Abstract
Motivation: New sequencing technologies have accelerated research on prokaryotic genomes and have made genome sequencing operations outside major genome sequencing centers routine. However, no off-the-shelf solution exists for the combined assembly, gene prediction, genome annotation and data presentation necessary to interpret sequencing data. The resulting requirement to invest significant resources into custom informatics support for genome sequencing projects remains a major impediment to the accessibility of high-throughput sequence data.
Results: We present a self-contained, automated high-throughput open source genome sequencing and computational genomics pipeline suitable for prokaryotic sequencing projects. The pipeline has been used at the Georgia Institute of Technology and the Centers for Disease Control and Prevention for the analysis of Neisseria meningitidis and Bordetella bronchiseptica genomes. The pipeline is capable of enhanced or manually assisted reference-based assembly using multiple assemblers and modes; gene predictor combining; and functional annotation of genes and gene products. Because every component of the pipeline is executed on a local machine with no need to access resources over the Internet, the pipeline is suitable for projects of a sensitive nature. Annotation of virulence-related features makes the pipeline particularly useful for projects working with pathogenic prokaryotes.
Availability and implementation: The pipeline is licensed under the open-source GNU General Public License and available at the Georgia Tech Neisseria Base (http://nbase.biology.gatech.edu/). The pipeline is implemented with a combination of Perl, Bourne Shell and MySQL and is compatible with Linux and other Unix systems.
Contact: king.jordan@biology.gatech.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Genome sequencing projects, pioneered in the 1990s (Fleischmann et al., 1995), require large-scale computational support in order to make their data accessible for use and interpretation by biologists. Large sequencing centers have traditionally employed or collaborated with teams of software engineers and computational biologists to develop the software and algorithms for sequencing hardware interfaces, enterprise data storage, sequence assembly and finishing, genome feature prediction and annotation, database mining, comparative analysis and database user interface development. While many of the components developed by these teams are now available online under open-access terms, the development of new, high-throughput sequencing technologies has necessitated updates to these tools and development of even more sophisticated algorithms to address the challenges raised by the new data. These new technologies—454 pyrosequencing (Margulies et al., 2005), ABI SOLiD (Shendure et al., 2005) and Illumina (Bentley et al., 2008)—are now collectively referred to as second generation sequencing technologies. Similar updates will be needed as the third generation of sequencing technologies, such as Pacific Biosciences' SMRT sequencing (Eid et al., 2009), enter production use. New and improved tools released for these technologies on a monthly basis include assemblers, mapping algorithms, base calling and error correction tools, and a multitude of other programs. Because of this fast pace of development, few experts are able to keep up with the state of the art in the field of computational genomics. Accordingly, the rate limiting step in genome sequencing projects is no longer the experimental characterization of the data but rather the availability of experts and resources for computational analysis.
At the same time, the increased affordability of these new sequencing machines has spawned a new generation of users who were previously unable to perform their own genome sequencing, and thus collaborated with large sequencing centers for genome sequencing and subsequent computational analysis. While these users are now able to experimentally characterize genomes in house, they often find themselves struggling to take full advantage of the resulting data and to make it useful to the scientific community since the informatics support for their genome projects is not sufficient.
Several large sequencing consortia (Aziz et al., 2008; Markowitz et al., 2009; Seshadri et al., 2007) have produced comprehensive, centralized web-based portals for the analysis of genomic and metagenomic data. While extremely useful for many types of projects and collaborations, these solutions inherently result in a loss of data processing flexibility compared to locally installed resources and may be unsuitable for projects dealing with sensitive data. Recently, another group (Stewart et al., 2009) has published DIYA, a software package for gene prediction and annotation in bacterial genomes with a modularized, open source microbial genome processing pipeline. However, DIYA does not include a genome assembly component, and does not provide for the combination of complementary algorithms for genome analysis.
To address the outstanding challenges for local computational genomics support, we have developed a state of the art, self-contained, automated high-throughput open source software pipeline for computational genomics in support of prokaryotic sequencing projects. To ensure the relevance of our pipeline, we checked the latest developments in computational genomics software for all stages of the pipeline, such as new versions of assembly and gene prediction programs and comparative surveys, and selected what we deemed to be the most suitable software packages. The pipeline is self-contained; that is, we used locally installable versions of all third-party tools instead of web-based services provided by many groups. We chose to do so for three reasons: first, because some of the applications we envision for this pipeline are of sensitive nature; second, to enhance robustness to external changes (e.g., online API changes or website address changes); and third, to improve the ability of developers to customize and derive from our pipeline. The pipeline is also automated and high-throughput: all components are organized in a hierarchical set of readily modifiable scripts, and the use of safe programming practices ensures that multiple copies of the pipeline can be run in parallel, taking advantage of multiple processors where possible.
Importantly, by using and combining the outputs of competitive, complementary algorithms for multiple stages of genome analysis, our pipeline allows for substantial improvement upon single-program solutions. The use of multiple algorithms also provides a way to improve robustness and conduct more comprehensive quality control when the output of one program is significantly different from that of another.
Computational support provided to prokaryotic genome projects by our pipeline can be subdivided into three stages: first, sequencing and assembly; second, feature prediction; and third, functional annotation. For the assembly stage, we developed a custom protocol specific to 454 pyrosequenced data, which resulted in a significant improvement to assembly quality of our test data compared to the baseline assembler bundled by the manufacturer. Other assemblers can be plugged in if necessary, and data from other sequencing technologies such as ABI SOLiD, Illumina and Sanger capillary-based machines can be used. For the prediction stage, we again included a custom combination of feature prediction methods for protein-coding genes, RNA genes, operon and promoter regions, which improves upon the individual constituent methods. The annotation stage includes several types of protein functional prediction algorithms. We also developed components for comparative analysis, interpretation and presentation (a web-based genome browser), which can be used downstream of our pipeline.
We have tested the pipeline on the bacterium Neisseria meningitidis, which is a human commensal of the nasopharanx and which can sometimes cause meningitis or septicemia (Rosenstein et al., 2001). When N.meningitidis does cause disease, it can be devastating with an ∼10% fatality rate and 15% sequelae rate. Neisseria meningitidis is a highly competent organism with a high recombination rate, and large chromosomal changes are common (Jolley et al., 2005; Schoen et al., 2008). This complicates computational genome analysis and makes N.meningitidis an appropriately challenging test for our pipeline. To demonstrate the general applicability of the pipeline, we have also tested it on a different pathogen, Bordetella bronchiseptica. Bordetella bronchiseptica is a Gram-negative bacterium that can cause bronchitis in humans, although it is more commonly found in smaller mammals (Parkhill et al., 2003). Much like Neisseria, Bordetella has extensive plasticity, likely due to the large number of repeat elements (Gerlach et al., 2001). Here, we analyze the first two complete genome sequences of B.bronchiseptica strains isolated from human hosts.
The rest of this article is organized as follows. The ‘System and Methods’ section describes the genomes which we used to test our pipeline, overall organization of the pipeline, and details of the algorithms used to perform tasks in the pipeline. In the ‘Discussion’ section, we discuss the objectives of our work on the pipeline and how these relate to larger developments in computational biology for next-generation sequencing.
2 SYSTEM AND METHODS
2.1 Genome test data
Neisseria meningitidis genomes were characterized via 454 pyrosequencing (Margulies et al., 2005) using either half or one quarter plate runs on the Roche 454 GS-20 or GS Titanium instrument (Table 1). For each genome, a random shotgun library was produced using Roche protocols for nebulization, end-polishing, adaptor ligation, nick repair and single-stranded library formation. Following emulsion PCR, DNA bound beads were isolated and sequenced using long-read (LR) sequencing kits. The number of reads produced in the experiments ranged from 200 000 to 600 000, and the average read lengths were between 100 and 330 bases. These data yielded 47.6–94.3 million bases per genome amounting to 20–40× coverage for the ∼2.2 Mb N.meningitidis genomes. After read trimming and re-filtering to recover short quality reads, the data were passed to the first stage of the pipeline—genome assembly.
Table 1.
Summary of sequencing projects used in the pipeline development
| Strain ID | Sequence typea | Serogroupb | Geographic originc | Date collected | Genome size | Closest referenced | Substitutions per position versus ref.e | Total reads | Total bases sequenced | Average read length | Coveragef | Instrument standardg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Neisseria meningitidis | ||||||||||||
| NM13220 | ST-7 | A | Philippines | 2005 | 2.2M | Z2491 | 0.076 | 197 067 | 47 569 493 | 241 | 21× | GS-20 |
| NM10699 | ST-32 | B | Oregon, USA | 2003 | 2.2M | MC58 | 0.053 | 418 751 | 81 775 264 | 195 | 37× | GS-20 |
| NM15141 | ST-11 | C | New York, USA | 2006 | 2.2M | FAM18 | 0.028 | 378 773 | 94 288 660 | 249 | 42× | GS-20 |
| NM9261 | ST-11 | W135 | Burkina Faso | 2002 | 2.2M | FAM18 | 0.030 | 206 634 | 69 957 473 | 338 | 31× | GS Ti |
| NM18575 | ST-2859 | A | Burkina Faso | 2003 | 2.2M | Z2491 | 0.033 | 283 888 | 84 013 571 | 296 | 38× | GS Ti |
| NM5178 | ST-32 | B | Oregon, USA | 1998 | 2.2M | MC58 | 0.050 | 270 332 | 88 664 981 | 328 | 40× | GS Ti |
| NM15293 | ST-32 | B | Georgia, USA | 2006 | 2.2M | MC58 | 0.054 | 276 733 | 90 951 566 | 329 | 41× | GS Ti |
|
Bordetella bronchiseptica | ||||||||||||
| BBE001 | N/Ah | N/A | Georgia, USA | 1956 | 5.3M | RB50 | 0.056 | 566 834 | 229 098 141 | 404 | 43× | GS Ti |
| BBF579 | N/A | N/A | Mississippi, USA | 2007 | 5.3M | RB50 | 0.104 | 533 099 | 228 467 710 | 429 | 43× | GS Ti |
Data for each strain are presented in rows.
aSequence type denotes the allelic profile assigned by multilocus sequence typing (MLST; Holmes et al., 1999; Maiden et al., 1998) on the basis of seven loci within well-conserved house-keeping genes.
bNeisseria meningitidis isolates are divided into serogroups by immunochemistry of polysaccharides present in their antiphagocytic capsule.
cThe region in which each strain was originally collected.
dStrain ID of the closest complete genome available in GenBank, as determined by 16S RNA phylogeny as well as whole-genome sequence identity, which agreed in all cases.
eInsertions, deletions and substitutions per position of genome as compared against the closest reference.
fCoverage denotes the average number of sequencing reads overlapping at a given position in the genome, calculated as the total number of bases sequenced divided by the estimated length of the genome.
gThe standard of the 454 pyrosequencing instrument and reagents used to sequence the data.
hSequence typing and serotyping was not performed on B.bronchiseptica.
2.2 Pipeline organization
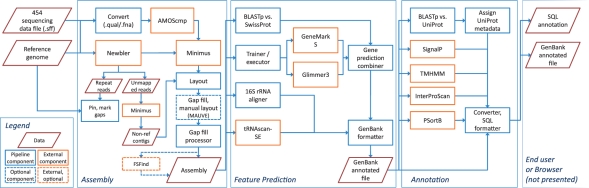
The analytical pipeline consists of three integrated subsystems: genome assembly, feature prediction and functional annotation. Each subsystem consists of a top-level execution script managing the input, output, format conversion and combination of results for a number of distinct software components. A hierarchy of scripts and external programs then performs the tasks required to complete each stage of analysis (Fig. 1).
Fig. 1.
Chart of data flow, major components and subsystems in the pipeline. Three subsystems are presented: genome assembly, feature prediction and functional annotation. Each subsystem consists of a top-level execution script managing the input, output, format conversion and combination of results for a number of components. A hierarchy of scripts and external programs then performs the tasks required to complete each stage. The legend for the flowchart indicates the identities of the distinct pipeline components: data, pipeline component, optional component, external component and external, optional component.
2.3 Assembly
Genome assembly was performed by evaluating multiple configurations of assemblers including the standard 454 assembler, Newbler (version 2.3), as well the Celera Assembler (Miller et al., 2008), the Phrap assembler (http://www.phrap.org/) and the AMOScmp mapped assembler (Pop et al., 2004). Several other assemblers were evaluated but ultimately excluded from the pipeline due to use limitations: for instance, the ALLPATHS 2 assembler (MacCallum et al., 2009) required paired-end reads to operate; our evaluation data contained no paired-end reads, and such a requirement unnecessarily constrains the user's options. The widely used Velvet assembler (Zerbino and Birney, 2008) was originally developed as a de novo assembler for Illumina sequencing technology, but its capability has been extended to accommodate 454 data as well. However, we were unable to configure the Velvet assembler to produce a usable assembly or take advantage of reference genomes using 454 data alone.
Evaluation of the results indicated that mapped assemblies of N.meningitidis genomes using previously finished strains were of superior quality to de novo assemblies. Using the most appropriate reference strains, it was found that Newbler and AMOScmp complement each other's performance in the assembly stage, with Newbler being able to join some contigs AMOScmp left gapped and vice versa. As a result, we decided to use a combination of these two assemblers' outputs for the final assembly. Then, the Minimus assembler (Sommer et al., 2007) from the AMOS package, a simple assembler for short genomes, was used to combine the constituent assemblies.
We also evaluated alternative base calling algorithms for 454 pyrosequencing data (Quinlan et al., 2008) but detected no improvement. Over the course of our project, accuracy of base calling in the Newbler assembler was reported to be significantly improved. We used the latest version of the assembler available at publication time (Section 2.3).
An optional component of the pipeline was created for frameshift detection using FSFind (Kislyuk et al., 2009). Frameshifts in protein-coding sequences are a known result of pyrosequencing errors caused by undercalls and overcalls in homopolymer runs (Kuo and Grigoriev, 2009). Briefly, this package creates a GeneMark model of the genome, makes gene predictions, and then scans the genome for possible frameshift positions on the basis of ORF configuration and coding potential. Once the possible frameshift sites are identified, a putative translation of the protein possibly encoded by the broken gene is compared against a protein database (SwissProt by default). The predicted frameshift site is also scanned for adjacent homopolymers. A heuristic set of confidence score cutoffs is then used to provide a set of frameshift predictions while minimizing the false positive rate. The predicted frameshift sites can then be verified experimentally or corrected speculatively. The user can inspect the dataset to decide whether locations predicted to contain frameshifts break gene models, and patch the sequences to fix up these positions. The prediction stage can then be re-run to correct the gene predictions. While further experimental analysis to address such errors is desirable (e.g. targeted PCR of predicted error locations or a recently popular choice of combining sequencing technologies such as 454 and Illumina), it incurs extra costs which we aim to avoid.
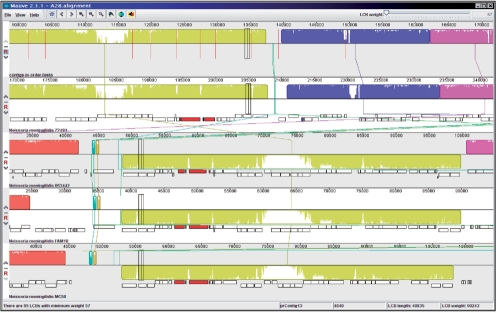
Unfinished assemblies produced in this stage contained 90–300 contigs each. No paired-end libraries or runs were available for the strains analyzed, and therefore scaffolding of the contigs was a challenge. Manual examination of the assemblies using the MAUVE (Darling et al., 2004) multiple whole-genome alignment and visualization package revealed numerous locations where contigs could be scaffolded with a small gap or minimal overlap (Fig. 2). As an optional step, we produced a table of such positions and a script which would scaffold contigs joined by the gap.
Fig. 2.
Comparative analysis of draft assembly with MAUVE. The top pane represents the active assembly; vertical lines indicate contig boundaries (gaps). The reference genomes are arranged in subsequent panes in order of phylogenetic distance. Blocks of synteny (LCBs) are displayed in different colors (an inversion of a large block is visible between panes 1–2 and 3–5). Most gaps within LCBs were joined in the manually assisted assembly, while considering factors such as sequence conservation on contig flanks and presence of protein-coding regions.
Then, a manual gap joining stage used the layout of the contigs according to their aligned positions on the reference using the AMOS package and manual examination of each gap, adjacent contig alignments and reference annotation in the MAUVE visualization tool. Although there is a possibility that rearrangements exist in those gaps as mapped to the closest reference genome, joining was only done after manual examination on a case-by-case basis in positions of high homology and full consensus between four of the reference strains, to minimize this possibility. While we provide the scripts and data format definitions necessary to complete this stage of the pipeline, it involves manual processing of the assembly and is therefore optional. This component is similar in function to Mauve Contig Mover (Rissman et al., 2009) but expands upon it in several ways. An option is provided in the pipeline to use Mauve Contig Mover.
The manually assisted genome assembly procedure resulted in an order-of-magnitude decrease in the number of gaps in comparison to the Newbler assembler (which in turn performed the best out of all standalone assemblers evaluated). In addition, the fully automated assembly metrics (N50 and contig count at equal minimal size) are an ∼20–50% improvement upon baseline Newbler performance (Table 2).
Table 2.
Summary of assembler performance
| Strain ID | Newbler statistics |
AMOScmp statistics |
Automatic combined assembly |
Manual combined assembly |
||||
|---|---|---|---|---|---|---|---|---|
| Contigs >500 nt, total size | N50a, longest contig | Contigs >500 nt, total size | N50, longest contig | Contigs >500 nt, total size | N50, longest contig | Contigs >500 nt, total size | % gapfill, longest contig | |
| NM13220 | 175 2.07M | 22K 106K | 202 2.06M | 21K 77K | 195 2.25M | 31K 107K | 57 2.30M | 1.8% 398K |
| NM10699 | 102 2.10M | 52K 143K | 116 2.10M | 43K 113K | 83 2.17M | 59K 143K | 40 2.18M | 1.1% 435K |
| NM15141 | 147 2.06M | 33K 171K | 190 2.05M | 22K 115K | 139 2.21M | 36K 171K | 50 2.28M | 2.0% 759K |
| NM9261 | 99 2.09M | 51K 184K | 133 2.07M | 37K 170K | 128 2.16M | 64K 231K | 27 2.21M | 1.6% 866K |
| NM18575 | 133 2.09M | 30K 172K | 147 2.09M | 29K 88K | 220 2.40M | 53K 231K | N/Ac | N/A |
| NM5178 | 89 2.13M | 56K 136K | 107 2.12M | 42K 131K | 104 2.17M | 59K 136K | N/A | N/A |
| NM15293 | 92 2.08M | 52K 144K | 110 2.06M | 42K 132K | 107 2.10M | 59K 144K | N/A | N/A |
| BBE001 | 146 5.05M | 70K 212K | 178 5.04M | 61K 173K | 214 5.03M | 80K 252K | N/A | N/A |
| BBF579 | 272 4.84M | 57K 88K | 321 4.84M | 46K 94K | 272b 4.84M | 57K 88K | N/A | N/A |
Data for each strain are presented in rows. Statistics from standalone assemblers (Newbler and AMOScmp) are presented together with results of the combining protocol (default output of the pipeline) and an optional, manually assisted predictive gap closure protocol.
aN50 is a standard quality metric for genome assemblies that summarizes the length distribution of contigs. It represents the size N such that 50% of the genome is contained in contigs of size N or greater. Greater N50 values indicate higher quality assemblies.
bNo improvement was detected from the combined assembly in strain BBF579, and the original Newbler assembly was automatically selected.
cThe manual combined assembly protocol was not performed for these projects.
The contigs in the assembly stage output were named according to the following format: prefix_contig#, where the prefix represents a unique strain identifier and # represents the zero-padded sequential number indicating the contig's predicted order on the chromosome. For example, the 25th contig for the N. meningitidis strain M13220 assembly would be named as CDC_NME_M13320_025. The prefix used in the pipeline is configurable by the user with a command line option.
2.4 Feature prediction
Feature prediction was performed in the genome using a suite of several programs. To predict genes, we used a combination of de novo and comparative methods. The Glimmer (Delcher et al., 1999) and GeneMark (Besemer et al., 2001) microbial gene predictors were used for de novo prediction, and BLASTp alignment (Altschul et al., 1997) of putative proteins was used for comparative prediction. Self-training procedures were followed for both de novo predictors, and the results, while highly concordant, were different enough (Table 3) to justify the inclusion of both algorithms. BLASTp alignment of all open reading frames (ORFs) at least 90 nt long was performed using the Swiss-Prot protein database (Boeckmann et al., 2003).
Table 3.
Prediction algorithm performance comparison and statistics
| Strain ID | Gene predictions by GeneMark | Gene predictions by Glimmer3 | Gene predictions by BLAST | ORFs with full consensusa | ORFs with partial consensusb | Total gene predic- tions reportedc | tRNAs predicted by tRNAScan-SE |
|---|---|---|---|---|---|---|---|
| NM13220 | 2530 | 2725 | 1353 | 1325 | 974 | 2299 | 52 |
| NM10699 | 2366 | 2494 | 1317 | 1284 | 826 | 2110 | 51 |
| NM15141 | 2411 | 2578 | 1369 | 1343 | 841 | 2184 | 57 |
| NM9261 | 2370 | 2553 | 1341 | 1308 | 802 | 2110 | 51 |
| NM18575 | 2751 | 2927 | 1495 | 1448 | 1023 | 2471 | 63 |
| NM5178 | 2377 | 2510 | 1315 | 1281 | 816 | 2097 | 52 |
| NM15293 | 2062 | 2040 | 1285 | 1261 | 802 | 2063 | 51 |
| BBE001 | 4793 | 4793 | 2744 | 2732 | 2067 | 4799 | 48 |
| BBF579 | 4649 | 4646 | 2652 | 2635 | 2021 | 4656 | 48 |
Data for each strain are presented in rows. Prediction counts from the three standalone gene prediction methods are presented. Counts of protein-coding gene predictions reported by our algorithm and tRNA genes are also shown. Data presented are based on the automatic combined assemblies from Table 2.
aNumber of ORFs with protein-coding gene predictions where all three predictors agreed exactly or with a slight difference in the predicted start site.
bORFs where only two of the three predictors made a prediction.
cTotal protein-coding gene predictions reported by the pipeline.
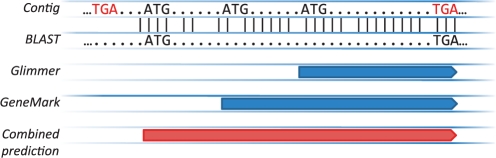
The results of these three methods were combined together using a combiner strategy outlined in Figure 3. In this strategy, we first check that at least half of the predictors report a gene in a given ORF—in our configuration, 2 of the 3 predictors. Then, the Met (putative translation start) codon closest to the beginning of the BLAST alignment is found and declared to be the gene start predicted by BLAST. We then find the gene start coordinate reported by the majority of the three predictors and report the resulting gene prediction. If no majority exists, we select the most upstream gene start predicted.
Fig. 3.
Schematics of combining strategy for prediction stage. BLAST alignment start, which may not coincide exactly with a start codon, is pinned to the closest start codon. Then, a consensus or most upstream start is selected.
In addition to protein-coding gene prediction, ribosomal genes were predicted using alignment to a reference database of ribosomal operons, and tRNA genes were predicted using the tRNAScan-SE package (Lowe and Eddy, 1997). The results are summarized in Table 3.
Results of the feature prediction stage are saved in a multi-extent GenBank formatted file. Features were named according to the following convention: contig-name_feature-id, where contig-name is as described earlier, and feature-id is a sequential zero-padded number unique to the feature across all contigs. For example, a gene with feature ID 1293 on contig 25 might have the name CDC_NME_M13320_025_1293.
To validate the overall accuracy of the gene prediction stage of the pipeline, we ran our gene prediction tools on the genome of Escherichia coli K12, one of the best-annotated bacterial genomes (analysis described in the Supplementary Material). Our pipeline was able to detect 95.7% of the annotated E.coli K12 protein-coding genes, and exactly predict starts in 85.5% of those. Fifty percent of the E.coli predictions that report incorrect start codons start within 35 nt of the true start, and all reported starts are within 200 nt of the true start.
2.5 Functional annotation
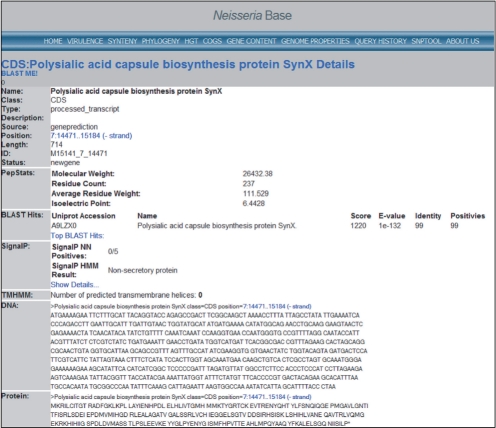
Functional annotation of genome features was also performed using a combination of tools. Annotation of protein coding genes was based on an integrated platform that makes use of six distinct annotation tools, four of which employ intrinsic sequence characteristics for annotation and two that use extrinsic homology-based approaches to compare sequences against databases of sequences and structures with known functions. Information on Gene Ontology (GO) terms, domain architecture and identity, subcellular localization, signal peptides, transmembrane helices and lipoprotein motifs is provided for each protein-coding gene (Fig. 4).
Fig. 4.
Example functional annotation listing of a N.meningitidis gene in the Neisseria Base. Draft genome data are shown including gene location, prediction and annotation status, peptide statistics, BLAST hits, signal peptide properties, transmembrane helix presence, DNA and protein sequence. All names, locations, functional annotations and other fields are searchable, and gene data are accessible from GBrowse genome browser tracks.
BLASTp alignment of predicted proteins was performed against the UniProt database (Uniprot, 2009). Homology-based searches were also made across thirteen sequence and protein domain databases with the InterProScan suite (Mulder and Apweiler, 2007). Parsing of the results was carried out against the corresponding InterPro database. The pipeline also stores the top five hits for each gene against the NCBI non-redundant protein database, to provide potentially useful information. All homology searches were run locally. Signal peptides were annotated using the SignalP package (Bendtsen et al., 2004) and transmembrane domains were annotated with the TMHMM package (Krogh et al., 2001). State of the art in subcellular localization algorithms was examined to ensure the best performance given our operational requirements. Insertion sequences (transposases) and proteins reported as virulence factors by VFDB (Chen et al., 2005; Yang et al., 2008) were also annotated. These annotations of virulence-related features make the pipeline particularly useful for projects working with pathogenic prokaryotes. Results of this analysis are summarized in Table 4.
Table 4.
Feature annotation statistics
| Strain ID | Total number of CDSa | Signal peptidesb | Transmembrane helicesc | Conserved hypothetical proteins | Putative uncharacterized proteins | Functional assignment inferred from homology | Virulence factorsd |
|---|---|---|---|---|---|---|---|
| NM13220 | 2299 | 326 (14.2%) | 184 (8.0%) | 10 (0.4%) | 708 (30.8%) | 603 (26.2%) | 36 (1.6%) |
| NM10699 | 2110 | 310 (14.7%) | 180 (8.5%) | 5 (0.2%) | 652 (30.9%) | 577 (27.3%) | 45 (2.1%) |
| NM15141 | 2184 | 317 (14.5%) | 173 (7.9%) | 16 (0.7%) | 590 (27.0%) | 583 (26.7%) | 50 (2.3%) |
| NM9261 | 2110 | 303 (14.4%) | 166 (7.9%) | 13 (0.6%) | 591 (28.0%) | 558 (26.4%) | 37 (1.8%) |
| NM18575 | 2471 | 349 (14.1%) | 193 (7.8%) | 13 (0.5%) | 725 (29.3%) | 668 (27.0%) | 48 (1.9%) |
| NM5178 | 2097 | 298 (14.2%) | 177 (8.4%) | 3 (0.1%) | 646 (30.8%) | 572 (27.3%) | 45 (2.1%) |
| NM15293 | 2063 | 304 (14.7%) | 168 (8.1%) | 6 (0.3%) | 613 (29.7%) | 567 (27.5%) | 47 (2.3%) |
| BBE001 | 4799 | 977 (20.4%) | 368 (7.7%) | 9 (0.2%) | 807 (16.8%) | 1184 (24.7%) | 54 (1.1%) |
| BBF579 | 4656 | 934 (20.1%) | 339 (7.3%) | 9 (0.2%) | 739 (15.9%) | 1171 (25.2%) | 45 (1.0%) |
Data for each strain are presented in rows. Data presented are based on the automatic combined assemblies from Table 2 and the gene predictions from Table 3.
aTotal putative protein-coding sequences analyzed.
bAs predicted by SignalP (Bendtsen et al., 2004); percentage of total CDS indicated in parentheses.
cAs predicted by TMHMM (Krogh et al., 2001).
dAs predicted by BLASTp alignment against VFDB (Chen et al., 2005; Yang et al., 2008); http://www.mgc.ac.cn/VFs/.
After the functional annotations were determined, a naming scheme was employed for each locus to conform to standard annotation terminology. Specific gene names were assigned according to homology-based results. For genes that had a Uniprot result with a best hit at >91% amino acid sequence identity and an e-value <1e-9, the gene assumed the best hit's name. If the best hit had the keyword “hypothetical’, then we used a domain name from InterPro to name the gene. For example, if a gene was given the name ‘hypothetical’ from Uniprot and a domain name of ‘transferase’ from InterPro, then the final name was ‘hypothetical transferase protein’. Therefore, most genes that were given ‘hypothetical’ or ‘putative’ prefixes could then be given a more compre-hensive name based on further information such as domain names or protein functions. Genes with unknown functions found across many genomes were given the name ‘conserved hypothetical protein’, and all other putative genes with unknown functions were given the name ‘putative uncharacterized protein’.
2.6 Availability
The pipeline software package is available at our website (http://nbase.biology.gatech.edu). The package contains detailed instructions and scripts for installation of the pipeline and all external programs, documentation on usage of the pipeline and its organization. Components which require large biological databases automatically download local copies of those databases upon installation.
All of the N.meningitidis genomes reported here, along with custom annotations and tools for searching and comparative sequence analysis, are available for researchers online at our genome browser database (http://nbase.biology.gatech.edu).
3 DISCUSSION
3.1 Genome biology of N.meningitidis and B.bronchiseptica
We have used the pathogen N.meningitidis for the majority of developmental and production testing of our pipeline. Although N.meningitidis gains no fitness advantage from virulence, it occasionally leaves its commensal state and causes devastating disease (Meyers et al., 2003). Several recent studies have used whole-genome analysis to determine the basis of virulence in this species but none have been conclusive (Hotopp et al., 2006; Perrin et al., 2002; Schoen et al., 2008). With the recent advent of next-generation sequencing and the application of an analytical pipeline, such as presented here, this problem and other problems like it can be addressed in individual laboratories on a genome-wide scale. Here, we briefly speculate on a few of the implications of our findings for the genome biology of N.meningitidis to underscore the potential utility of our pipeline.
Whole-genome analysis of microbes has led to the development of the ‘pan-genome’ concept (Tettelin et al., 2005). A pan-genome refers to the collection of all genes found within different strains of the same species. An open pan-genome means that the genome of any given strain will contain unique genes not found within the genomes of other known strains of the same species. The extent to which microbial pan-genomes are open is a matter of debate (Lapierre and Gogarten, 2009). Recent studies have suggested that the N.meningitidis pan-genome is essentially open (Schoen et al., 2008), consistent with the fact that it is known to be a highly competent species (Chen and Dubnau, 2004; Kroll et al., 1998). We evaluated this hypothesis by finding the number of unique genes in each of the seven strains reported here along with seven previously published strains, using the results of our analytical pipeline. Our findings are consistent with Schoen et al. (2008), in the sense that every genome sequence was found to contain at least 43 unique genes not found in any other strain. Thus, the N.meningitidis pan-genome does appear to be open.
N.meningitidis is a human commensal that most often does not cause disease, and avirulent strains of the species are referred to as carriage strains. Results of previous comparative genomic analyses have been taken to suggest that carriage strains represent a distinct evolutionary group that is basal to a group of related virulent strains of N.meningitidis (Schoen et al., 2008). We tested this hypothesis using the results of our analytical pipeline applied to three carriage strains and eight virulent strains of N.meningitidis. Whole-genome sequences were aligned and pairwise distances between genomes, based on nucleotide diversity levels, were compared within and between groups of carriage and virulent strains. We found that average of the pairwise genome sequence distances within (w) the carriage and virulent groups of strains was not significantly different from the average pairwise distances between (b) groups (w=0.074±0.027, b=0.090±0.014, t=0.693, P=0.491). This result is inconsistent with the previously held notion that carriage and virulent strains represent distinct evolutionary groups based on whole-genome analysis. However, our findings are consistent with earlier work that found little genetic differentiation between carriage and virulent strains of N.meningitidis (Jolley et al., 2005).
Currently, there is no unambiguous molecular assay to distinguish B.bronchiseptica from other Bordetella species. One reason the two B.bronchiseptica genomes reported here were characterized was to discover genes unique to the species (i.e. not present in any other Bordetella species) to facilitate the development of a B.bronchiseptica-specific PCR assay. To identify such genes, we performed BLASTn with B.bronchiseptica query genes uncovered by our pipeline against other B.bronchiseptica strain genomes along with four genomes of closely related Bordetella species. We uncovered a total of 223 genes that are present in all B.bronchiseptica strains and absent in all other Bordetella species. To narrow down this set of potential PCR assay targets, we searched for the most conserved B. bronchiseptica-specific genes. As a point of reference, we determined the sodC gene used in the N.meningitidis-specific PCR assay (Kroll et al., 1998) to be 99.6% identical among all six completely sequenced strains of N.meningitidis. There are seven B. bronchiseptica-specific genes with ≥99.6% sequence identity; these genes represent a prioritized list of potential PCR assay targets.
3.2 Computational genomics pipeline
We have presented our computational genomics pipeline, a local solution for automated, high-throughput computational support of prokaryotic genome sequencing projects. While the revolution in sequencing technology makes possible the execution of genome projects within individual laboratories, the computational infrastructure to fully realize this possibility does not yet exist. We made a comprehensive effort to put the tools required for this infrastructure into the hands of biologists working with next-generation sequencing data. Our aim in the course of this project was to facilitate decentralized biological discoveries based on affordable whole-genome prokaryotic sequencing, a mode of science termed ‘investigator-initiated genomics’. For example, one project enabled by the pipeline in our laboratory is a platform for SNP detection and analysis in groups of bacterial genomes.
One of our major goals was to provide full automation of our pipeline's entire workflow, and this has been achieved. On the other hand, to allow computationally savvy users to realize the power of customizability, a semi-automated process is desirable. We have made an effort to strike a balance between these objectives, and provide a modular, hierarchically organized structure to permit maximum customization when so desired.
The state of the art in prokaryotic computational genomics moves at a formidable pace. The modular organization of our pipeline, along with the emphasis on integration of complementary software tools, allows us to continually update our platform to keep pace with developments in computational genomics. For instance, if a new, better assembler becomes available, we can include its results in the assembly stage with a simple change to the pipeline code.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all participants of the Georgia Tech Computational Genomics class; to Leonardo Mariño-Ramírez for valuable guidance and input; and to Joshua S. Weitz for his support.
Funding: Defense Advanced Research Projects Agency (HR0011-05-1-0057 to A.O.K.); The Alfred P. Sloan Foundation (BR-4839 to I.K.J.); Georgia Research Alliance (GRA.VAC09.O to I.K.J., P.J., S.A.); Centers for Disease Control and Prevention (1 R36 GD 000075-1 to L.S.K.); Bioinformatics program, Georgia Institute of Technology (to J.H., P.J., V.N., S.A.).
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JDV, et al. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bentley D, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, et al. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- Chen L, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, et al. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, et al. Real-Time DNA Sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Gerlach G, et al. Evolutionary trends in the genus Bordetella. Microbes Infect./Institut Pasteur. 2001;3:61–72. doi: 10.1016/s1286-4579(00)01353-8. [DOI] [PubMed] [Google Scholar]
- Holmes EC, et al. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 1999;16:741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- Hotopp JD, et al. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152:3733–3749. doi: 10.1099/mic.0.29261-0. [DOI] [PubMed] [Google Scholar]
- Jolley KA, et al. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 2005;22:562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- Kislyuk A, et al. Frameshift detection in prokaryotic genomic sequences. Int. J. Bioinform. Res. Appl. 2009;5:458–477. doi: 10.1504/IJBRA.2009.027519. [DOI] [PubMed] [Google Scholar]
- Krogh A, et al. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kroll JS, et al. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl Acad. Sci. USA. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A, Grigoriev V. Challenges in whole-genome annotation of pyrosequenced fungal genomes. 2009 Available at: http://dx.doi.org/10.1038/npre.2009.3191.1. [Google Scholar]
- Lapierre P, Gogarten JP. Estimating the size of the bacterial pan-genome. Trends Genet. 2009;25:107–110. doi: 10.1016/j.tig.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum I, et al. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 2009;10:R103. doi: 10.1186/gb-2009-10-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci.USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2009;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers LA, et al. Epidemiology, hypermutation, within-host evolution and the virulence of Neisseria meningitidis. Proc. Biol. Sci./Roy. Soc. 2003;270:1667–1677. doi: 10.1098/rspb.2003.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- Parkhill J, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Perrin AS, et al. Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species. Infect. Immun. 2002;70:7063–7072. doi: 10.1128/IAI.70.12.7063-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M, et al. Comparative genome assembly. Brief Bioinform. 2004;5:237–248. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- Quinlan A, et al. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat. Methods. 2008;5:179–181. doi: 10.1038/nmeth.1172. [DOI] [PubMed] [Google Scholar]
- Rissman A, et al. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein NE, et al. Meningococcal disease. N. Engl. J. Med. 2001;344:1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- Schoen C, et al. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl Acad. Sci. 2008;105:3473–3478. doi: 10.1073/pnas.0800151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, et al. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Sommer D, et al. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, et al. DIYA: a bacterial annotation pipeline for any genomics lab. Bioinformatics. 2009;25:962–963. doi: 10.1093/bioinformatics/btp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc. Natl Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniprot Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008;36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.