Abstract
Increased mitogen-activated protein kinase (MAPK) activity correlates with a more malignant hepatocellular carcinoma (HCC) phenotype. There is a reciprocal regulation between p44/42 MAPK (extracellular signal-regulated kinase [ERK]1/2) and the dual-specificity MAPK phosphatase MKP-1/DUSP1. ERK phosphorylates DUSP1, facilitating its proteasomal degradation, whereas DUSP1 inhibits ERK activity. Methionine adenosyltransferase 1a (Mat1a) knockout (KO) mice express hepatic S-adenosylmethionine (SAM) deficiency and increased ERK activity and develop HCC. The aim of this study was to examine whether DUSP1 expression is regulated by SAM and if so, elucidate the molecular mechanisms. Studies were conducted using Mat1a KO mice livers, cultured mouse and human hepatocytes, and 20S and 26S proteasomes. DUSP1 messenger RNA (mRNA) and protein levels were reduced markedly in livers of Mat1a KO mice and in cultured mouse and human hepatocytes with protein falling to lower levels than mRNA. SAM treatment protected against the fall in DUSP1 mRNA and protein levels in mouse and human hepatocytes. SAM increased DUSP1 transcription, p53 binding to DUSP1 promoter, and stability of its mRNA and protein. Proteasomal chymotrypsin-like and caspase-like activities were increased in Mat1a KO livers and cultured hepatocytes, which was blocked by SAM treatment. SAM inhibited chymotrypsin-like and caspase-like activities by 40% and 70%, respectively, in 20S proteasomes and caused rapid degradation of some of the 26S proteasomal subunits, which was blocked by the proteasome inhibitor MG132. SAM treatment in Mat1a KO mice for 7 days raised SAM, DUSP1, mRNA and protein levels and lowered proteosomal and ERK activities.
Conclusion
DUSP1 mRNA and protein levels are lower in Mat1a KO livers and fall rapidly in cultured hepatocytes. SAM treatment increases DUSP1 expression through multiple mechanisms, and this may suppress ERK activity and malignant degeneration.
Human hepatocellular carcinoma (HCC) is associated with increased expression and activity of mitogen-activated protein kinase (MAPK) signaling intermediates. MAPKs are a family of serine/threonine kinase with three main components: extracellular signal-regulated kinase (ERK), cjun NH2-terminal kinase, and p38.1,2 MAPK dephosphorylation and inactivation is carried out by several phosphatases, including the dual-specificity MAPK phosphatases (MKPs or DUSPs), which simultaneously dephosphorylate both serine/threonine and tyrosine residues.3,4 DUSP1 is the first member of this family that has been characterized. It is controlled by an early response gene, which is transiently induced by mitogens and stress signals such as serum, cytokines, ultraviolet radiation, heat shock, and hypoxia.5
Prolonged activation of ERK promotes phosphorylation at the Ser296 residue of its inhibitor, DUSP1.6,7 Phosphorylation of this specific residue renders the DUSP1 protein susceptible to proteasomal degradation by two substrate recognition protein belonging to a large S-phase kinase-associated protein/cullin/F box ubiquitin ligase: the S-phase kinase-associated protein 2 (SKP2) and CDC28 protein kinase b1 complex. In contrast, transient activation of ERK leads to catalytic activation of DUSP1 followed by inactivation of ERK.7 This body of evidence indicates that DUSP1 feedback inhibits ERK and that DUSP1 might be a crucial regulation of ERK activity in the cell.
Proteasome inhibitors act in part through down-regulation of MAPK signaling cascade. The eukaryotic proteasome is a large multicatalytic, multisubunit proteasome complex possessing at least three distinct activities, which are associated with three different β-subunits, respectively: chymotrypsin-like activity (with the β5 subunit), trypsin-like activity (with the β2 subunit), and peptidyl-glutamyl peptide-hydrolyzing-like (caspase-like) activity (with the β1 subunit).8 Inhibition of the chymotrypsin-like activity, but not of the trypsin-like activity, of the proteasome has been associated with induction of tumor cell apoptosis.9 In HCC cell lines, ERK or MAPK kinase inhibitors increased apoptosis and inhibited tumorigenicity and cell cycle progression.10
Patients with cirrhosis of various etiologies have decreased hepatic methionine adenosyltransferase (MAT) activity and S-adenosylmethionine (SAM) biosynthesis. 11 SAM, which is generated in the first reaction in the metabolism of methionine, catalyzed by MAT, is the principal biological methyl donor and a precursor for glutathione and polyamines. The catalytic subunit of MAT is the product of two different genes: MAT1A, which is expressed mainly in the adult liver, and MAT2A, which is widely distributed, expressed in the fetal liver, and replaces MAT1A in HCC.11 Consequences of hepatic SAM deficiency as illustrated by the Mat1a knockout (KO) mouse model include increased susceptibility to steatosis and oxidative liver injury, spontaneous development of steatohepatitis and HCC.12,13 In addition, ERK is hyperphosphorylated and cyclin D1 expression higher in Mat1a KO mice at baseline.14 Given the known reciprocal regulation of ERK and DUSP1, the aims of the present study were to investigate whether DUSP1 expression is altered when SAM is depleted and if so, elucidate the molecular mechanisms.
Materials and Methods
Primary Cultures and Reagents
Mouse hepatocytes from 2- to 3-month-old male C57/B6 mice were isolated by way of collagenase perfusion as described15 by the Cell Culture Core of the USC Research Center for Liver Diseases. Primary human hepatocytes were obtained in suspension culture in cold preservation medium (24 hours after livers were harvested) from CellzDirect (Pittsboro, NC). Cells were plated at a density of 0.5 × 106 cells per well in 6-well plates. Cultures were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 mM penicillin, and 50 mg/mL streptomycin sulfate. After 2 hours, the medium was changed to contain 2.5% fetal bovine serum.16 SAM in the form of disulfate p-tolue-nesulfonate dried powder was generously provided by Gnosis SRL (Cairate, Italy). All other reagents were of analytical grade and were obtained from commercial sources.
Cell Treatments
Hepatocytes were treated with SAM (1 or 2 mM) for different time points. When indicated, cells were pretreated with actinomycin D (5 µg/mL) for 5 minutes or MG132 (20 µM) for 1 hour. Medium was changed after MG132 pretreatment. Treatments with actinomycin D and MG132 under these conditions did not cause any toxicity as evaluated by way of trypan blue exclusion.17
Mat1a KO Mice and Treatment
The husbandry of male Mat1a KO and wild-type (WT) littermates has been described.12 Mice were given 100 mg/kg/day SAM dissolved in water or equal volume of water by way of oral gavage and were sacrificed after 7 days. Liver tissues were removed, snap frozen in liquid nitrogen, and stored at −80°C until analysis. Animals were treated humanely and all procedures were in compliance with institutional guidelines for the use of laboratory animals.
RNA Extraction and Real-Time Polymerase Chain Reaction Analysis
Total RNA isolated from hepatocytes and livers as described18 was subjected to reverse-transcription using M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA). One microliter of reverse transcription product was subjected to quantitative real-time polymerase chain reaction (PCR) analysis. The primers and TaqMan probes for DUSP1, Skp2, and Universal PCR Master Mix were purchased from ABI (Foster City, CA). 18SrRNA was used as a housekeeping gene as described.19 The ΔCt obtained was used to find the relative expression of genes according to the following formula:
where ΔΔCt = ΔCt of respective genes in experimental groups −ΔCt of the same genes in control group.
Protein Extraction and Western Blot Analysis
Protein extracts from primary hepatocytes and liver samples were prepared as described15 and resolved by electrophoresis on 12%–13% sodium dodecyl sulfate–polyacrylamide gels. Western blotting was performed following standard protocols (Amersham BioSciences, Piscataway, NJ) using primary antibodies for DUSP1 (Santa Cruz Biotechnologies, Santa Cruz, CA), phosphorylated DUSP1 (Ser359), ERK, phosphorylated ERK, actin (from Cell Signaling, Danvers, MA), and SKP2 (from Santa Cruz Biotechnologies).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed according to the ChIP assay kit protocol provided by Upstate with antibodies against p53 (Santa Cruz Biotechnologies) and actin (Cell Signaling). PCR of the DUSP1 promoter region across one relevant p53 site within −476 to −18820 was performed with the forward primer 5′-CAGGGCAGGTGTCCTCAG-3′ (bp −476 to −458 [all sequences are relative to the ATG start codon]) and the reverse primer 5′-CGCGGTTTTATGTAGCCTCT-3′ (bp −208 to −188). All PCR products were run on 2% (wt/vol) agarose gels prestained with ethidium bromide.
Proteasome Chymotrypsin-Like and Caspase-Like Activity Assays
Cells and mouse liver tissues were harvested, washed twice with phosphate-buffered saline, and homogenized in a lysis buffer [50 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (pH 7.5), 5 mM ethylene diamine tetraacetic acid, 150 mM NaCl, 1% Triton X-100, and 2 mM adenosine triphosphate]. After 30 minutes of rocking at 4°C, the mixtures were centrifuged at 12,000g for 15 minutes and the supernatants were collected. Mouse 20S proteasome (Boston Biochem, Cambridge, MA) was diluted in 10 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (pH 7.6) and incubated with SAM (1 or 2 mM) and MG132 (20 µM) for different durations. Chymotrypsin-like activity and caspase-like activity were measured in whole-cell (5 µg) and mouse liver tissue extracts (2 µg) or mouse 20S proteasome using a Proteasome-Glo assay system (Promega, Madison, WI) following the manufacturer’s instructions.
Human 26S Proteasome Stability Assay
Human 26S proteasomes were extracted from HeLa cells as described.21 26S proteasomes were incubated with SAM (1 or 2 mM) and MG132 (250 or 500 µM) for different durations. The samples are resolved by way of electrophoresis on 10% sodium dodecyl sulfate–polyacrylamide gels. Western blotting was performed following standard protocols (Amersham BioSciences) using antibodies to proteasomal subunits α4, Rpn10/S5a, Rpn7/S10a, β2, and β3 (ENZO Life Sciences, Switzerland).
Determination of SAM Levels
SAM levels were measured from liver samples using high-performance liquid chromatography as described.15
Statistical Analysis
Data are expressed as the mean ± standard error of the mean (SEM). Statistical analysis was performed using analysis of variance and Fisher’s exact test. For messenger RNA (mRNA) and protein levels, ratios of genes and proteins to respective housekeeping densitometric values were compared. Significance was defined by P < 0.05.
Results
DUSP1 Expression Is Decreased While ERK Activity Is Increased in Mat1a KO Mice Livers
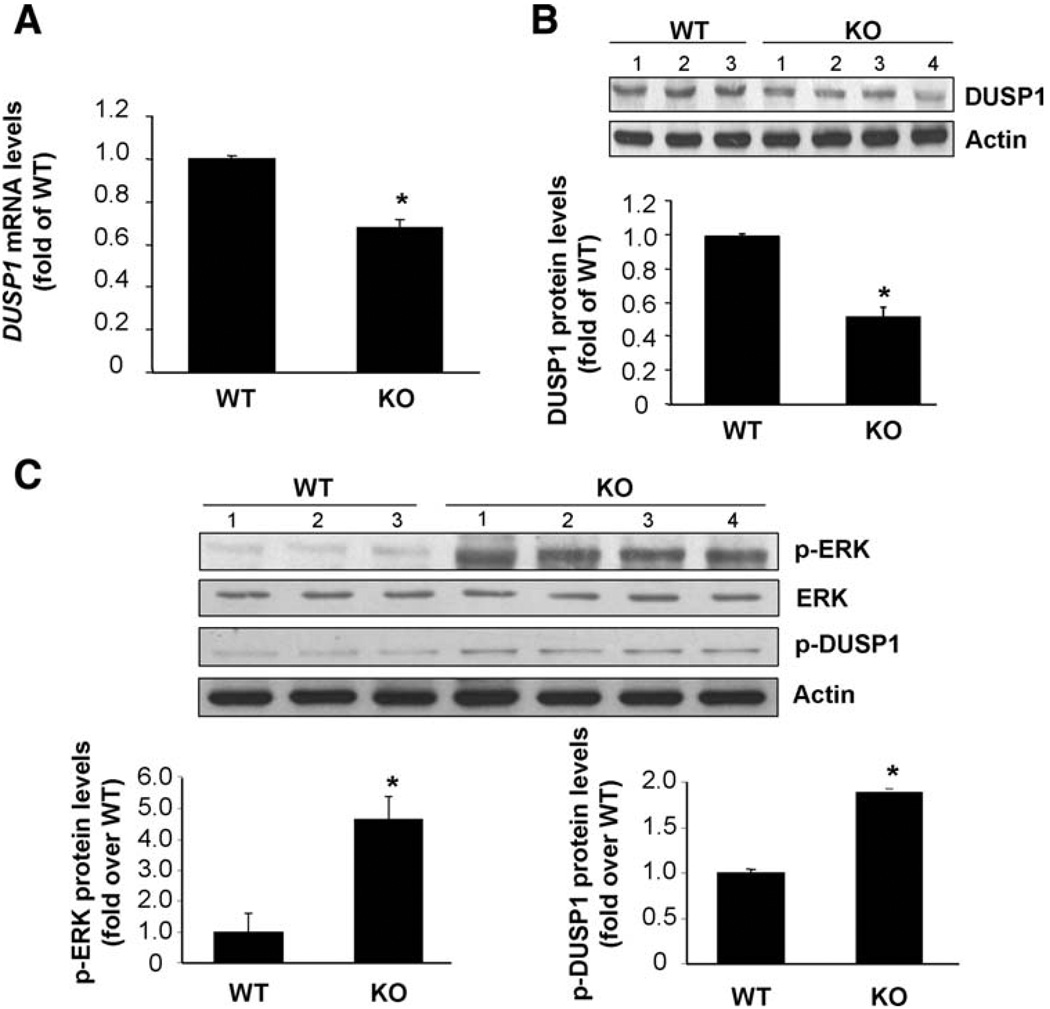
DUSP1 mRNA and protein levels were decreased by 35% and 55%, respectively, in livers of 4- to 6-month-old Mat1a KO mice compared with age-matched WT controls (Fig. 1A,B). This correlated with an increase in ERK activity as indicated indirectly by the amount of phosphorylated ERK as well as phosphorylated DUSP-1 (Fig. 1C).
Fig. 1.
DUSP1 expression is reduced while ERK is activated in Mat1a KO mice. (A) DUSP1 mRNA levels as measured by way of real-time PCR. Results are expressed as fold of age-matched WT mice (mean ± SEM) from four WT and nine KO mice. *P < 0.05 versus WT. (B,C) DUSP1 protein levels as measured by western blot analysis. Results are expressed as fold of age-matched WT mice (mean ± SEM) from three WT and four KO mice. *P < 0.001 versus WT.
Effect of SAM on DUSP1 Expression in Primary Cultures of Mouse and Human Hepatocytes
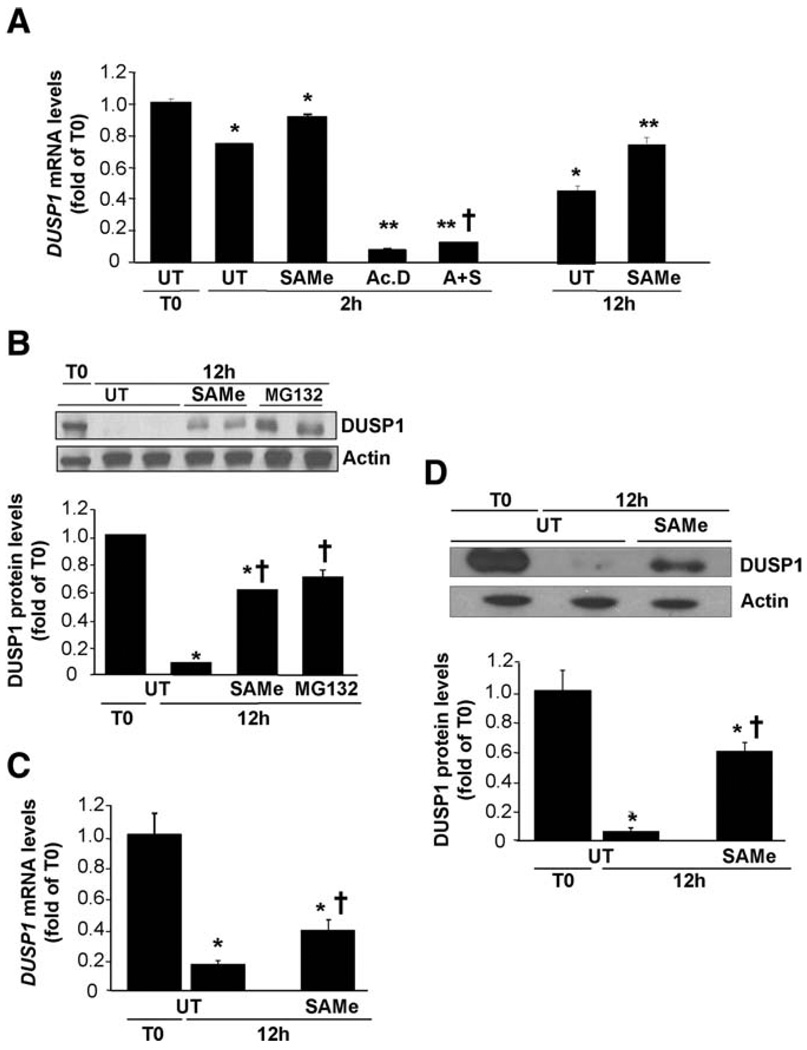
Hepatocytes in primary culture rapidly lose MAT1A expression and SAM levels fall.22 Figure 2 shows that 12 hours after culture of mouse hepatocytes, DUSP1 mRNA levels fell by 60% (Fig. 2A) and protein levels fell by 90% (Fig. 2B). Adding SAM (2 mM) to culture medium blunted the fall in DUSP1 mRNA and protein levels (Fig. 2A,B). DUSP1 mRNA levels fell very rapidly, and by 2 hours after culture were already 35% lower (Fig. 2A). Treatment with actinomycin D for 2 hours reduced DUSP1 mRNA levels to less than 6% of baseline, but SAM addition blunted the effect of actinomycin D slightly (Fig. 2A). Loss of DUSP1 protein can be significantly prevented by pretreatment with the proteasome inhibitor MG132, which exerted an effect comparable to that of SAM (Fig. 2B). Similar to mouse hepatocytes, human hepatocytes in primary culture also rapidly lose DUSP1 expression at both mRNA and protein levels (Fig. 2C,D). SAM treatment protected against decreases in both levels, but protection against decreased DUSP1 protein levels was greater (Fig. 2C,D).
Fig. 2.
Effect of SAM treatment on DUSP1 expression in cultured hepatocytes. Mouse (A, B) or human (C, D) hepatocytes were treated with SAM (2 mM) for 2 or 12 hours. Where indicated, cells were pretreated with actinomycin D (Ac.D, 5 µg/mL) for 5 minutes. (A) Effect of SAM and actinomycin D treatment on DUSP1 mRNA levels in mouse hepatocytes measured by way of real-time PCR. Cells were pretreated with Ac.D for 5 minutes followed by Ac.D alone or with SAM (A+S) for another 2 hours. Results are expressed as fold of T0 cells (mean ± SEM) from three to five independent experiments. *P < 0.01 versus T0; **P < 0.001 versus T0 and UT at 2 or 12 hours; †P < 0.05 versus Ac.D alone. (B) Effect of SAM treatment on DUSP1 protein levels in cultured mouse hepatocytes on western blot analysis. Mouse hepatocytes were cultured and treated with SAM (2 mM) for 12 hours. When indicated, cells were pretreated with MG132 (20 µM) for 1 hour. Results are expressed as fold of T0 cells (mean ± SEM) from three independent experiments. *P < 0.001 versus T0; †P < 0.002 versus UT at 12 hours. (C) Effect of SAM treatment on DUSP1 mRNA levels in human hepatocytes. Results are expressed as fold of T0 cells (mean ± SEM) from three independent donors. *P < 0.0001 versus T0; †P < 0.02 versus UT at 12 hours. (D) Effect of SAM treatment on DUSP1 protein levels in human hepatocytes. Results are expressed as fold of T0 cells (mean ± SEM) from three independent donors. *P < 0.0001 versus T0; †P < 0.02 versus UT at 12 hours.
Effects of SAM on Proteasomal Activity
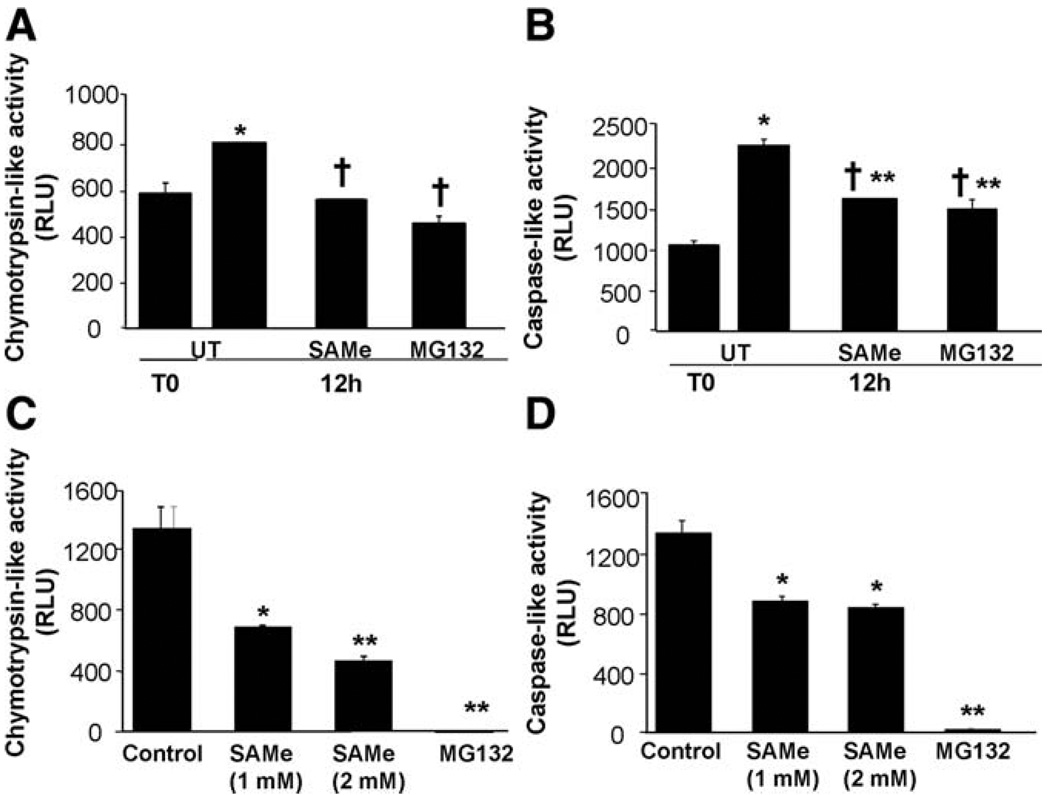
Because SAM protected against decreases in DUSP1 protein levels much more than in mRNA levels, and that the magnitude of protection is similar to MG132, a well-known inhibitor of proteasomal activity, we next investigated whether proteasomal activity (measured by chymotrypsin-like and caspase-like activities) was altered during the culture of hepatocytes. Figure 3A shows that 12 hours after culturing, chymotrypsin-like activity increased, and this can be blocked completely by either SAM or MG132. Similarly, caspase-like activity also increased, and this was partially blocked to equal extent by SAM and MG132 (Fig. 3B). To make sure that SAM’s inhibitory effect is direct, we measured proteasomal activity using 20S proteasomes. Figure 3C shows that SAM was able to inhibit both chymotrypsin-like (Fig. 3C) and caspase-like (Fig. 3D) activities in 20S proteasomes. Inhibition was maximal after a 1-hour incubation with 20S proteasomes (data not shown).
Fig. 3.
Effect of SAM treatment on proteasomal chymotrypsin-like and caspase-like activity in cultured mouse hepatocytes (A, B) and 20S proteasome (C, D). Mouse hepatocytes were cultured and treated with SAM (2 mM) for 12 hours. When indicated, cells were pretreated with MG132 (20 µM) for 1 hour. (A) Effect of SAM treatment on proteasomal chymotrypsin-like activity. Results are expressed as relative light units (RLU) (mean ± SEM) from three independent experiments. *P < 0.001 versus T0; †P < 0.0001 versus UT at 12 hours. (B) Effect of SAM treatment on proteasomal caspase-like activity. Results are expressed as RLU (mean ± SEM) from three independent experiments. *P < 0.0001 versus T0; **P < 0.001 versus T0; †P < 0.0001 versus UT at 12 hours. (C, D) Effect of SAM treatment on proteasomal chymotrypsin-like (C) or caspase-like (D) activities in 20S proteasome. Five µg of 20S proteasome treated with SAM (1 or 2 mM) or MG132 (20 µM) for 12 hours were analyzed. Results are expressed as RLU (mean ± SEM) from three independent experiments. *P < 0.001 versus control; **P < 0.0001 versus control.
Effects of SAM on Human 26S Proteasome Stability
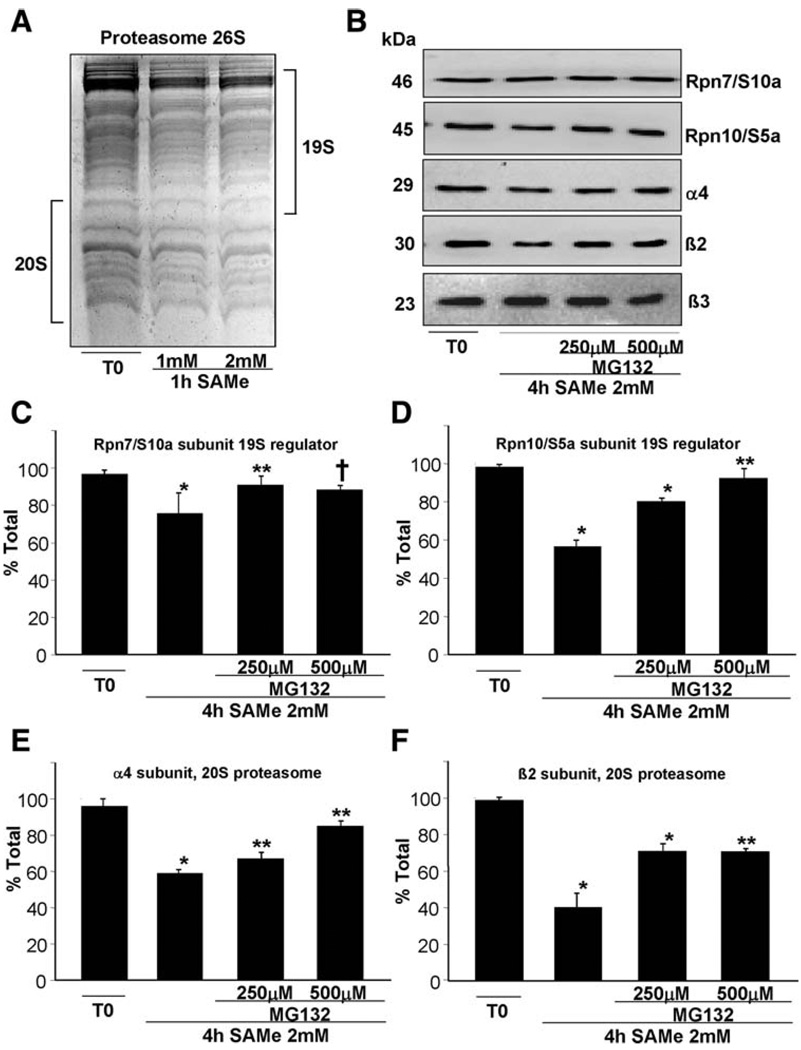
We next examined whether SAM can affect proteosomal stability. Figure 4 shows that the stability of purified human 26S proteasome in vitro is compromised after treatment with SAM. This effect can be easily observed by Coomassie Blue staining of 26S proteasome treated 1 hour at 37°C with SAM (Fig. 4A). Subunits of the 20S and 19S subunits appear to be affected. To better estimate the effect of SAM on the stability of the various subunits of the proteasome, western blot detection was performed using antibodies specifically recognizing subunits of both 20S and 19S (Fig. 4B). The α4 and β2 subunits of the 20S and Rpn10/S5a of 19S are severely destabilized after 4 hours of 2 mM SAM treatment, with a decrease of up to 60% (Fig. 4D–F). Effects on stability of these subunits can already be seen after 1 hour (data not shown). In contrast, the stability of Rpn7/S10a is only partially affected, and β3 is unaffected. Degradation of these proteasome subunits can be prevented by MG132 in a concentration-dependent manner as reported.23
Fig. 4.
In vitro effects of SAM on 26S proteasome stability. (A) Purified human 26S proteasomes were treated with SAM at the indicated times and concentrations. Reactions were analyzed by Coomassie Blue staining of acrylamide gels. (B) Similar reactions in the presence or absence of 2 mM SAM with or without MG132 at the indicated concentrations for 4 hours were analyzed by way of western blotting using antibodies specifically recognizing the α4, β2, and β3 subunits of the 20S and Rpn10/S5a and Rpn7/S10a of the 19S. (C-F) Quantitative results on the subunits from four different experiments using the Quantity One program (Bio-Rad). Results are expressed as fold of T0 (mean ± SEM). (C) *P < 0.002; **P < 0.03; †P < 0.004 versus T0. (D) *P < 0.0001; **P < 0.05 versus T0. (E) *P < 0.0001; **P < 0.003 versus T0. (F) *P < 0.002; **P < 0.0001 versus T0.
Effects of SAM Treatment on Proteasomal Activity, DUSP1 Degradative Pathway, and DUSPs Expression in Mat1a KO Mice
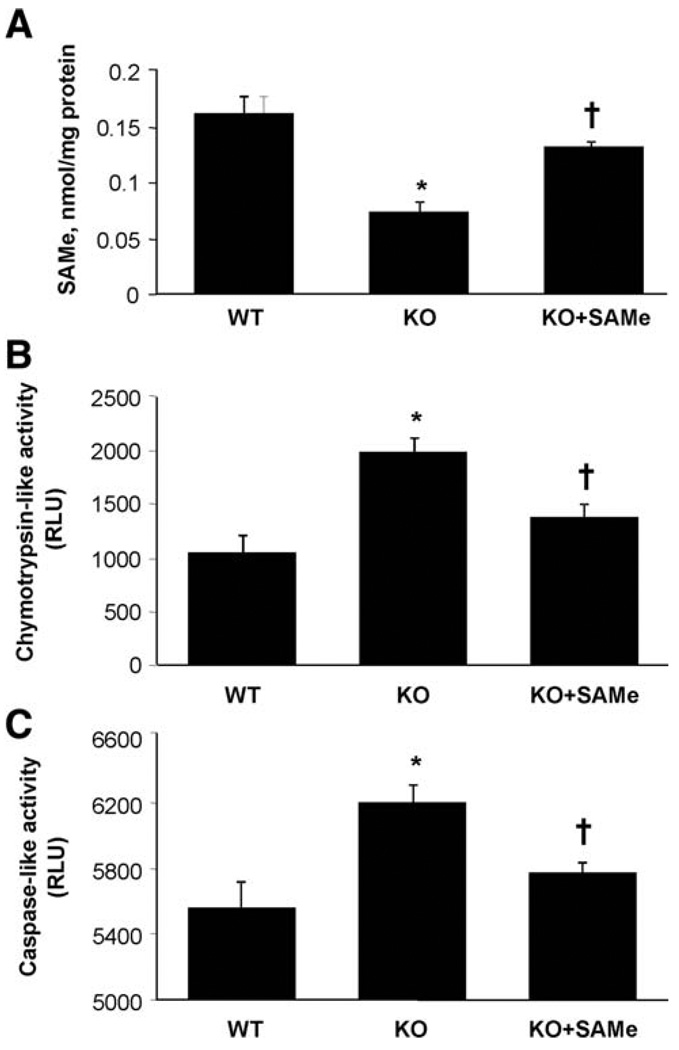
To see if SAM treatment in Mat1a KO mice can restore DUSP1 expression, we treated KO mice with oral SAM administered daily for 7 days. This dose was based on our recent work showing that it was well tolerated and led to a change in gene expression and glutathione levels.24 Figure 5A shows that hepatic SAM levels, which were 55% lower in untreated KO mice, normalized after 1 week of SAM treatment. Chymotrypsin-like (Fig. 5B) and caspase-like (Fig. 5C) activities were higher in untreated KO mice compared with WT, and this was nearly corrected by SAM treatment.
Fig. 5.
Effect of SAM treatment on SAM levels, proteasomal chymotrypsin-like and caspase-like activity in Mat1a KO mice. Protein extracts from livers of 4- to 6-month old Mat1a KO, Mat1a KO treated with SAM (100 mg/kg by gavage once daily for 7 days) or age-matched WT mice were analyzed for SAM levels and proteasomal chymotrypsin-like and caspase-like activity in tissue extracts as described in Materials and Methods. (A) Effect of SAM treatment on hepatic SAM levels in Mat1a KO mice. Results are expressed as nmol of SAM per milligrams of protein (mean ± SEM) from four to nine mice per group. *P < 0.01 versus WT; †P < 0.001 versus KO. (B) Effect of SAM treatment on proteasomal chymotrypsin-like activity in Mat1a KO mice. Results are expressed as RLU (mean ± SEM) from four to nine mice per group. *P < 0.01 versus WT; †P < 0.001 versus KO. (C) Effect of SAM treatment on proteasomal caspase-like activity. Results are expressed as RLU (mean ± SEM) from four to nine mice per group. *P < 0.016 versus WT; †P < 0.02 versus KO.
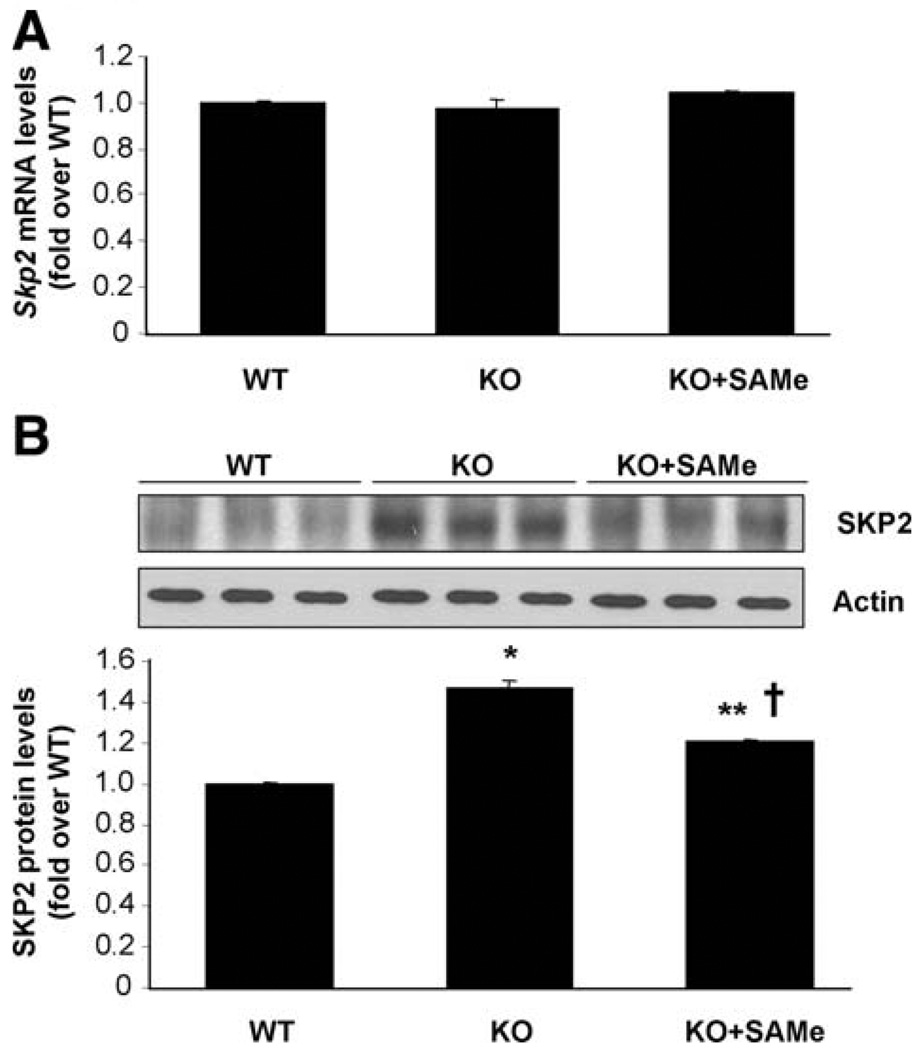
In addition to proteasomal activity, enhanced activity of SKP2, an E3 ligase responsible for DUSP1 ubiquitination, can also increase DUSP1 degradation.6 To examine this possibility, SKP2 expression was measured in Mat1a KO livers with and without SAM treatment. Figure 6 shows that while Skp2 mRNA levels were unchanged (Fig. 6A), SKP2 protein levels were higher in Mat1a KO livers, and this was partially corrected after SAM treatment (Fig. 6B).
Fig. 6.
Effects of SAM treatment on SKP2 expression in Mat1a KO mice. (A) Real-time PCR analysis of Skp2 mRNA levels. Results are expressed as fold of age-matched WT mice (mean ± SEM) from three to four mice in each group. (B) SKP2 protein levels measured by way of western blot analysis. Results are expressed as fold of the data from age-matched WT mice (mean ± SEM) from three WT, four untreated, and four SAM-treated KO mice. *P < 0.001 versus WT; **P < 0.03 versus WT; †P < 0.02 versus KO.
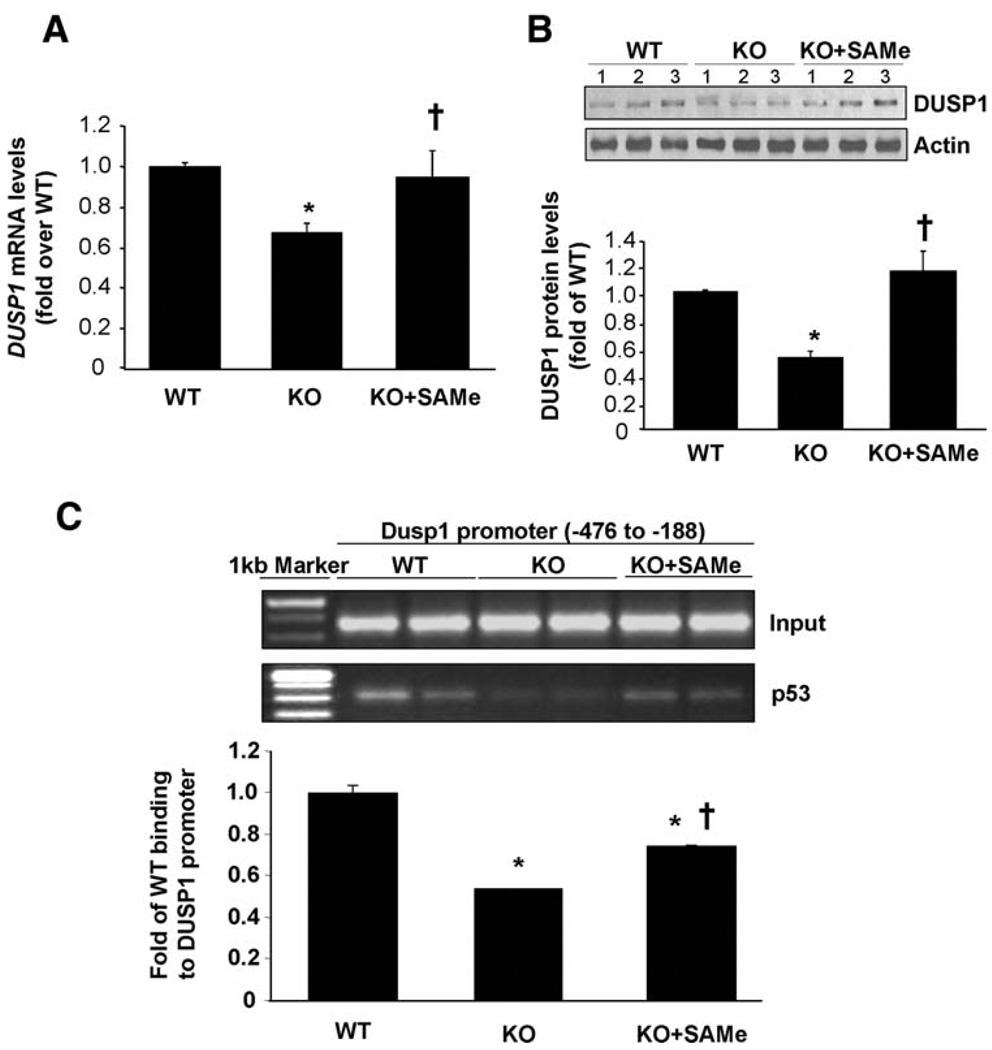
DUSP1 mRNA levels, which were about 40% lower in untreated KO mice, normalized after SAM treatment (Fig. 7A). Similarly, SAM treatment also restored DUSP1 protein levels in Mat1a KO mice, to even higher than WT levels (Fig. 7B). Because p53 can trans-activate the regulatory region of DUSP1,20 it is possible that SAM can influence this interaction in vivo. Consistent with this, ChIP assay showed that binding of p53 to the DUSP1 promoter region was lower in untreated KO mice compared with WT, and this was partially corrected by SAM treatment (Fig. 7C).
Fig. 7.
Effects of SAM treatment on DUSP1 expression in Mat1a KO mice. (A) Real-time PCR analysis measured DUSP1 mRNA levels. Results are expressed as fold of age-matched WT mice (mean ± SEM) from four to nine mice per group. *P < 0.001 versus WT; †P < 0.01 versus KO. (B) DUSP1 protein levels measured by way of western blot analysis. Results are expressed as fold of WT mice (mean ± SEM) from three to nine mice per group. *P < 0.001 versus WT; †P < 0.001 versus KO. (C) SAM increased the physical association of p53 with chromatin containing the regulatory region of DUSP1. Thirty milligrams of tissue from livers of 4- to 6-month-old untreated, SAM-treated Mat1a KO, or age-matched WT mice were used for ChIP assay. Input genomic DNA (Input) was used as a loading control, and immunoprecipitation with an antibody against actin was used as a negative control (not shown). Results were normalized to the input and expressed as fold of control binding to the DUSP1 promoter (mean ± SEM) from three WT, four untreated, and four SAM-treated mice. *P < 0.001 versus WT; †P < 0.02 versus KO.
Effects of SAM Treatment on ERK Activity in Mat1a KO Mice
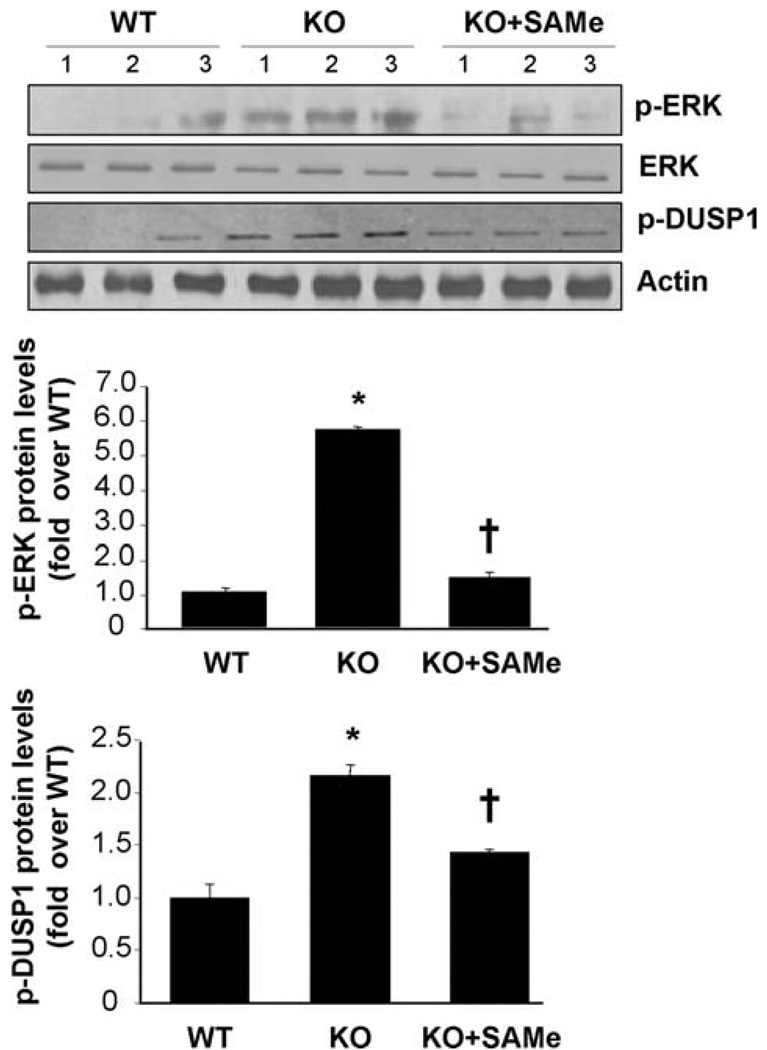
One week of SAM treatment resulted in lowering of ERK activity as indicated by phosphorylated ERK and phosphorylated DUSP1 levels (Fig. 8).
Fig. 8.
Effects of SAM treatment on activity of ERK in Mat1a KO mouse livers. Phosphorylation of ERK and DUSP1 was used as an indirect measure of ERK activity using western blot analyses for phosphorylated ERK, phosphorylated DUSP1, total ERK, DUSP1, and actin as described in Materials and Methods in untreated mice, SAM-treated Mat1a KO mice, and WT controls. Results are expressed as fold of WT mice (mean ± SEM) from three to nine mice per group. *P < 0.001 versus WT; †P < 0.001 versus KO.
Discussion
DUSP1 encodes a dual threonine/tyrosine phosphatase that specifically dephosphorylates and inactivates MAPKs.5,20 DUSP1, also referred to as MKP-1, CL100, or 3CH134, is a prototypic member of DUSPs family.25 Thus far, at least 10 family members have been cloned.5 Whereas the expression of some DUSPs is tissue-specific, DUSP1 is widely distributed, and its expression is responsive to various environmental stimuli, including mitogens, oxidative stress, heat shock, ultraviolet radiation, high osmotic pressure, and hormones.26 DUSP1 is encoded by an immediate-early gene, and its transcription is rapidly induced in response to many of the stimuli that activate MAPKs, which is a key mechanism to control the activity of MAPKs.5,27 After induction, DUSP1 is in turn rapidly degraded by the ubiquitin-proteasome pathway.6
MAPKs are components of intracellular signaling pathways that control a vast array of cellular processes such as cell growth, immune response, infection, and injury. The ERK pathway is phosphorylation cascades, in which the activation of each MAPK requires dual phosphorylation by specific MAPK kinase at the threonine and tyrosine residues of the T x Y sites (where x represents glutamate in ERK) in the activation loop, and dephosphorylation of these residues by DUSP1 terminates such activation.25–27 Each activated MAPK specifically targets a variety of proteins such as downstream kinases and transcription factors that regulate expression of particular genes, many of which serve to inhibit MAPK feedback.28 In addition, physical interaction of MAPKs with DUSP1 activates the phosphatase activity to inhibit MAPKs.6 These are some of the regulatory mechanisms to keep MAPK activity in check.
In many human cancers, DUSP1 expression was found to be increased in the early carcinogenic stage but fell progressively with higher histologic grade and metastasis.29 In human HCC, DUSP1 expression has been reported to be inversely correlated with proliferation index and microvessel density, and directly correlated with apoptotic index and survival.30 Decreased DUSP1 expression in HCC was one of the independent prognosticators for the survival of HCC patients.31 The down-regulation of DUSP1 occurs predominantly as the tumor progresses from localized to advanced disease. However, the mechanisms by which DUSP1 expression decreases are not entirely clear.30
Recently, we reported that the protein level of apurinic/apyrimidinic endonuclease 1 (APEX1), a key protein involved in DNA repair, is lower in Mat1a KO livers and cultured hepatocytes.22 We further showed that SAM treatment stabilized APEX1 protein in vitro.22 These results support the notion that SAM regulates proteasomal activity, but proof is lacking. Because DUSP1 is a protein well known to undergo proteasomal degradation, which is induced with sustained ERK activation,7 and the observation that ERK1 activity is increased in Mat1a KO livers,14 we anticipated lower levels of DUSP1 protein in Mat1a KO livers and that SAM is likely to regulate its expression. In the present study, we confirmed our speculation and provided definitive proof that SAM can stabilize proteins by inhibiting proteasomal activity. Furthermore, our data support the scenario that a chronically decreased SAM level can lower DUSP1 expression and allow higher ERK activity to persist.
We found that hepatic DUSP1 expression is lower at both the mRNA and protein levels in Mat1a KO mice. We confirmed higher ERK activity as we have reported previously.14 Using cultured hepatocytes as an in vitro model for loss of MAT1A and SAM depletion, we observed the same changes in DUSP1, though with an even more profound depletion in its protein level compared with Mat1a KO livers. DUSP1 mRNA is very labile, because its levels fell by 94% in 2 hours when de novo RNA synthesis was inhibited. SAM treatment in cultured hepatocytes and in Mat1a KO mice protected against decreased mRNA and protein levels. SAM exerts its protective effect on the DUSP1 mRNA at the transcriptional as well as posttranscriptional levels. SAM is likely to increase DUSP1 transcription by way of p53, which is a transcriptional activator of DUSP1.20 This is because SAM stabilizes APEX1, which in turn enhances p53-dependent transcriptional activity.22,32 This notion is further supported by the ChIP findings, because p53 binding to the DUSP1 promoter region containing the p53 element was lower in Mat1a KO livers and partially corrected after SAM treatment. SAM treatment also increased mRNA stability slightly but the molecular mechanism remains to be elucidated. SAM had a more protective effect on the DUSP1 protein level, which was equal to that of the proteasomal inhibitor MG132. Indeed, proteasomal activity was higher in Mat1a KO livers and in cultured hepatocytes, and SAM treatment largely prevented or corrected these changes. The evidence that SAM can directly inhibit proteasomal activity is its ability to inhibit both chymotrypsin-like and caspase-like activities in pure 20S proteasomes in as little as 1 hour. In addition, incubating human 26S proteasome with SAM led to a rapid (1-hour) degradation of some but not all of the subunits, and this effect was blocked by MG132. This result is consistent with one of two possibilities: either SAM activates the intrinsic protease activity of the proteasome, leading to degradation of some of its subunits, or it modifies the proteasome posttranslationally (such as methylation), which makes the protein more susceptible to protease degradation. These possibilities will be investigated in the future.
In addition to enhanced proteasomal degradation, chronic SAM depletion also resulted in increased protein levels of SKP2, an E3 ligase responsible for ubiquitination of DUSP1. This increase can further contribute to destabilization of DUSP1 and can be partially corrected by SAM treatment. How SAM level regulates SKP2 protein level is unclear at the present time and will require additional study.
Because DUSP1 and ERK exert reciprocal regulation on each other, it is possible that SAM protects DUSP1 protein degradation by inhibiting ERK activity. However, SAM treatment for up to 27 hours had no influence on ERK activity in cultured hepatocytes, which makes it unlikely.33 We also did not observe a change in ERK activity after SAM treatment in cultured hepatocytes (data not shown). However, SAM treatment in Mat1a KO mice reduced ERK activity, which likely reflects different durations of treatment (7 days versus 12 hours in culture). With lowering of ERK activity, there is less phosphorylation of DUSP1 and less proteasomal degradation. This can then feed forward to further reduce ERK.
Our findings have clear relevance to many chronic human liver diseases that are associated with increased risk for HCC. Most chronic liver diseases have lower MAT1A expression and decreased MAT activity, culminating in reduced SAM biosynthesis.11 Results from the present study suggest that chronic SAM depletion facilitates DUSP1 proteasomal degradation and allows ERK (and other MAPK) activation to persist. SAM has been demonstrated to exert chemopreventive effect in several rodent models of HCC.34,35 There are many potential mechanisms, including preventing hypomethylation and increased expression of proto-oncogenes, 34 exerting a selectively proapoptotic effect in liver cancer cells36 and inhibiting angiogenesis.35 With the new finding that SAM can stabilize proteins such as APEX1 and DUSP1 by inhibiting the proteasomal pathway, it greatly widens our understanding of how SAM can influence so many cellular activities.
In conclusion, DUSP1 expression falls when SAM is depleted and SAM treatment can increase DUSP1 transcription and stabilize its mRNA and protein. SAM stabilizes DUSP1 protein by inhibiting the proteasomal pathway. Given that patients with chronic liver disease have impaired hepatic SAM biosynthesis, our findings are highly relevant to the molecular mechanisms of the predisposition to HCC and how SAM may function as a chemopreventive agent.
Acknowledgments
Supported by National Institutes of Health Grants DK51719 (to S. C. Lu) and AT1576 (to S. C. Lu and J. M. Mato); the Spanish Ministry of Education and Science Ramón y Cajal Program Grant BFU 2008-01108/BMC (to M. S. Rodríguez); and the Spanish National Plan for Research, Development, and Innovation Grants SAF 2005-00855 and HEPADIP-EULSHM-CT-205 (to J. M. Mato). Primary hepatocytes were provided by the Cell Culture Core of the USC Research Center for Liver Diseases (DK48522). CIBERehd is funded by the Carlos III Health Institute. M. L. Tomasi is a postdoctoral fellow partly funded by the University of Sassari.
Abbreviations
- APEX1
apurinic/apyrimidinic endonuclease 1
- ChIP
chromatin immunoprecipitation
- ERK
extracellular signal-regulated kinase
- HCC
hepatocellular carcinoma
- KO
knockout
- MAPK
mitogen-activated protein kinase
- MAT
methionine adenosyltransferase
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- RLU
relative light units
- SAM
S-adenosylmethionine
- SEM
standard error of the mean
- SKP2
S-phase kinase-associated protein 2
- WT
wild type
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 2.Cano E, Mahadevan LC. Parallel signal processing among mammalian MAPKs. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 3.Tonks NK, Neel BG. From form to function: signaling by protein tyrosine phosphatases. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 4.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Bil. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylate MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 6.Lin YW, Yang JL. Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. J Biol Chem. 2006;281:915–926. doi: 10.1074/jbc.M508720200. [DOI] [PubMed] [Google Scholar]
- 7.Li YW, Chuang SM, Yang JL. ERK1/2 achieves sustained activation by stimulating MAPK phosphatase 1 degradation via the ubiquitin-proteasome pathway. J Biol Chem. 2003;278:21534–21541. doi: 10.1074/jbc.M301854200. [DOI] [PubMed] [Google Scholar]
- 8.Groll M, Ditzel L, Lo̊we J, Stock D, Stock D, Bochtler M, Bartunik HD, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;368:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 9.An B, Goldfarb RH, Siman R, Dou QP. Novel dipeptidyl proteaseome inhibitors overcome Bcl-2 protective function and selectively accumulate the cyclin-dependent kinase inhibitor p27 and induce apoptosis in transformed, but not normal, human fibroblast. Cell Death Differ. 1998;5:1062–1075. doi: 10.1038/sj.cdd.4400436. [DOI] [PubMed] [Google Scholar]
- 10.Wiesenauer CA, Yip-Schneider MT, Wang Y, Schmidt CM. multiple anticancer effects of blocking MEK-ERK signaling in hepatocellular carcinoma. J Am Coll Surg. 2004;198:410–421. doi: 10.1016/j.jamcollsurg.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45:1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 12.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, García-Trevijano ER, Huang ZZ, Chen L, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Zeng Y, Yang H, Lee TD, French SW, Corrales FJ, et al. Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J. 2004;18:914–916. doi: 10.1096/fj.03-1204fje. [DOI] [PubMed] [Google Scholar]
- 15.Ramani K, Yang H, Xia M, Ara AI, Mato JM, Lu SC. Leptin’s mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;47:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Trevijano ER, Latasa MU, Carretero MV, Berasain C, Mato JM, Avila MA. S-adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: a new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J. 2000;14:2511–2518. doi: 10.1096/fj.00-0121com. [DOI] [PubMed] [Google Scholar]
- 17.Freshney R. Culture of Animal Cells: A manual of Basic Technique. NewYork: Alan R. Liss Inc.; 1987. [Google Scholar]
- 18.Yang H, Ara AI, Magilnick N, Xia M, Ramani K, Chen H, et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veal N, Hsieh CL, Xiong S, Mato JM, Lu S, Tsukamoto H. Inhibition of lipopolysaccharide-stimulated TNF-alpha promoter activity by S-adenosylmethionine and 5′-methylthioadenosine. Am J Physiol Gastrointest Liver Physiol. 2004;287:G352–G362. doi: 10.1152/ajpgi.00316.2003. [DOI] [PubMed] [Google Scholar]
- 20.Liu YX, Wang J, Guo J, Wu J, Lieberman HB, Yin Y. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol Cancer Res. 2008;6:624–633. doi: 10.1158/1541-7786.MCR-07-2019. [DOI] [PubMed] [Google Scholar]
- 21.Coux O, Goldberg AL. Enzymes catalyzing ubiquitination and proteolytic processing of the p105 precursor of nuclear factor kappaB1. J Biol Chem. 1998;273:8820–8828. doi: 10.1074/jbc.273.15.8820. [DOI] [PubMed] [Google Scholar]
- 22.Tomasi ML, Iglesias-Ara A, Yang H, Ramani K, Feo F, Pascale MR, et al. S-adenosylmethionine regulates apurinic/apyrimidinic endonuclease 1 stability: implication in hepatocarcinogenesis. Gastroenterology. 2009:1025–1036. doi: 10.1053/j.gastro.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang HP, Ramani K, Xia M, Ko KS, Li TWH, Oh P, et al. Dysregulation of glutathione synthesis during cholestasis in mice: molecular mechanisms and therapeutic implications. Hepatology. 2009;49:1982–1991. doi: 10.1002/hep.22908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 26.Franklin CC, Kraft AS. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J Biol Chem. 1997;272:16917–16923. doi: 10.1074/jbc.272.27.16917. [DOI] [PubMed] [Google Scholar]
- 27.Keyse SM, Emslie EA. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359:644–647. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- 28.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 29.Magi-Galluzzi C, Mishra R, Fiorentino M, Montironi R, Yao H, Capodieci P, et al. Mitogen-activated protein kinase phosphatase 1 is overexpressed in prostate cancers and is inversely related to apoptosis. Lab Invest. 1997;76:37–51. [PubMed] [Google Scholar]
- 30.Calvisi DF, Pinna F, Meloni F, Ladu S, Pellegrino R, Sini M, et al. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res. 2008;68:4192–4200. doi: 10.1158/0008-5472.CAN-07-6157. [DOI] [PubMed] [Google Scholar]
- 31.Tsujita E, Taketomi A, Gion T, Kuroda Y, Endo K, Watanabe A, et al. Suppressed MKP-1 is an independent predictor of outcome in patients with hepatocellular carcinoma. Oncology. 2005;69:342–347. doi: 10.1159/000089766. [DOI] [PubMed] [Google Scholar]
- 32.Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Trevijano ER, Martínez-Chantar ML, Latasa MU, Mato JM, Avila MA. NO sensitizes rat hepatocytes to hepatocyte growth factor-induced proliferation through the modulation of S-adenosylmethionine levels. Gastroenterology. 2002;122:1355–1363. doi: 10.1053/gast.2002.33020. [DOI] [PubMed] [Google Scholar]
- 34.Pascale RM, Simile MM, De Miglio MR, Nufris A, Daino L, Seddaiu MA, et al. Chemoprevention by S-adenosyl-L-methionine of rat liver carcinogenesis initiated by 1,2-dimethylhydrazine and promoted by orotic acid. Carcinogenesis. 1995;16:427–430. doi: 10.1093/carcin/16.2.427. [DOI] [PubMed] [Google Scholar]
- 35.Lu SC, Ramani K, Ou XP, Lin M, Yu V, Ko K, et al. S-adenosylmethionine in the chemoprevention and treatment of hepatocellular carcinoma in a rat model. Hepatology. 2009;50:462–471. doi: 10.1002/hep.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Sadda MR, Li M, Zeng Y, Chen L, Bae W, et al. S-adenosylmethionine and its metabolite induce apoptosis in HepG2 cells: role of protein phosphatase 1 and Bcl-xS. Hepatology. 2004;40:221–231. doi: 10.1002/hep.20274. [DOI] [PubMed] [Google Scholar]