, etc.) and mutant alleles. The nomenclature rules are applicable to both mouse and rat.
Tools to identify rat models of disease
Strain selection strategies can be a challenging step in designing experiments to study the pathophysiological mechanisms involved in disease development, progression and amelioration. Public health relevance is a key component of many research programs and often a key requirement for successfully obtaining funding; thus, most research programs focus on understanding human disease mechanisms and design studies to investigate alterations that result in disease traits. Identification of disease models that mimic human disease traits can be done in using several approaches. Physiologists frequently focus on phenotypes, or traits, that have been measured under control and experimental conditions. The control conditions can be considered equivalent to a ‘non-diseased’ model while the experimental conditions could equate to a ‘diseased’ model. The experimental condition could be induced by a surgical procedure, administration of a drug or chemical, or could be an animal model with a different genetic makeup. The unique phenotypes that resemble disease traits can be used to study disease mechanisms, but if different animal models are incorporated, identification of genomic regions or genes implicated in the disease can follow. The advantage to identifying one or many rat models of disease lies in the ability to extend the studies to a variety of experimental conditions (atmospheric condition, diet, environmental enrichment, substance administration) or time course studies, including developmental stages and ageing. A second method used to create and identify rat models of disease is by identifying a genomic region that has been linked to an altered phenotype. Genetic linkage studies have been used to map chromosomal regions that segregate with phenotypes and report quantitative trait loci (QTL) [8]. QTL studies have been used to identify altered genes or genomic regions that result in altered phenotypes and can then be used to narrow the region to identify or target genes for follow up studies. Genetic linkage studies require a large population, but resources exist that can narrow the genomic region associated with phenotypes. One such resource is the QTL report section at the RGD. These reports provide summaries of phenotypes, disease annotations, map positions and strains. The QTL summary at RGD can provide insight into genetic background effect and gene–gene interactions. To further narrow the list of causative genes, development of congenic strains or molecular approaches to target candidate genes can be used to begin the gene discovery phase. Finally, there is growing need to identify rat models in which verification studies can be designed for genes found to be significantly associated to disease in genome wide association studies (GWAS). Rat models can be used to study the disease mechanisms for pathways in which GWAS genes have been shown to play a role or to study the effect of genome background. Inbred strains, or transgenic strains, can be used for targeted validation studies. Specific gene knockouts can now be created to directly test the role of genes in molecular or physiological pathways known to be altered in disease states.
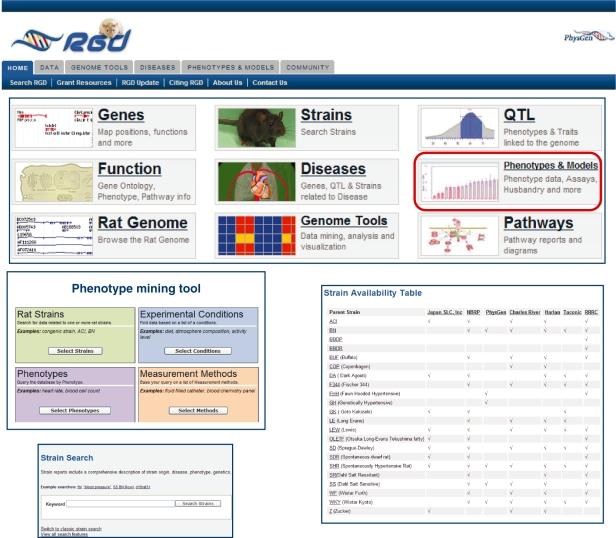
The convincing evidence that a rat strain is a good model for study of disease mechanism is physiological data, i.e. significantly different in the disease model as compared to a control, non-diseased strain. The primary source of rat physiological data remains published manuscripts. These manuscripts contain the hypothesis, a detailed description of the methods, and results for a small number of phenotypes that point to a step in a disease pathway. The studies might involve a single strain under varied experimental conditions, before and after a surgical manipulation, or before, during and after the delivery of a drug. Other studies may include multiple strains to compare genetic background, but still typically only focus on a small number of phenotypes per study. Another source of physiological data is from textbooks, via personal communications or through oral presentations at conferences or seminars, and through review articles. Recently, several large programs have generated large physiological data sets associated with a wide range of rat strains and made these data freely available. The PhysGen PGA measured more than 200 phenotypes in more than 50 different rat strains (http://pga.mcw.edu). The phenotype measurements include cardiovascular, pulmonary, renal, vascular, respiratory and biochemical traits. The results can be viewed, analyzed and downloaded by querying by strain or by phenotype. The National BioResource Program in Japan (NBRP) is another valuable resource for physiological data for strains by providing a catalogue of standard phenotype measurements for a set of well characterized rat strains (http://www.anim.med.kyoto-u.ac.jp/nbr/). The rat research community has been in need of a single repository that provides tools to integrate data collected and presented in multiple formats. To address this need, the RGD has developed a Phenotypes and Strain portal that provides an entryway for physiologist to access both phenotype data as well as strain information [23]. Within this portal, a phenotype data mining tool is under development that provides a single access point to query physiological data by strain, phenotype, experimental conditions, or measurement method (Figure 4). The prototype of this tool has been released and currently includes a small subset of the cardiovascular data available through the PhysGen PGA, some NBRP data, and some results from select publications. As additional data is added to the database, the phenotype data mining tool will become a powerful tool to assist researchers in the selection of appropriate strains, experimental conditions that exacerbate or ameliorate the disease traits, and will allow researchers to compare traits measured using different methods.
Figure 4:
The RGD provides entryways to a wide variety of genetic, genomic, phenotype and strain information. Physiologist can access data through the ‘Phenotypes & Models’ portal providing entry to a new phenotype mining tool (middle left), a strain search (lower left), and a resource for strain availability (lower right).
Resources for physiological studies
Physiological studies often require weeks of planning, including pre-treatment of animals prior to the actual experiments that produce the publishable results. Successful experimental designs include many details that begin weeks prior to the actual protocol. Researchers must develop a breeding strategy, consider animal husbandry, methods for strain preservation and conditioning of the rats prior to the study with altered environmental conditions, diet, acute and chronic drug administration, surgical procedures, and timeline for data collection. In the past, husbandry was often overlooked as an aspect of the study that could have a significant impact on the results. However, many studies have demonstrated that diet, environment and level of enrichment can impact physiological data [24–26]. Colony management and techniques to improve breeding efficiency is another aspect of research design strategies that must be adopted by researchers if new strain development and maintenance is included in the experimental design. After new strains are developed and studied, the colonies cannot always be maintained within the laboratory due to financial and/or space limitations. Cryopreservation or methods to reduce the cost of colony maintenance are useful resources. The Phenotypes and Models portal at RGD has developed a section that includes access to detailed information or links to other resources that can provide this valuable laboratory information.
CONCLUSIONS AND FUTURE DIRECTIONS
The use of rats in studies directed at unraveling physiological and pathophysiological pathways has been ongoing for many decades. With the advent of new technologies to target genes, the use of rat models will likely see a dramatic increase in the next decade as candidate disease genes are tested for functional role in the development of disease. The growth in available rat models to study disease is necessitating a need to create new tools and resources to integrate and mine physiological data. As the existing resources grow and new tools are created, biomedical researchers will have the ability to meet the existing challenges facing translational research. High-throughput experimental techniques and large scale data analysis, combined with data standards and methods for data integration, are producing unified resources that facilitate physiological research in the rat with a focus on distinct disease traits, risk of development and disease prevention strategies.
Key Points.
Rat strains are used as models to study physiological mechanisms, as models for human disease, and for testing drug candidates.
Integrating physiological data from a wide variety of rat strains will provide the foundation for tools to select ideal disease and non-disease (control) models.
The need to link rat physiological data with the genome is growing rapidly as candidate genes from human population studies need to be tested for biological function in animal models with disease susceptibility.
FUNDING
National Heart, Lung and Blood Institute at the National Institutes of Health [PhysGen Program for Genomic Applications, grant number U01 HL066579; Rat Genome Database, grant number R01 HL064541].
Acknowledgements
The author would like to thank the committed members of the PhysGen Program for Genomic Application and RGD teams that contributed to the development of the resources and tools that each of these programs provide to the scientific community.
Biography
Melinda Dwinell is an Assistant Professor of Physiology in the Human & Molecular Genetics Center at the Medical College of Wisconsin. Her interests are physiological genomics and integrating phenotype and genotype data.
References
- Hübner N. Expressing physiology. Nat Genet. 2006;38:140–1. doi: 10.1038/ng0206-140. [DOI] [PubMed] [Google Scholar]
- Gill TJ, III, Smith GJ, Wissler RW, et al. The rat as an experimental animal. Science. 1989;245:269–76. doi: 10.1126/science.2665079. [DOI] [PubMed] [Google Scholar]
- Aitman TJ, Critser JK, Cuppen E, et al. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–22. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
- Owens DR. Spontaneous, surgically and chemically induced models of disease. In: Suckow MA, Weisbroth SH, Franklin CL, editors. The Laboratory Rat. Amsterdam: Elsevier; 2006. pp. 679–92. [Google Scholar]
- Mashimo T, Serikawa T. Rat resources in biomedical research. Curr Pharm Biotechnol. 2009;10:214–20. doi: 10.2174/138920109787315105. [DOI] [PubMed] [Google Scholar]
- Koch LG, Britton SL. Development of animal models to test the fundamental basis of gene-environment interactions. Obesity. 2008:S28–32. doi: 10.1038/oby.2008.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikawa T, Mashimo T, Takizawa A, et al. National BioResource Project – Rat and Related Activities. Exp Amin. 2009;58:333–41. doi: 10.1538/expanim.58.333. [DOI] [PubMed] [Google Scholar]
- Moreno C, Jacob HJ. Integrating biology with rat genomic tools. In: Suckow MA, Weisbroth SH, Franklin CL, editors. The Laboratory Rat. Amsterdam: Elsevier; 2006. pp. 679–92. [Google Scholar]
- Cowley AW, Jr, Liang M, Roman RJ, et al. Consomic rat model systems for physiological genomics. Acta Physiol Scand. 2004;181:585–92. doi: 10.1111/j.1365-201X.2004.01334.x. [DOI] [PubMed] [Google Scholar]
- Johnson MD, He L, Herman D, et al. Dissection of chromosome 18 blood pressure and salt-sensitivity quantitative trait loci in the spontaneously hypertensive rat. Hypertension. 2009;54:639–45. doi: 10.1161/HYPERTENSIONAHA.108.126664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneas I, Rodrigues MV, Pauletti BA, et al. Congenic strains provide evidence that four mapped loci in chromosomes 2, 4 and 16 influence hypertension in the SHR. Physiol Genomics. 2009;37:52–7. doi: 10.1152/physiolgenomics.90299.2008. [DOI] [PubMed] [Google Scholar]
- Olofsson P, Holmberg J, Tordsson J, et al. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat Genet. 2003;33:25–32. doi: 10.1038/ng1058. [DOI] [PubMed] [Google Scholar]
- Rintisch C, Forster M, Holmdahl R. Detection of arthritis-susceptibility loci, including Ncf1, and variable effects of the major histocompatibility complex region depending on genetic background in rats. Arthritis Rheum. 2009;60:419–27. doi: 10.1002/art.24292. [DOI] [PubMed] [Google Scholar]
- Garrett MR, Rapp JP. Defining the blood pressure QTL on Chromosome 7 in Dahl rats by a 177-kb congenic segment containing Cyp11b1. Mamm Genome. 2003;14:268–73. doi: 10.1007/s00335-002-2245-9. [DOI] [PubMed] [Google Scholar]
- Joe B, Saad Y, Lee NH, et al. Positional identification of variants of Adamts16 linked to inherited hypertension. Hum Mol Genet. 2009;18:2825–38. doi: 10.1093/hmg/ddp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LJ, Robledo RF, Bult CJ, et al. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–98. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, et al. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–9. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Liao J, Cui C, Chen S, et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–5. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009:325–433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzi J, Fraichard A, Thiam K. Use of genetically modified rat models for translational medicine. Drug Discov Today. 2008;13:488–94. doi: 10.1016/j.drudis.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Dwinell MR, Worthey EA, Shimoyama M, et al. The Rat Genome Database 2009: variation, ontologies and pathways. Nuc Acids Res. 2009;37:D744–9. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson DL, Kunert MP, Kaldunski ML, et al. Influence of diet and genetics on hypertension and renal disease in Dahl salt-sensitive rats. Physiol Genomics. 2004;16:194–203. doi: 10.1152/physiolgenomics.00151.2003. [DOI] [PubMed] [Google Scholar]
- Bavis RW, Mitchell GD. Long-term effects on the perinatal environment on respiratory control. J Appl Physiol. 2008;104:1220–9. doi: 10.1152/japplphysiol.01086.2007. [DOI] [PubMed] [Google Scholar]
- Snow JB, Kanagy NL, Walker BR, et al. Rat strain differences in pulmonary artery smooth muscle Ca(+2) entry following chronic hypoxia. Microcirculation. 2009;16:603–14. doi: 10.1080/10739680903114268. [DOI] [PMC free article] [PubMed] [Google Scholar]