Abstract
Summary: We present SVDetect, a program designed to identify genomic structural variations from paired-end and mate-pair next-generation sequencing data produced by the Illumina GA and ABI SOLiD platforms. Applying both sliding-window and clustering strategies, we use anomalously mapped read pairs provided by current short read aligners to localize genomic rearrangements and classify them according to their type, e.g. large insertions–deletions, inversions, duplications and balanced or unbalanced inter-chromosomal translocations. SVDetect outputs predicted structural variants in various file formats for appropriate graphical visualization.
Availability: Source code and sample data are available at http://svdetect.sourceforge.net/
Contact: svdetect@curie.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The identification of genomic structural variations is a key step in understanding human genetic diversity and evolution as well as disease etiology. Numerous genetic diseases, including cancer, have been associated with structural variants (SVs; Futreal et al., 2004). Although array-based techniques have been successful in many studies for detecting SVs, the relatively low resolution in the detection of breakpoints and the characterization of small SVs remained challenging. With the arrival of high-throughput sequencing technologies such as the Illumina Genome Analyzer or the Applied Biosystems SOLiD system, using short-insert paired-end or mate-paired reads (referred here as paired-ends) has improved our ability to detect SVs (Korbel et al., 2007). With a priori information from paired-ends such as order, orientation and insert size of pairs as constraints during read alignment to the reference genome, anomalously mapped pairs indicate potential genomic variations from the reference. The need for algorithms specifically designed for SV prediction from paired-end mapping (PEM) data has recently led to the development of new software packages, including GASV (Sindi et al., 2009), BreakDancer (Chen et al., 2009) and others (see review, Medvedev et al., 2009).
Here, we present a new freely available program called SVDetect for SV detection and type prediction from PEM data. SVDetect identifies different types of SVs, e.g. large insertions–deletions and inversions, with both clustering and sliding-window strategies, and helps to visualize them at the genomic scale. Compared to other tools, the novelty of our method consists in its multiple ability to: (i) analyze both paired-end and mate-pair sequencing data; (ii) use unique PEM constraints to improve SV detection; (iii) predict various types of tandem duplication and to distinguish between balanced and unbalanced rearrangements; (iv) compare SVs across multiple samples; (v) construct copy number profiles; and (vi) create various output file formats for graphical views of SV.
2 METHODS
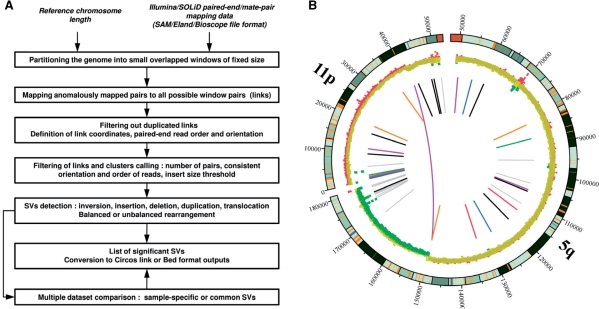
The first step in SVDetect is to regroup all pairs that are suspected to originate from the same SV. The input consists of paired-ends mapped to the reference genome, so that either the orientation of pairs is incorrect and/or the distance between them is out of the typical range. Starting from a list of such paired-end anomalous mapping, SVDetect uses a sliding-window strategy to identify all groups of pairs sharing a similar genomic location. The reference genome is divided into overlapping windows of fixed size, and each pair of windows can possibly form a link if at least one pair anchors them by its ends (Fig. 1A). To each link connecting two genomic fragments, we assigned a certain number of features, such as chromosomal location, number of pairs, orientation and order of the involved paired-ends. After removing redundant links, these features are further used in the filtering step to call clusters of anomalous paired-end reads and detect the type of corresponding SVs.
Fig. 1.
Overview of SVDetect algorithm and output. (A) The workflow. (B) Graphical visualization of predicted SVs. Genomic locations of inter- and intra-chromosomal links are shown using the Circos software. Starting from outside of the circle, the following features are displayed: chromosome ideograms, scatter plot of the copy-number profile and color-coded spans of chromosomal links.
The filtering procedure of SVDetect takes as input all links previously identified and uses user-defined filtering parameter values to call PEM clusters. The minimum number of paired-ends is one of the most important filtering parameters to call a cluster. Use of such a threshold improves confidence in the detection of SVs. Another option is filtering of pairs whose ends are not oriented in the same way as the ends of the majority of pairs in the two linked regions. If one of the two ends of remaining pairs has an unexpected strand orientation, the cluster is annotated as a potential inversion. The order of paired-ends is used to annotate an inter-chromosomal cluster both as a balanced or an unbalanced translocation, and to estimate a genomic coordinate range of predicted breakpoints (the resolution depending on the insert size). To achieve this goal, SVDetect filters out any pair for which the order of a read and its mate is inconsistent with the majority of pairs in the cluster. The read order is also used to characterize balanced SV affecting only one chromosome, e.g. to predict the two breakpoints of an inversion (see Supplementary Material for details of PEM signatures and the order filtering procedure).
Constraints on the distance between mapped ends in a pair are used to filter intra-chromosomal PEM clusters when calling insertion–deletion events. Assuming the normality of insert size distribution of aligned reads, we generally use 2 to 3 SD from the mean insert size to detect potential deletions and insertions. The mean insert size for pairs in a called cluster is then provided. By combining the information about strand, order and insert size of paired-end reads for each cluster, the type of rearrangement can be predicted. Lastly, predicted SVs coming from multiple paired-end datasets can be compared to identify common or sample-specific variants.
SVDetect provides additional functionality to analyze paired-end coverage complementary to the previously described strategy, to improve SV characterization. From all pairs correctly mapped with the expected insert size, we calculate the log-ratio of depth-of-coverage between a sample and a control dataset in a sliding window along the genome. This ratio is used to characterize copy-number profiles by identifying potential loss or gain events, and can be compared to the previously predicted SV positions.
SVDetect offers different output formats to facilitate the subsequent analysis of reported paired-end clusters. Data conversion to the BED format or the Circos link format (Krzywinski et al., 2009) is available for the graphical representation of pairs and/or copy-number profiles (Fig. 1B). A user-defined color-code related to the number of pairs can be chosen for better visualization of potentially significant SVs.
3 RESULTS
To illustrate the use of SVDetect, we tested the program to predict SVs from two different types of mate-pair sequencing data: Illumina GAII 50 bp reads of neuroblastoma cell lines, and Applied Biosystems SOLiD v2 25 bp reads of a wild-type and mutant pif1Δ strains in yeast. First, from 2070 anomalous Illumina read pairs in the chromosome arms 5q and 11p, we found 37 clusters specific to the neuroblastoma sample compared with a reference sample. Only one significant cluster (seven pairs) suggests an inter-chromosomal SV, predicted to be an unbalanced translocation that is also supported by the corresponding copy-number profile (Fig. 1B). Close to the predicted breakpoint location in 5q, we found an intra-chromosomal cluster predicted to be an inverted duplication, suggesting a more complex genome rearrangement. The yeast mate-paired datasets were used to compare SVDetect with the variant detection tool GASV. From approximately 1 million of aberrantly mapped read pairs in the wild-type and mutant strains, both tools retrieved all five known SVs in the mutant strain. With respect to GASV, the specific filtering procedures introduced by SVDetect discard hypothetical rearrangements supported with inconsistent orientation or order of their read pairs (see Supplementary Material for tool comparison and data analysis).
Funding: ‘Projet Incitatif Collaboratif Bioinformatique et Biostatistiques’ of the Institut Curie; Ligue Nationale Contre le Cancer; Institut National du Cancer.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;18:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P, et al. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6(Suppl. 11):S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- Sindi S, et al. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:222–230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.