Abstract
Plant viral vectors have great potential in rapid production of important pharmaceutical proteins. However, high-yield production of heterooligomeric proteins that require the expression and assembly of two or more protein subunits often suffers problems due to the “competing” nature of viral vectors derived from the same virus. Previously we reported that a bean yellow dwarf virus (BeYDV)-derived, three-component DNA replicon system allows rapid production of single recombinant proteins in plants (Huang et al. 2009). In this article, we report further development of this expression system for its application in high-yield production of oligomeric protein complexes including monoclonal antibodies (mAbs) in plants. We showed that the BeYDV replicon system permits simultaneous efficient replication of two DNA replicons and thus, high-level accumulation of two recombinant proteins in the same plant cell. We also demonstrated that a single vector that contains multiple replicon cassettes was as efficient as the three-component system in driving the expression of two distinct proteins. Using either the non-competing, three-vector system or the multi-replicon single vector, we produced both the heavy and light chain subunits of a protective IgG mAb 6D8 against Ebola virus GP1 (Wilson et al. 2000) at 0.5 mg of mAb per gram leaf fresh weight within 4 days post infiltration of Nicotiana benthamiana leaves. We further demonstrated that full-size tetrameric IgG complex containing two heavy and two light chains was efficiently assembled and readily purified, and retained its functionality in specific binding to inactivated Ebola virus. Thus, our single-vector replicon system provides high-yield production capacity for heterooligomeric proteins, yet eliminates the difficult task of identifying non-competing virus and the need for co-infection of multiple expression modules. The multi-replicon vector represents a significant advance in transient expression technology for antibody production in plants.
Keywords: Replicon, Monoclonal antibody, Transient expression, Plant-made Pharmaceuticals, replicon, viral vector, therapeutics
INTRODUCTION
The use of plants as factories to produce important pharmaceutical proteins, including vaccine antigens and therapeutic monoclonal antibodies (mAbs), is attractive for several reasons. First, plants can produce large volumes of proteins efficiently and sustainably and, under certain conditions, can have significant advantages in manufacturing costs (Giddings 2001; Hood et al. 2002). Second, the growth of plants does not require animal- or human-derived nutrients and therefore has minimal risks of contamination with animal or human pathogens and toxins. Third, plants possess an endomembrane system and secretory pathway that are similar to mammalian cells (Vitale and Pedrazzini 2005), permitting appropriate post-translational modification of recombinant proteins, which are often critical to their proper functions.
Since the first demonstration of mAb expression in transgenic plants in 1989 (Hiatt et al. 1989), many different forms of mAbs have been produced in plant systems (reviewed in (Chen 2008; Ma 2003; Ma et al. 2005; Stoger et al. 2004)) using either transient, viral based expression systems (Canizares et al. 2005; Yusibov et al. 2006) or stably transgenic plants (Floss et al. 2007; Giddings et al. 2000; Twyman et al. 2005). The latter strategy suffers from the long time frame required to generate transgenic plants and the generally low protein yields (<40 µg/g of fresh biomass). In contrast, plant viral vectors have the potential to rapidly produce high-levels of foreign proteins owing to their efficient replication and the resulting high copy numbers of gene of interest (Lico et al. 2008). However, until recently it has been difficult to efficiently express multi-component mAbs with plant viral vectors, because co-delivery of viral vectors built on the same virus backbone always results in early segregation and subsequent preferential amplification of one of the vectors in one cell – a common scenario of “competing replicons” (Dietrich and Maiss 2003; Diveki et al. 2002; Hull and Plaskitt 1970). This problem has been recently overcome by utilizing two sets of vectors derived from non-competing tobacco mosaic virus (TMV) and potato virus X (PVX), to produce full-size IgG at levels as high as 0.5 mg of mAb per gram leaf fresh weight (Giritch et al. 2006). However, it will be a difficult task to identify additional viruses compatible with both TMV and PVX expression systems to allow efficient co-expression and assembly of three or more distinct subunit proteins such as secretory IgA and IgM in same cells. Thus, there is a need to develop an advanced transient expression system which consists of a minimum number of vectors but still permits high-level co-expression and assembly of multiple subunit proteins.
Previously we reported that a bean yellow dwarf virus (BeYDV)-derived, three-component DNA replicon system allows rapid high-yield production of single recombinant proteins in plants (Huang et al. 2009). In the present study, we showed that this three component replicon system permits simultaneous efficient replication of two separate DNA replicons and high-level accumulation of two proteins encoded by the replicons. Moreover, we constructed a single vector that contains multiple replicon cassettes and demonstrated that it was as efficient as the three-component system in directing expression of two distinct protein molecules. Using either the non-competing, three-vector system or the multi-replicon single vector, we demonstrated co-expression of green fluorescent protein (GFP) and Discosoma sp. red fluorescent protein (DsRed). Using proteins that fluoresce at different wavelengths, it was possible to show that two different foreign proteins were expressed in the same cells. Moreover, we produced both the heavy and light chain molecules of a protective IgG mAb 6D8 against Ebola virus GP1 (Wilson et al. 2000) at ~0.5 mg of mAb per gram leaf fresh weight (LFW) within 4 to 8 days post infiltration (dpi) of Nicotiana benthamiana leaves. We further demonstrated that full-size IgG complex containing two heavy chains and two light chains was efficiently assembled, readily purified, and functioned properly to bind inactivated Ebola virus. Thus, our single-vector replicon system provides high-yield production capacity, eliminates the difficult task of identifying non-competing virus, and obviates the need for co-infection of multiple expression modules. The multi-replicon vector represents a significant advance in transient expression technology for antibody production in plants.
MATERIALS AND METHODS
Vector construction
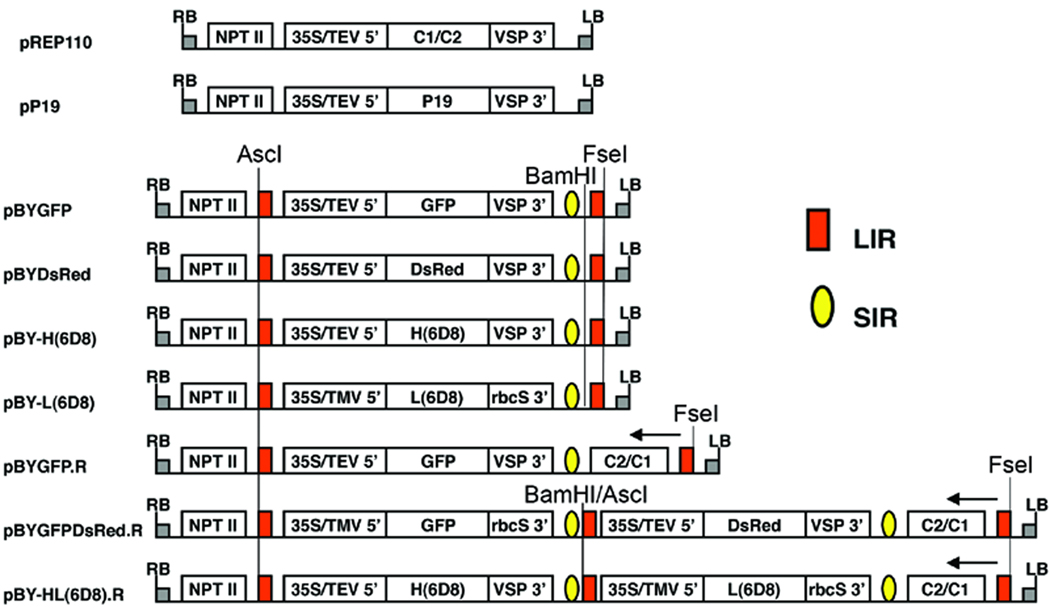
The construction of plasmids pREP110, pP19, pBYGFP and pBYGFP.R (Fig. 1) has been previously described (Huang et al. 2009).
Figure 1.
Schematic representation of the T-DNA regions of the vectors used in this study. 35S/TEV 5’, CaMV 35S promoter with tobacco etch virus 5’ UTR; 35S/TMV 5’, CaMV 35S promoter with tobacco mosaic virus 5’ UTR; VSP 3’, soybean vspB gene 3’ element; rbcS 3’, pea rbcS gene 3’ element, NPT II, expression cassette encoding nptII gene for kanamycin resistance; LIR (red box), long intergenic region of BeYDV genome; SIR (yellow oval), short intergenic region of BeYDV genome; C1/C2 (pREP110) or C2/C1 (pBYGFP.R, pBYGFPDsRed.R, and pBY-HL(6D8).R), BeYDV ORFs C1 and C2, encoding replication initiation protein (Rep) and RepA; LB and RB, the left and right borders of the T-DNA region. Arrows at right-most LIR in pBYGFP.R, pBYGFPDsRed.R, and pBY-HL(6D8).R indicate direction of transcription, from the LIR promoter, of the complimentary sense genes C1/C2. In the dual replicon constructs pBYGFPDsRed.R and pBY-HL(6D8).R, the middle LIR functions in concert with the left-side LIR to release the left-side replicon, and it also functions in concert with the right-side LIR to release the right-side replicon. In pBYGFPDsRed.R and pBY-HL(6D8).R, BamHI/AscI marked at the middle LIR is the site of fusion of two replicons via the Klenow-filled BamHI site of the left-side replicon and the Klenow-filled AscI site of the right-side replicon.
For the construction of pBYDsRed (Fig. 1), the DsRed gene was amplified from pDsRed1-1 (Clontech cat# 6922-1) with primers 5’-ATCGTCTAGAACCATGGTGCGCTCCTCCAAG and 5’-ATTAGAGCTCCTACAGGAACAGGTGGTG, digested with XbaI and SacI, and ligated into pIBT210 to make pIBT-DsRed, from which the XhoI-SacI fragment was substituted into pBYGFP to make pBYDsRed (Fig. 1). Tandem dual replicon constructs (e.g. pBYGFPDsRed.R, Fig. 1) were designed to contain two replicons in tandem, linked by a LIR element. The first replicon contains LIR-35S cassette-SIR-LIR, and the second contains LIR-35S cassette-SIR-C2/C1-LIR, with the bold/underlined LIR element of each cassette overlapping to generate LIR-35S cassette-SIR-LIR-35S cassette-SIR-C2/C1-LIR. Each replicon is thus defined by the LIR borders. We used CaMV 35S promoters with a single enhancer element, obtained by amplification of the expression cassettes in pBYDsRed and pIBT210.3 (Judge et al. 2004) with primers 35S-Sda (5’-TGACCTGCAGgCATGGTGGAGCACGACA) and VSPHT (5’-TGAATAGTGCATATCAGCATACCTTA). The 35S promoter and 5’-UTR of TMV in the fragment amplified from pIBT210.3 (SdaI-NcoI), the GFP gene from pBYGFP (NcoI-KpnI), and the pea ribulose1,5-bisphosphate carboxylase small subunit (rbcS) terminator (Friedrichsen et al. 2000) on a KpnI-EcoRI fragment were ligated together into the PstI and EcoRI sites of pBY024 (Mor et al. 2003) to make pBYGFP210.3. The fragment containing the GFP partial replicon (AscI)-LIR-35S/TMV/GFP-SIR-(blunt), obtained by digestion of pBYGFP210.3 with BamHI and filling with Klenow enzyme, then digestion with AscI, was ligated with the 35S-Sda primer-amplified pBYDsRed partial replicon (blunt)-LIR-35S/TEV/DsRed-SIR-(SacI) digested with AscI and filled with Klenow enzyme, and then digested with SacI, into the vector pBYHBc.R (Huang et al. 2009) that had been digested with AscI and SacI, to make pBY-GFPDsRed.R (Fig. 1).
The gene sequences for heavy (H2) and light (K3) chains of mouse monoclonal antibody 6D8 (Wilson et al. 2000) were de-immunized for humans by substitution of human constant region sequences for gamma type 1 and kappa chains (Biovation, Edinburgh, Scotland). The resulting sequences were used to design plant codon-optimized genes, and synthesized commercially (Retrogen, San Diego, CA). The H2 gene in pCHF4-6D8-H2 (Mapp Biopharmaceutical, Inc.) was end-tailored to add a C-terminal ‘SEKDEL’ hexapeptide by PCR with the primer H2-SEKDEL-Kpn (5’-GCGGTACCTTAAAGCTCATCCTTCTCTGATTTACCCGGAGACAAGGAGAG), digested with NcoI and KpnI, and inserted into pPS1 (Huang and Mason 2004) to make p6D8-H2, from which the XhoI-EcoRI fragment containing TEV5’-UTR-H2-VSP3’ was substituted into pBYGFP to make pBYH(6D8) (Fig. 1). The K3 gene in pCHF4-6D8-K3 (Mapp Biopharmaceutical, Inc.) was obtained as an NcoI-KpnI fragment, ligated with the 35S promoter-TMV5’ and the rbcS3’ elements, and substituted into pBYGFP vector to make pBY-L(6D8) (Fig. 1). The replicon (LIR to SIR) from pBY-L(6D8) was substituted into pBYGFP.R to make pBYK3R. The H2-SEKDEL fragment was amplified from pBY-H(6D8) with primers H2-Xba (5’- ACGATCTAGAACAATGGGATGGTCTTGCATC) and VSPHT, digested with XbaI and KpnI, and substituted into the vector pBY027 (Mor et al. 2003) to make pBY-H2K210. The replicon from pBY-H2K210 was then inserted into pBYK3R to make the tandem dual replicon construct pBY-HL(6D8).R (Fig. 1).
Agroinfiltration of Nicotiana benthamiana leaves
Binary vectors were separately introduced into Agrobacterium tumefaciens LBA4404 by electroporation. The agroinfiltration procedure was performed as previously described (Huang et al. 2009).
Plant DNA extraction and Southern blotting
Total DNA extraction and Southern blotting was performed as previously described (Huang et al. 2009). Digoxygenin (DIG)-labeled gene-specific probes were synthesized by PCR with primers (5’-GTCACCATGGTGAGCAAGGGCGAG -) and (5’-CTCAGGATCCTTACTTGTACAGCTCGTC -) for the GFP gene, and with primers (5’-ATCGTCTAGAACCATGGTGCGCTCCTCCAAG-) and (5’-ATTAGAGCTCCTACAGGAACAGGTGGTG-) for the DsRed gene, respectively, according to the manufacturer’s instructions (Roche Applied Science, Indianapolis, IN).
Protein analysis
Plant tissue harvest
For expression time-course experiments, Agroinfiltrated Nicotiana benthamiana leaves were harvest on days 2, 4, 6, and 8 dpi. For other protein analysis, plant leaves were harvest 4 dpi.
Protein extraction
Total protein extract was obtained by homogenizing leaves with extraction buffer (25mM sodium phosphate, pH 6.6, 100mM NaCl, 1mM EDTA, 0.1% TritonX-100, 10mg/ml sodium ascorbate, 10µg/ml leupeptin, 0.3mg/ml PMSF) using a FastPrep machine (Bio101, Vista, CA) following the manufacture’s instruction. The crude plant extract was clarified by centrifugation at 14,000g for 10 min at 4°C.
ELISA
The clarified total protein extract was analyzed by ELISA designed to detect the assembled form of mAb (with both light and heavy chains) as described previously (Giritch et al. 2006). Briefly, plates were coated with a goat anti-human IgG specific to gamma heavy chain (Southern Biotech, AL). After incubation with plant protein extract, a HRP-conjugated anti-human-kappa chain antibody (Southern Biotech, AL) was used as the detection antibody, using human IgG reference standard (Southern Biotech, AL).
SDS-PAGE and western blot
SDS-PAGE and western blotting were performed as previously described (Santi et al. 2008). Briefly, plant protein example or human IgG standard was mixed with sample buffer either under reducing or nonreducing condition and then subjected to SDS-PAGE. PAGE gels were either stained with Coomassie blue or used to transfer proteins onto PVDF membranes. The anti-human gamma and kappa specific antibodies described above (ELISA) were used for western blot analysis.
Antigen binding assay
Polyvinyl chloride 96-well ELISA plates were coated with 50µl of 1:1000-diluted irradiated Ebola virus (a gift from Dr. William Pratt, USAMRIID) and incubated at 4°C for 12 hr. Plates were blocked with 5% skim milk in PBST (10.6mM Na2HPO4, 2.9mM KH2PO4, 104.7mM NaCl, 0.05% Tween-20) at room temperature for 2 hrs. Subsequently, the plates were incubated with different concentrations of the plant-derived 6D8 mAb or human IgG (Southern Biotech, AL) diluted in 1% skim milk in PBST for 1 hr at 37°C. HRP-conjugated goat anti-human IgG (Southern Biotech) was then added and incubated for 1 hr at 37°C. The plates were developed with TMB substrate (KPL inc., MD) and the OD was read at 450nm.
6D8 antibody purification
N. benthamiana leaves infiltrated with 6D8 mAb constructs were harvest on 4 dpi and were homogenized with the extraction buffer (25mM sodium phosphate, pH 6.6, 100mM NaCl). Crude extract was filtered through Miracloth and centrifuged at 17,700g for 15 min at 4°C to remove cell debris. Ammonium sulfate was slowly added to the clarified plant extracts to 35% saturation with thorough mixing at 4°C. The sample was centrifuged at 17,700g for 15 min at 4°C and the pellet was saved for analysis. The 35% ammonium sulfate supernatant was further processed by adding ammonium sulfate to 60% saturation. The sample was again centrifuged at 17,700g for 15 min and the supernatant was discarded. The 60% ammonium sulfate pellet was resuspended in the extraction buffer and was then applied to a Protein G column (Pierce, IL). After sample loading, the resin was washed with the extraction buffer and the column was eluted with 50mM sodium citrate, pH2.5. The elution fraction was neutralized immediately with 1M Tris-base to a final pH of 7.5. The purity of 6D8 mAb was determined by quantitating Coomassie blue–stained protein bands on SDS-PAGE using a densitometer.
Isolation of N. benthamiana mesophyll protoplasts and fluorescence microscopy
Protoplasts were released from infiltrated leaf tissue by incubation for 10–16 hours at room temperature in a solution containing 0.4M mannitol, 8mM CaCl2, 0.4% cellulase and 0.4% Macerozyme R-10. Released protoplasts were filtrated through a nylon mesh with a 62-µm pore diameter.
Visualization of GFP and DsRed
Leaves co-expressing GFP and DsRed were viewed under a UVGL-25 lamp (UVP, Upland, CA). Protoplasts were viewed with a Nikon inverted microscope with GFP filter sets (Chroma #41028; excitation filter, 500/20 nm; emission filter, 535/30 nm) and DsRed filter sets (Chroma #42005; excitation filter, 540/40nm; emission filter, 600/50 nm).
RESULTS
The three-vector BeYDV replicon system is non-competing
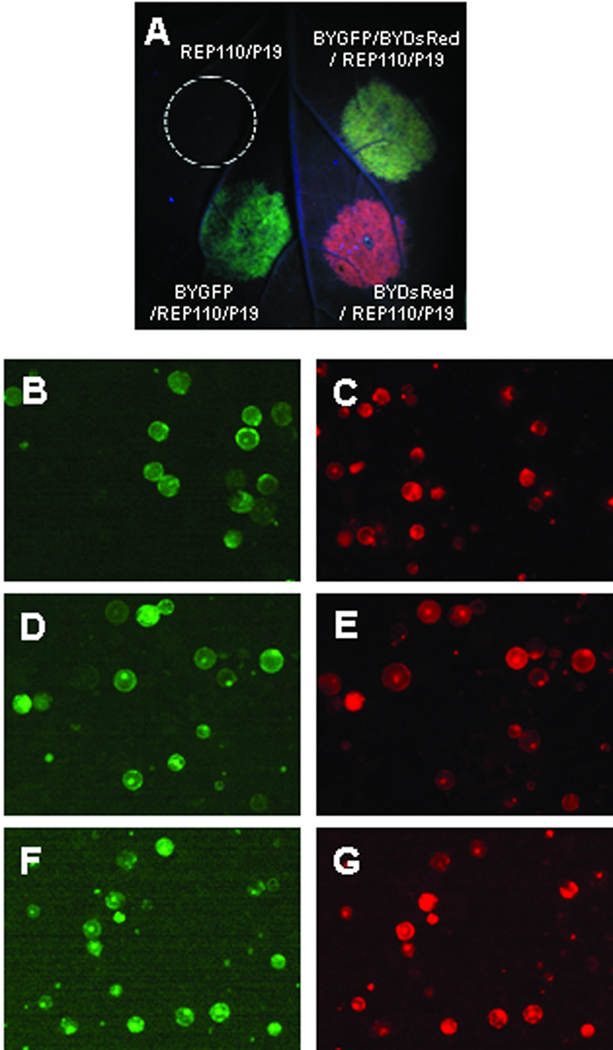
We previously showed (Huang et al. 2009) that a BeYDV-derived three-vector (e.g., pBYGFP, pREP110 and pP19 in Fig. 1) transient expression system provides extremely efficient replication of a single DNA replicon and high-level expression in plant cells of protein encoded by the single replicon vector (e.g., pBYGFP). In the present study, we first tested whether two replicon vectors encoding different proteins can be co-expressed. We infiltrated N. benthamiana leaves with BYGFP and/or BYDsRed in combination with REP110 and P19 vectors (Fig. 1). BYGFP/REP110/P19 and BYDsRed/REP110/P19 infiltrated areas displayed green and red fluorescence, respectively, while the fluorescence of BYGFP/BYDsRed co-infiltrated area appeared yellow (Fig. 2A), probably due to overlapping of both green and red fluorescence. Mesophyll protoplasts were subsequently isolated from infiltrated areas and observed under fluorescence microscope. When two populations of protoplasts that individually expressed BYGFP or BYDsRed were mixed, no overlapping fluorescent protoplast was observed (Fig. 2B and 2C). In contrast, the majority (>80%) of the fluorescent protoplasts from co-infiltration of BYGFP/BYDsRed exhibited both green and red fluorescence (Fig. 2D and 2E), indicating the efficient co-expression of GFP and DsRed in the same cells.
Figure 2.
Expression of GFP and/or DsRed replicons in plant leaf cells. A: N. benthamiana leaves infiltrated with Agrobacterium strain harboring expression vectors as indicated were examined and photographed at 5 days post infiltration (dpi) under a handheld UV lamp as described in Materials and Methods. Co-infiltration of a replicon (pBYGFP or pBYDsRed) with pREP110 allows replication of the DNA lying between the LIR regions (Fig. 1). Co-infiltration with pP19 suppresses post-transcriptional gene silencing. B – G: Fluorescence microscopy of mesophyll protoplasts expressing GFP and/or DsRed replicons. Mesophyll protoplasts were prepared from infiltrated leaf tissue at 5 dpi and subsequently viewed under a Nikon inverted microscope with both GFP and DsRed filters. B and C: mixture of protoplasts from leaves infiltrated separately with BYGFP/REP110/P19 or BYDsRed/REP110/P19. D and E: protoplasts from leaves infiltrated with BYGFP/BYDsRed/REP110/P19. F and G: protoplasts from leaves infiltrated with BYGFPDsRed.R. Panels B, D and F are viewed with a GFP filter; and C, E and G are viewed with a DsRed filter. Almost all cells in panels D, E, F, and G show both green and red fluorescence, indicating co-expression of GFP and DsRed.
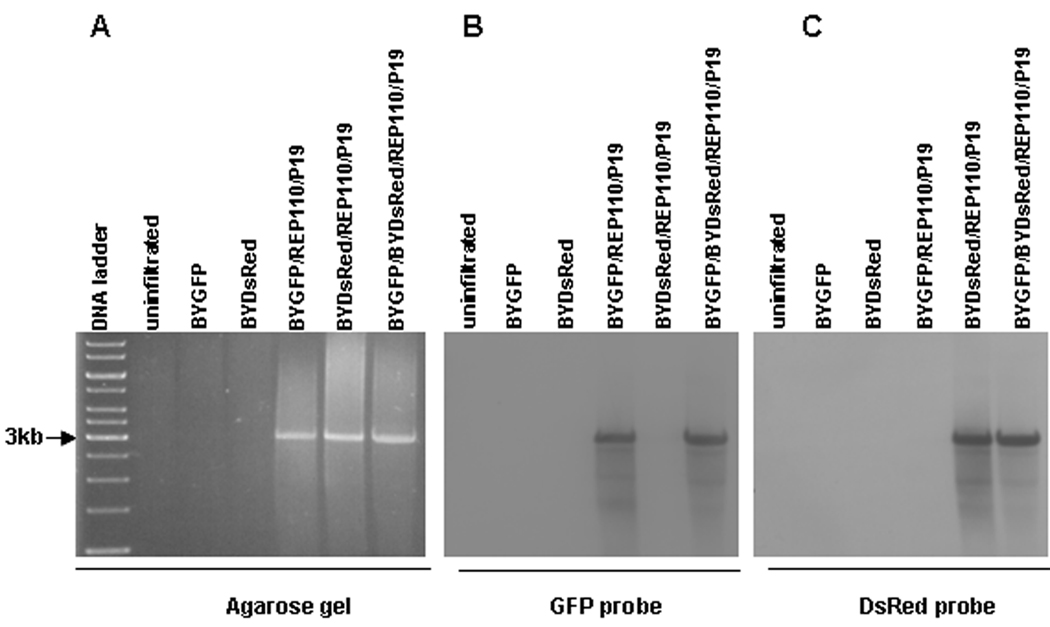
To determine whether amplification of both replicons is efficient, we performed Southern blot of DNA extracted from co-infiltrated leaves. As shown in Fig. 3A, the ethidium bromide-stained gel showed the expected ~3kbp band in the BYGFP/REP110/P19, the BYDsRed/REP110/P19, and the BYGFP/BYDsRed/REP110/P19 samples, indicating efficient formation of DNA replicons. No discrete bands were observed in DNA samples from uninfiltrated, BYGFP alone, or BYDsRed alone leaves. Southern blotting with both GFP and DsRed probes showed that the BYGFP/REP110/P19 replicon reacted with only the GFP probe, while the BYDsRed/REP110/P19 bound only the DsRed probe (Fig. 3B and 3C). However, the BYGFP/BYDsRed/REP110/P19 sample produced positive signals for both GFP and DsRed probes, with the signal intensity similar to those representing the corresponding single replicons, suggesting that the BYGFP and BYDsRed replicons are near-equally co-expressed. Together, these results demonstrate that the BeYDV replicon system is non-competing, which will enable its application in producing multiple-subunit proteins of pharmaceutical importance, such as mAbs.
Figure 3.
Southern blot showing non-competing formation of two replicons. Leaf DNA was extracted at 5dpi from leaves infiltrated with the indicated vector combinations, digested with XhoI, and run on a 1% agarose gel. The resulting gel was stained with ethidium bromide (A), blotted onto membrane, and detected with a GFP probe (B). The same membrane was subsequently stripped and re-hybridized with a DsRed probe (C). The preparation of GFP- and DsRed-specific probes is described in Materials and Methods. Arrow at left (3kb) marks the size of the major DNA species observed on both stained gel and Southern blots. Both GFP and DsRed replicons are ~3kbp, but note that the GFP probe does not bind to the DsRed replicon (B), and vice versa (C).
High-yield transient expression of a protective mAb against Ebola virus using non-competing replicon vectors
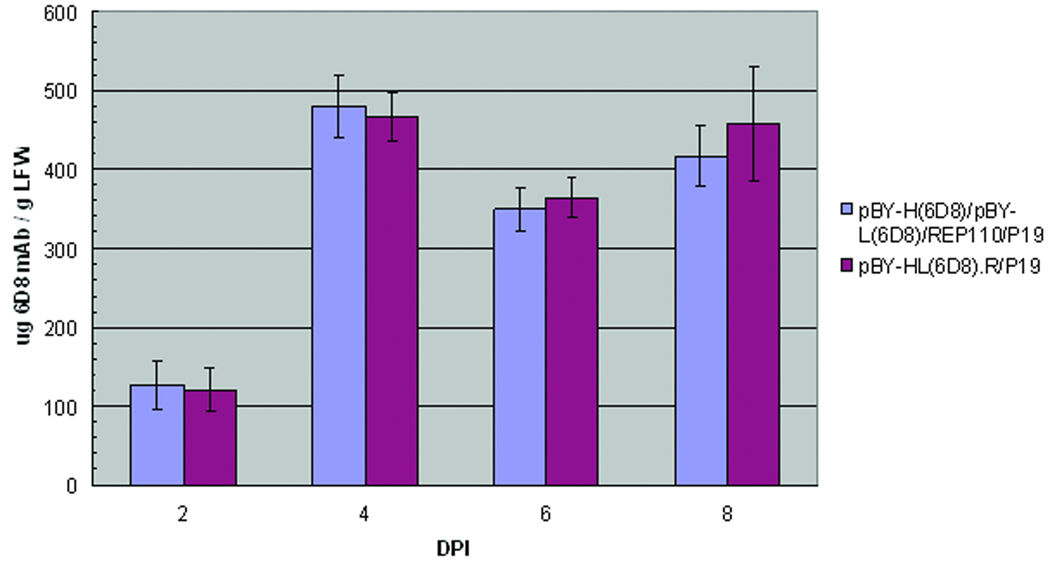
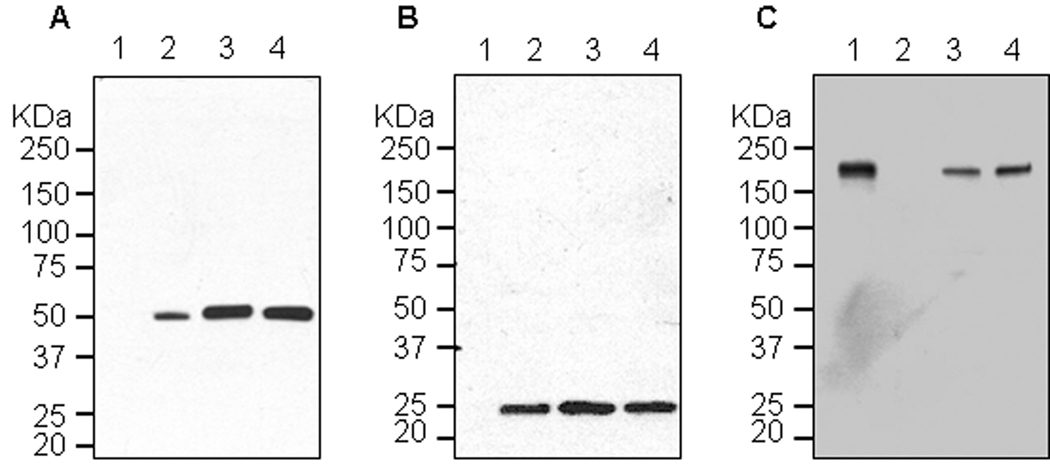
To demonstrate the utility of our non-competing expression system, we expressed a protective mAb against Ebola virus glycoprotein GP1 (6D8) by co-infiltration of N. benthamiana leaves with two replicon vectors, pBY-H(6D8) and pBY-L(6D8) (Fig. 1), encoding the heavy chain and light chain subunits of 6D8 mAb, respectively, along with vectors for REP110 and P19. ELISA showed that 6D8 mAb accumulated within 2 dpi and rapidly reached a peak in 4 dpi at 0.4–.5 mg/g LFW (Fig. 4). Western blot with SDS-PAGE under reducing condition confirmed the presence of both heavy and light chains with the expected molecular weights of 50 kDa and 25 kDa, respectively (Fig. 5A and 5B, lane 3). When the same samples were analyzed under non-reducing condition, a ~170 kDa band was observed, which represents the fully assembled hetero-tetrameric form of 6D8 mAb (Fig. 5C, lane 3). The slightly higher molecular weight (~170 kDa vs. the calculated 150 kDa) suggests that 6D8 is glycosylated as predicted for proteins with potential N-link glycosylation sites targeted to the endomembrane system of plant cells. The western blots also indicated that no proteolytic clipping of the light or heavy chains occurred, since only the expected size bands were observed (Fig. 5A–5C, lane 3).
Figure 4.
Expression of 6D8 mAb in N. benthamiana plants. N. benthamiana leaves were either co-infiltrated with separate vectors for light chain (pBY-L(6D8)),heavy chain (pBY-H(6D8)), REP110 and P19 (blue bars), or infiltrated with a single vector harboring replicons for light, heavy chain and Rep (pBY-HL(6D8).R, red bars). Leaves were also infiltrated with a construct encoding the mRNA silencing inhibitor P19 as a negative control. Agroinfiltrated leaves were harvest on days 2, 4, 6, and 8 dpi and protein extracts were obtained by homogenizing leaves with extraction buffer (Materials and Methods). Clarified leaf protein extracts were analyzed with an ELISA that detects the assembled form of 6D8 mAb with a goat anti-human gamma heavy chain specific antibody as the capture antibody and a HRP-conjugated anti-human-kappa chain antibody as the detecting antibody. Data are means ± SD of samples from three independent infiltration experiments.
Figure 5.
Western blot analysis of plant-derived 6D8. Protein samples were separated on a 4–12% SDS-PAGE gradient gel under denaturing and reducing condition (A and B) or under non-reducing condition (C) and blotted onto a PVDF membrane. The membrane was incubated with a goat anti-human gamma chain antibody or goat anti-human kappa chain antibody to detect heavy chain (A) or light chain (B and C). Lane 1, Protein samples extracted from un-infiltrated leaves (A and B), or human IgG as a reference standard (C); lane 2, human IgG as a reference standard (A and B), or Protein samples from un-infiltrated leaves (C); lane 3, Protein sample extracted from the leaves co-infiltrated with separate replicons for light chain (pBY-L(6D8)) and heavy chain (pBY-H(6D8)) ; lane 4, Protein extracted from the single vector pBY-HL(6D8).R infiltrated leaves.
Two replicons can be delivered in a single vector
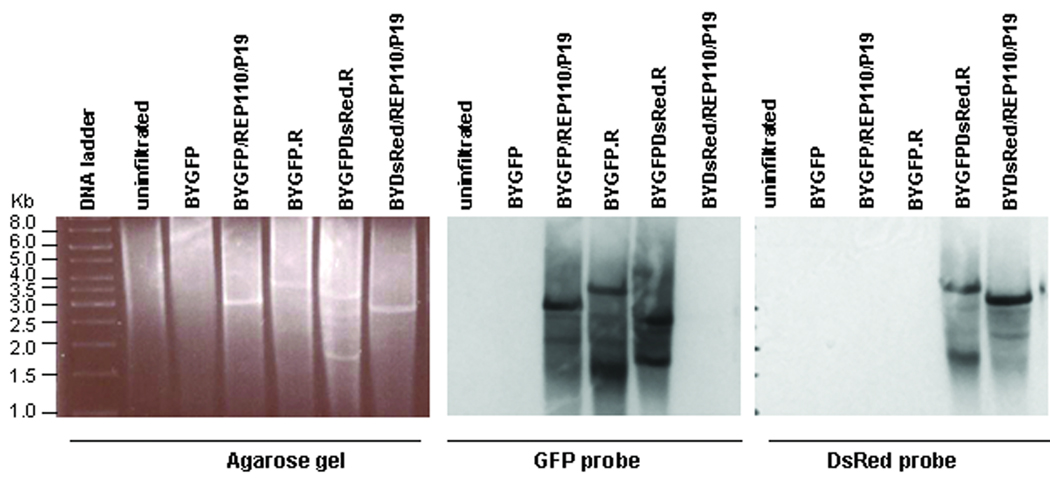
We previously showed that the three-vector replicon system can be simplified without compromising yields (Huang et al. 2009). A single vector replicon containing a native REP expression cassette (C1/C2 coding region under the control of the viral LIR promoter, e.g. pBYGFP.R in Fig. 1) works as well as the three-vector system for the expression of GFP (Huang et al. 2009). To test if the single-vector replicon system can be used to simultaneously express multiple proteins, we constructed pBYGFPDsRed.R for co-expression of GFP and DsRed (Fig. 1). Examination of protoplasts prepared from pBYGFPDsRed.R infiltrated leaves showed that both GFP and DsRed fluorescence were simultaneously detected in ~95% of the fluorescent protoplasts (Fig. 2F and 2G), indicating high-efficiency co-expression of both fluorescent proteins within same cell. Analyses of DNA from infiltrated leaves reveals the BYGFPDsRed.R sample produced replicons of different sizes which also reacted differently with GFP or DsRed probes (Fig. 6), indicating the simultaneous presence of GFP and DsRed replicons. For the BYGFPDsRed.R sample, the strongest DsRed-probe-reacting band is ~3.5kb (Fig. 6), which is similar to the size of the BYGFP.R replicon, suggesting this band represents the DsRed replicon comprising the 35S/TEV 5’-DsRed-VSP 3’-SIR-C2/C1 sequence within the two LIRs (see Fig. 1); while the top GFP-probe-reacting band is ~2.6kb (Fig. 6), which is expected for a smaller replicon consisting of the 35S/TMV 5’-GFP-rbcS 3’-SIR region between two adjacent LIR elements (see Fig. 1). These results demonstrate that the two tandemly linked replicons can be released and amplified independently, leading to high-level proliferation of two distinct double-stranded DNA replicons and efficient expression of both proteins. We also observed that in the BYGFPDsRed.R lanes a ~1.6kb band reacted with the GFP probe and a slightly higher ~1.7kb band less strongly with the DsRed probe (Fig. 6), probably representing the single-strand DNA forms of the corresponding replicons. Overall, the data (Fig. 2F and 2G and Fig.6) indicate that the multi-replicon single vector system produces high-level co-expression of two target proteins.
Figure 6.
Formation of two separate replicons by a single vector. DNA from leaves infiltrated with various vector combinations as indicated was digested with XhoI, run on 1% agarose gel and stained with ethidium bromide (A), blotted onto membrane, and detected with a GFP probe (B) or a DsRed probe (C). The preparation of GFP- and DsRed-specific probes is described in Materials and Methods. Numbers at left indicate sizes in kbp of DNA molecular weight markers. Note that pBYGFP produced a replicon of ~3kbp (consistent with data in Fig. 3), while the replicon from pBYGFP.R is ~4kbp due to the presence of the C1/C2 gene. The dual replicon construct pBYGFPDsRed.R produces a GFP replicon at ~3kbp and a DsRed replicon containing the C1C2 gene (Fig. 1) at ~4kbp.
Co-expression and assembly of full-size IgG by the multi-replicon single vector
To further demonstrate the effectiveness of the multi-replicon single-vector system, we used pBY-HL(6D8).R (Fig. 1), which harbors expression cassettes for the light and heavy chains of 6D8 mAb and REP (C1/C2). ELISA of pBY-HL(6D8).R leaf extracts showed that 6D8 mAb accumulated to a level comparable to that produced by the co-infiltration of four separate vectors (Fig. 4). The time course of the expression of 6D8 mAb with this single vector was also similar to that obtained with separate vectors (Fig 4). Furthermore, SDS-PAGE and western blot analyses demonstrate that the mAb produced by pBY-HL(6D8).R has the correct light and heavy chain components (Fig. 5A and 5B, Lane 4) and is fully assembled (Fig. 5C, Lane 4 ).
Purification and characterization of the replicon-expressed full-size IgG antibody
To further validate the multi-replicon single-vector expression system and to examine the structural and functional properties of the plant-made IgG, we used ammonium sulfate precipitation followed by protein G affinity chromatography to purify the single-vector-derived 6D8 mAb. SDS-PAGE Coomassie blue staining analysis of purification fractions showed that the combination of ammonium sulfate precipitation and Protein G affinity chromatography effectively removed most plant host proteins including the most abundant plant protein RuBisCo (Fig. 7, Lanes 2 and 4) and 6D8 mAb was purified to >90% purity with intact light and heavy chains (Fig. 7, Lane 5). A similar analysis under non-reducing condition indicated that the purified 6D8 antibody is in its fully assembled tetrameric form (data not shown). The purified mAb was further examined in a binding assay in which different concentrations of plant-derived 6D8 were incubated with irradiated Ebola virus coated on an ELISA plate. A generic human IgG was used as a negative control for the assay. Figure 8 shows the OD450 increased as more 6D8 mAb was applied in the reaction. In contrast, the OD450 for a generic human IgG remained at a basal level regardless the amount of this IgG used for the reaction (Fig. 8). This result indicates that plant produced 6D8 retains its specific affinity for Ebola virus GP1 protein. Analyses for 6D8 produced by co-infiltration of plant leaves with separate light chain and heavy chain replicons showed similar results in antibody purity, SDS-PAGE pattern, and its ability to specifically bind to Ebola virus (data not shown).
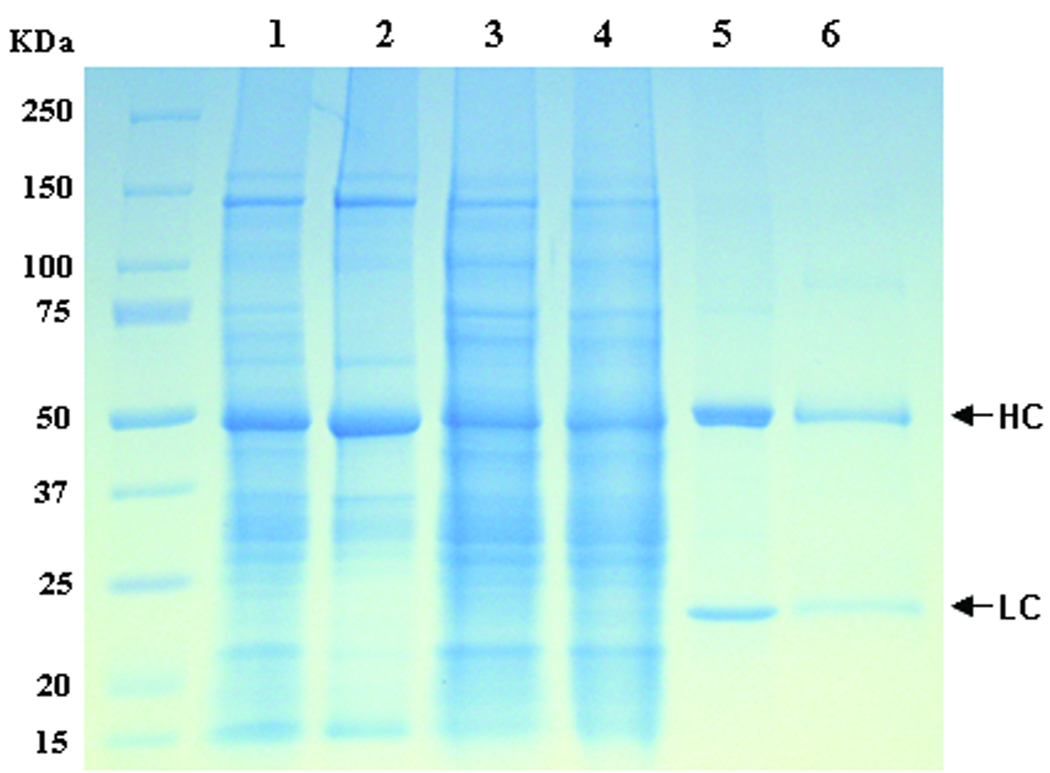
Figure 7.
Purification of 6D8 mAb from N. benthamiana leaves. 6D8 mAb from the multi-replicon single vector (pBY-HL(6D8).R) Infiltrated leaves was purified and analyzed on a SDS-PAGE under reducing conditions. Lane 1, Clarified plant extract ; lane 2, Plant proteins removed by 35% ammonium sulfate precipitation; lane 3, 60% ammonium sulfate pellet fraction resuspended for Protein G Chromatography; lane 4, Protein G flow-through fraction; lane 5, Purified 6D8 mAb in the Protein G eluate; lane 6, Human IgG as a reference standard.
Figure 8.
Plant-derived 6D8 mAb shows specific binding to Ebola virus. Plant-derived 6D8 mAb with concentrations between 1 to 2000 ng/ml was incubated with irradiated Ebola virus immobilized on a 96-well ELISA plate. 6D8 mAb bound to Ebola virus was detected by an anti-human IgG secondary antibody conjugated with horse radish peroxides. A generic human IgG was used a negative control. The OD450 (means ± SD) from three independent experiments are presented.
DISCUSSION
Plant production of mAbs employs two main approaches: plant virus-based transient expression and stable transgenic plants. For the latter strategy, the long time frame (months to years) required to create stable transgenic plants is impractical for rapid production of protein samples for pre-clinical studies. In addition, the lack of strong regulatory elements to drive high-level protein accumulation and the position effect associated with the randomness of transgene integration in the plant genome still presents challenges for stable transgenic technology (Chen 2008). In contrast, transient systems that are focused on production speed have greatly reduced the time needed to obtain material for testing the function of recombinant antibodies in preclinical trials (Chen et al. 2009; Lico et al. 2008). In addition, highly efficient viral replicon vectors have enabled high-level antibody expression in plants. For example, it has been recently reported that full-size mAbs can be rapidly produced at levels as high as 0.5 mg of mAb per gram leaf fresh weight (Giritch et al. 2006). However, the process requires two sets of noncompeting viral vectors derived from TMV and PVX (Giritch et al. 2006). For the production and assembly of more complex mAbs such as secretory IgA and IgM, it remains a difficult task to find additional viruses compatible with the existing TMV/PVX expression system to allow efficient co-expression of three or more distinct subunit proteins in same cells.
Unlike traditional or “deconstructed” viral expression systems (Giritch et al. 2006) which rely on in planta assembly and replication of near-full-length viral genome, the BeYDV-derived DNA replicon system requires only two viral elements, the LIR region and the Rep/RepA, for the initiation and amplification of episomal replicons (Mor et al. 2003). We hypothesized that this system could be used for non-competing co-expression of multiple protein subunits. We found that co-infiltration of replicons encoding two fluorescence proteins (GFP and DsRed) resulted in the exhibition of both fluorescences, with >80% of cells expressing both proteins in the same cell (Fig. 2), indicating our BeYDV replicon system is indeed noncompeting. Our subsequent effort focused on simplifying the system so that it can be feasible for practical production of multiple-subunit protein pharmaceutical candidates. Our group has previously showed that single replicon vectors containing a gene of interest and the Rep/RepA expression cassettes (such as pBYGFP.R in Fig. 1) were as efficient as their three-component counterparts in directing the expression of target proteins (Huang et al. 2009). In this study, we created single vectors containing two tandemly linked replicons expressing distinct genes of interest, with the hypothesis that the two replicons can efficiently replicate without interference. Transient leaf infiltration with one such vector, pBYGFPDsRed.R (Fig. 1), resulted in efficient formation of both GFP and DsRed replicons as evidenced by Southern blotting (Fig. 6) as well as co-expression of GFP and DsRed in ~95% cells (Fig. 2F and 2G), thus proving that our hypothesis is correct. Collectively, our results with model proteins GFP and DsRed indicate the feasibility of using our replicon system for high-level co-expression of two subunit proteins.
To evaluate the utility and capacity of our replicon systems for antibody production, we co-expressed both the heavy chain and light chain molecules of a mAb protective against Ebola virus GP1 using either the three-component noncompeting vectors or the single vector pBY-HL (6D8).R. Our results showed that both systems could rapidly (4 dpi) produce 6D8 mAb at ~0.5 mg of mAb per gram leaf fresh weight (Fig. 4), which is comparable to the highest level ever achieved with a plant-based expression system (Giritch et al. 2006). Furthermore, the mAb can correctly assemble into its tetrameric structure (Fig. 5C). We did not observe any non-assembled or partial assembled 6D8 mAb, suggesting that the assembly of the mAb using the DNA replicon system is highly efficient. We further demonstrated that 6D8 mAb produced with this single vector system not only has the correct assembled tetrameric structure, but also can be readily purified, and is functionally active in specific binding to inactivated Ebola virus. In addition to 6D8, we have also successfully expressed several other mAbs and mAb-fusion proteins twice as large as 6D8 with full assembly and functionality (Chen, unpublished data). Our success in producing the fully-assembled tetrameric functional IgG (two heterooligomeric subunits) with a two-replicon single vector strongly suggest that simultaneous expression of as many as four hetero-subunits can be easily achieved using two of such vectors, or by creating single vectors with three or more tandem linked replicons. To our best knowledge, this technology is the first and the only one that has such a potential, among all plant transient replicon expression systems.
While the BeYDV multi-replicon vector provided high expression levels, there may be additional ways to improve its performance. As suggested by others, molecular chaperones could be included in the expression cassette to accommodate the increasing protein folding and assembly demand by the high-level accumulation of oligomeric mAbs (Nuttall et al. 2002). Furthermore, the anti-silencing element P19 could be incorporated into the single vector system. We have not fully explored the effect of P19 with the multi-replicon single-vector. Finally, incorporation of translation enhancer elements from cowpea mosaic virus RNA-2 (Sainsbury and Lomonossoff 2008) have the potential to substantially increase expression of mRNA produced by the DNA replicon system.
In summary, the results presented in this paper collectively demonstrate that the BeYDV-derived DNA replicon expression system is capable of rapidly producing high levels of multi-subunit antibodies in plants. The ability of a simplified multi-replicon single vector to deliver multiple expression cassettes/replicons for production of mAbs and other hetero-oligomeric pharmaceuticals provides some key advantages. Once optimized, this expression system has the potential to provide a technology platform to produce a wide spectrum of pharmaceutical proteins with speed, efficiency, cost-effectiveness and safety.
ACKNOWLEDGEMENTS
This work was supported by NIH Grants # 5U01AI061253, 1U01AI075549, U01AI061270 and 5U19AI066332.
REFERENCES
- Canizares MC, Nicholson L, Lomonossoff GP. Use of viral vectors for vaccine production in plants. Immunol Cell Biol. 2005;83(3):263–270. doi: 10.1111/j.1440-1711.2005.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. Expression and Purification of Pharmaceutical Proteins in Plants Biological Engineering. 2008;1(4):291–321. [Google Scholar]
- Chen Q, Tacket CO, Mason H, Mor T, Cardineau GA, Arntzen C. Subunit Vaccines Produced Using Plant Biotechnology. In: Levine MM, editor. New Generation Vaccines. Fourth Edition. New York: Academic Press; 2009. p In press. [Google Scholar]
- Dietrich C, Maiss E. Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J Gen Virol. 2003;84(Pt 10):2871–2876. doi: 10.1099/vir.0.19245-0. [DOI] [PubMed] [Google Scholar]
- Diveki Z, Salanki K, Balazs E. Limited utility of blue fluorescent protein (BFP) in monitoring plant virus movement. Biochimie. 2002;84(10):997–1002. doi: 10.1016/s0300-9084(02)00007-x. [DOI] [PubMed] [Google Scholar]
- Floss D, Falkenburg D, Conrad U. Production of vaccines and therapeutic antibodies for veterinary applications in transgenic plants: an overview. Transgenic Research. 2007;16(3):315–332. doi: 10.1007/s11248-007-9095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123(4):1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings G. Transgenic plants as protein factories. Curr Opin Biotechnol. 2001;12(5):450–454. doi: 10.1016/s0958-1669(00)00244-5. [DOI] [PubMed] [Google Scholar]
- Giddings G, Allison G, Brooks D, Carter A. Transgenic plants as factories for biopharmaceuticals. Nat Biotechnol. 2000;18(11):1151–1155. doi: 10.1038/81132. [DOI] [PubMed] [Google Scholar]
- Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci U S A. 2006;103(40):14701–14706. doi: 10.1073/pnas.0606631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt A, Cafferkey R, Bowdish K. Production of antibodies in transgenic plants. Nature. 1989;342(6245):76–78. doi: 10.1038/342076a0. [DOI] [PubMed] [Google Scholar]
- Hood EE, Woodard SL, Horn ME. Monoclonal antibody manufacturing in transgenic plants--myths and realities. Curr Opin Biotechnol. 2002;13(6):630–635. doi: 10.1016/s0958-1669(02)00351-8. [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen Q, Hjelm B, Arntzen C, Mason H. A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnology and Bioengineering. 2009;103(4):706–714. doi: 10.1002/bit.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Mason HS. Conformational analysis of hepatitis B surface antigen fusions in an Agrobacterium-mediated transient expression system. Plant Biotechnology Journal. 2004;2:241–249. doi: 10.1111/j.1467-7652.2004.00068.x. [DOI] [PubMed] [Google Scholar]
- Hull R, Plaskitt A. Electron microscopy on the behavior of two strains of alfalfa mosaic virus in mixed infections. Virology. 1970;42(3):773–776. doi: 10.1016/0042-6822(70)90322-3. [DOI] [PubMed] [Google Scholar]
- Judge NA, Mason HS, O'Brien AD. Plant cell-based intimin vaccine given orally to mice primed with intimin reduces time of Escherichia coli O157:H7 shedding in feces. Infect Immun. 2004;72(1):168–175. doi: 10.1128/IAI.72.1.168-175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lico C, Chen Q, Santi L. Viral vectors for production of recombinant proteins in plants. J Cell Physiol. 2008;216(2):366–377. doi: 10.1002/jcp.21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Drake Pascal MW, Christou Paul. The production of recombinant pharmaceutical proteins in plants. Nature Reviews (Genetics) 2003;4:794–805. doi: 10.1038/nrg1177. [DOI] [PubMed] [Google Scholar]
- Ma JK, Drake PM, Chargelegue D, Obregon P, Prada A. Antibody processing and engineering in plants, and new strategies for vaccine production. Vaccine. 2005;23(15):1814–1818. doi: 10.1016/j.vaccine.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Mor TS, Moon YS, Palmer KE, Mason HS. Geminivirus vectors for high-level expression of foreign proteins in plant cells. Biotechnol Bioeng. 2003;81(4):430–437. doi: 10.1002/bit.10483. [DOI] [PubMed] [Google Scholar]
- Nuttall J, Vine N, Hadlington JL, Drake P, Frigerio L, Ma JK. ER-resident chaperone interactions with recombinant antibodies in transgenic plants. Eur J Biochem. 2002;269(24):6042–6051. doi: 10.1046/j.1432-1033.2002.03302.x. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Lomonossoff GP. Extremely High-Level and Rapid Transient Protein Production in Plants without the Use of Viral Replication. Plant Physiol. 2008;148(3):1212–1218. doi: 10.1104/pp.108.126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi L, Batchelor L, Huang Z, Hjelm B, Kilbourne J, Arntzen CJ, Chen Q, Mason HS. An efficient plant viral expression system generating orally immunogenic Norwalk virus-like particles. Vaccine. 2008;26(15):1846–1854. doi: 10.1016/j.vaccine.2008.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger E, Schillberg S, Twyman RM, Fischer R, Christou P. Antibody production in transgenic plants. Methods Mol Biol. 2004;248:301–318. doi: 10.1385/1-59259-666-5:301. [DOI] [PubMed] [Google Scholar]
- Twyman RM, Schillberg S, Fischer R. Transgenic plants in the biopharmaceutical market. Expert Opin Emerg Drugs. 2005;10(1):185–218. doi: 10.1517/14728214.10.1.185. [DOI] [PubMed] [Google Scholar]
- Vitale A, Pedrazzini E. Recombinant pharmaceuticals from plants: the plant endomembrane system as bioreactor. Mol Interv. 2005;5(4):216–225. doi: 10.1124/mi.5.4.5. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287(5458):1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- Yusibov V, Rabindran S, Commandeur U, Twyman RM, Fischer R. The potential of plant virus vectors for vaccine production. Drugs R D. 2006;7(4):203–217. doi: 10.2165/00126839-200607040-00001. [DOI] [PubMed] [Google Scholar]