Abstract
Glial cell line-derived neurotrophic factor (GDNF) plays a crucial role in regulating the proliferation of spermatogonial stem cells (SSC). The signaling pathways mediating the function of GDNF in SSC remain unclear. This study was designed to determine whether GDNF signals via the Ras/ERK1/2 pathway in the C18-4 cells, a mouse SSC line. The identity of this cell line was confirmed by the expression of various markers for germ cells, proliferating spermatogonia, and SSC, including GCNA1, PCNA, Oct-4, GFRα 1, and Ret. Western blot analysis revealed that GDNF activated Ret tyrosine phosphorylation. All three isoforms of Shc were phosphorylated upon GDNF stimulation, and GDNF induced the binding of Shc and Grb2 to the phosphorylated Ret as indicated by immunoprecipitation and Western blotting. The active Ras was induced by GDNF, which further activated ERK1/2 phosphorylation. GDNF stimulated the phosphorylation of CREB-1, ATF-1, and CREM-1, and c-fos transcription. Notably, the increase in ERK1/2 phosphorylation, c-fos transcription, bromodeoxyuridine incorporation, and metaphase counts induced by GDNF, was completely blocked by pretreatment with PD98059, a specific inhibitor for MEK1, the upstream regulator of ERK1/2. GDNF stimulation eventually up-regulated cyclin A and CDK2 expression. Together, these data suggest that GDNF induces CREB/ATF-1 family member phosphorylation and c-fos transcription via the Ras/ERK1/2 pathway to promote the proliferation of SSC. Unveiling GDNF signaling cascades in SSC has important implications in providing attractive treatment targets for male infertility and testicular cancers as well as for the potential de-differentiation of these cells to cells that mimic embryonic stem cells.
Keywords: GDNF, spermatogonial stem cells, Ret, Ras/ERK pathway, CREB/ATF-1 family, c-fos
INTRODUCTION
Spermatogenesis is a cellular process by which spermatogonial stem cells (SSC) divide and differentiate into spermatozoa. SSC are unique since they are the only stem cells in the body that undergo self-renewal throughout life and transmit genetic information to offspring [1, 2, 3]. A better understanding of the molecular mechanisms controlling self-renewal, differentiation, or apoptosis of SSC is essential for the regulation of spermatogenesis as well as for the potential use of the SSC to produce embryonic-like stem cells. Glial cell line-derived neurotrophic factor (GDNF) is the first molecule known to regulate the cell fate decision of SSC [4, 5]. In vivo data from transgenic mice indicate that GDNF mediates the renewal and differentiation of undifferentiated spermatogonia in a dose-dependent manner [4]. While GDNF-deficient mice show partial depletion of SSC, the mice over expressing GDNF display an accumulation of undifferentiated spermatogonia [4]. GDNF can also promote the proliferation of undifferentiated spermatogonia in vivo [6] and stimulates DNA synthesis in Ret-expressing spermatogonia [7]. In vitro, it has been demonstrated by our group and others that GDNF facilitates the expansion of SSC over a long culture period [8, 9, 10]. However, the signal transduction pathways that lead to the GDNF-induced DNA synthesis and proliferation of SSC remain largely an enigma.
GDNF signals through a multicomponent receptor complex comprised of the Ret receptor tyrosine kinase and a member of the GFRα family of glycosylphosphatidylinositol (GPI)-anchored receptors, which are required for GDNF binding to Ret [11, 12]. In nervous tissue, there are multiple-pathways described for GDNF signaling. GDNF can trigger intracellular signaling through a Ret-independent pathway via GPI-linked protein GFRα1, which leads to activation of Src family tyrosine kinase and mediates various downstream responses to promote cell survival [13, 14]. We have recently shown that GDNF uses Src tyrosine kinase and phosphatidylinositol 3-kinase activation to up-regulate N-myc expression in SSC [10]. In neuronal cell lines that co-express Ret and GFRα1, GDNF signals through the Ret-dependent pathway to induce intracellular signal cascades [11, 14, 15]. Within the seminiferous tubules, GDNF is secreted by Sertoli cells [4, 16]. We and others have shown that the SSC but not differentiating germ cells express its co-receptors, GFRα1 and Ret [9, 17, 18]. It has been suggested that GDNF mediates Ret signaling via GFRα1 to regulate the cell fate of undifferentiated spermatogonia [17]. Nevertheless, the downstream events triggered by GDNF/ GFRα1/Ret need to be elucidated.
The small guanosine triphosphatase protein Ras is known as a key mediator for proliferation and differentiation [19–21], and the extracellular signal-regulated kinases (ERK), an important member of the mitogen-activated protein kinases, is involved in modulating a variety of cellular functions, including cell proliferation, differentiation, and cell cycle progression [22, 23]. It has been suggested that ERK is essential for the proliferation of c-kit expressing type A1–A4 spermatogonia stimulated with stem cell factor (SCF) [22]. In the current study, we sought to ascertain whether GDNF signals through the Ras/ERK1/2 pathway in the C18-4 cells, a murine spermatogonial stem cell line. We found that GDNF activated the tyrosine phosphorylation of Ret and Shc, and that GDNF induced the binding of Shc and Grb2 to the phosphorylated Ret. GDNF stimulation subsequently activated the Ras/ERK1/2 pathway, which led to the phosphorylation of transcription factors cAMP responsive element-binding protein 1 (CREB-1), the activating transcription factor-1 (ATF-1), and cAMP response element modulation protein 1 (CREM-1), as well as the transcription of the immediate early gene c-fos. The active Ras and ERK1/2 phosphorylation induced by GDNF were further confirmed in freshly isolated type A spermatogonia from 6-day-old mice. The inhibitor PD98059 for the MEK1/ERK1/2 pathway completely blocked the up-regulation in ERK1/2 phosphorylation, c-fos transcription, bromodeoxyuridine incorporation, and metaphase counts induced by GDNF. Furthermore, GDNF increased the expression of cell cycle regulators, including cyclin A and cyclin-dependent kinase 2 (CDK2) but not cyclin D1, and PD98059 blocked the up-regulation of cyclin A and CDK2 expression. Our data demonstrate that GDNF/GFRα1/Ret signaling activates the Ras/ERK1/2 pathway to promote DNA synthesis and cell proliferation.
MATERIALS and METHODS
The C18-4 Cells
The C18-4 cells, a mouse male germline stem cell line, were generated by our group [24].
Animals
BALB/c mothers with 6-day-old male pups were obtained from the Charles River Laboratories, Inc. (Wilmington, MA, USA). All animal care procedures were carried out pursuant to the National Research Council’s Guide for the Care and Use of Laboratory Animals, and the experimental protocols were approved by the Animal Care and Use Committee of Georgetown University.
Immunofluorescence
The immunofluorescence procedures used on the C18-4 cells were described in our previous publication [25]. The antibodies included: GCNA1 from Dr. George C. Enders; PCNA (EMD Biosciences, Inc., San Diego, CA); Oct-4 (Santa Cruz Biotechnology Inc., Santa Cruz, CA); GFRα1 (Santa Cruz Biotechnology Inc.); Ret (R&D Systems, Minneapolis, MN). All the secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. DAPI was used to stain the cell nuclei, and the cells were observed for epifluorescence using an Olympus Fluoview 500 Laser Scanning Microscope (Olympus, Melville, NY). Immunofluorescence was also carried out to determine the expression change and subcellular localization of cyclin A in the C18-4 cells with 100 ng/ml of recombinant rat GDNF (R&D Systems) treatment for 6 h and 24 h and in the GDNF-untreated cells using anti-cyclin A (Santa Cruz Biotechnology Inc.).
RNA Extraction and RT-PCR
Total RNA was extracted from the GDNF-treated or -untreated C18-4 cells using Trizol reagent (Invitrogen, Carlsbad, CA). RT-PCR was performed according to the protocol we described previously [25]. The forward and reverse primers of the chosen genes, including PCNA, Oct-4, GFRα1, Ret, Gapdh, and c-fos were designed and listed in Supplementary Data - Table 1. PCR products were separated by electrophoresis on 1.2% agarose gels. The gels were exposed to a Transilluminator (Fisher Scientific, Pittsburg, PA), and pictures were taken with a Photo-Documentation Camera (Fisher Scientific). The data were scanned with an Epson Perfection 3200 PHOTO, and densitometric analyses were processed with Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Isolation of type A spermatogonia from 6-day-old mice
Type A spermatogonia and Sertoli cells were isolated from the testes of 6-day-old mice using mechanical dissociation and enzymatic digestion containing collagenase, trypsin, and hyaluronidase [26, 27]. Type A spermatogonia were further separated from the Sertoli cells by differential plating and the purity of type A spermatogonia was around 95% as evaluated by immunocytochemical staining using an antibody to GCNA1.
Immunoprecipitation and Western Blotting
The C18-4 cells were cultured in the DMEM/F12 supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 unit/ml penicillin, and 100 mg/ml streptomycin, in 75 cm2 cell culture flasks until cell confluence was up to 80%. The cells were starved in serum-free DMEM/F12 for 16 h prior to the incubation with 100 ng/ml GDNF at 34°C for 5 min and 15 min and lysed with 1 ml of RIPA lysis buffer (Santa Cruz Biotechnology Inc.). After 30 min lysis on ice, cell lysates were cleared by centrifugation at 12,000 g, and the concentration of protein was measured by the Bio-Rad Bradford assay (BioRad Laboratories, Hercules, CA).
Five micrograms of cell lysate from each sample were used for SDS-PAGE, and Western blots were performed according to the protocol we described previously [25]. The chosen antibodies included phospho-Ret (Tyr 1062) (Santa Cruz Biotechnology Inc.) and phospho-CREB-1 (Ser-133) (Santa Cruz Biotechnology Inc.). After extensive washes in PBS, the blots were detected using the Western Blotting Luminol Reagent (Santa Cruz Biotechnology Inc.) and exposed to autoradiography films. The membranes were stripped using the Restore™ Western Blot Stripping Buffer (Pierce Biotechnology, Inc., Rockford, IL) and re-probed for the detection of Ret (Santa Cruz Biotechnology Inc.) or β-actin (IMGENEX Corp, San Diego, CA). The films were scanned with an Epson Perfection 3200 PHOTO, and densitometric analyses were processed with Adobe Photoshop 7.0.
The C18-4 cells were pretreated with or without PD98059 (BIOSOURCE, Camarillo, CA) for 30 min and then treated with or without 100 ng/ml of GDNF for the indicated time periods. Western blot analysis was carried out using antibodies to phospho-ERK1/2 (Thr 202/Tyr 204), cyclin A, cyclin D1, and CDK2, respectively. All the antibodies were purchased from Santa Cruz Biotechnology Inc. The blots were re-probed for detecting ERK2 or β-actin using antibodies to ERK2 (Santa Cruz Biotechnology Inc.) or β-actin.
Type A spermatogonia were cultured in 100 mm Petri dishes with DMEM/F12 containing 10% FBS and 2 mM L-glutamine for 3 h to allow restoring and attaching to the dish. The cells were starved in serum-free DMEM/F12 for 8 h, and then 100 ng/ml GDNF was added to the medium for 5 min and 15 min. Western blotting was perform using antibody to phospho-ERK1/2 (Thr 202/Tyr 204). The blots were re-probed for detecting ERK2 using an antibody to ERK2.
For immunoprecipitations with anti-Shc, 5 µl of anti-Shc (Upstate Biotechnology Inc., Lake Placid, NY) was added to 500 µl of cell lysates from each sample overnight at 4°C. The immunocomplex was captured by adding 50 µl of the prepared Protein A agarose beads slurry (Upstate Biotechnology Inc.) for 2 h at 4°C. The beads were washed five times with RIPA lysis buffer and boiled for 5 min in protein sample buffer. Half the immunoprecipitates was fractionated by SDS-PAGE, and Western blots were performed using anti-phospho-Shc (Tyr 317) (Upstate Biotechnology Inc.) or anti-phospho-Ret (Tyr 1062). The blots were re-probed for the detection of Shc using an antibody to Shc.
Immunoprecipitations with anti-Grb2 (Upstate Biotechnology Inc.) were carried out using 2 µg of anti-Grb2 that was added to 300 µl of cell lysates from each sample overnight at 4°C. The immunocomplex was captured by adding 50 µl of the prepared Protein G MicroBeads (Miltenyi Biotec Inc., Auburn, CA) for 30 min on ice and proceeded to magnetic separation using µMACS Separator (Miltenyi Biotec Inc.). Half the immunoprecipitates was fractionated by SDS-PAGE, and Western blots were performed using anti-phospho-Ret (Tyr 1062). The blots were re-probed for the detection of Grb2 using an antibody to Grb2.
The Ras activation Assay
The C18-4 cells and type A spermatogonia were treated with 100 ng/ml of GDNF for 5 min and 15 min or without GDNF treatment and then lysed with 1 ml of Lysis/Binding/Washing Buffer (Assay Designs, Inc., Ann Arbor, MI). The cell lysates from each sample were incubated with GST-Raf1-RBD and an Immobilized Glutathione Disc for 1 h at 4°C using the StressXpress Ras Activation Kit (Assay Designs, Inc.). Half the eluted samples was separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with a specific antibody to Ras (Assay Designs, Inc.).
Bromodeoxyuridine (BrdU) Incorporation Assay
The C18-4 cells were grown on coverslips in 6-well culture plates with DMEM/F12 containing 10% FBS for 6 h to allow the cells to attach to coverslips. The cells were starved in serum-free DMEM/F12 for 16 h prior to pretreatment with or without 100 µM of PD98059 for 30 min, and then 100 ng/ml GDNF and 30 µg/ml of BrdU (Sigma, Saint Louis, MO) or 30 µg/ml of BrdU alone was added to the medium. After 16 h of culture, immunocytochemical staining was performed using a monoclonal anti-BrdU (Sigma, Saint Louis, MO) pursuant to the DBA kit (Zymed Laboratories Inc., South San Francisco, CA). The percentage of BrdU-positive cells was counted out of 500 cells, and the data were presented from three experiments.
Metaphase Counting
The C18-4 cells were cultured in 100 mm Petri dishes with DMEM/F12 containing 10% FBS for 6 h. The cells were starved in serum-free DMEM/F12 for 16 h prior to pretreatment with or without 100 µM of PD98059 for 30 min, and then 100 ng/ml GDNF were added to the medium. Colcemid was added to the culture medium for 1 h prior to cell harvest. After 24 h of culture, the cells were processed using standard metaphase counting procedures. The percentage of cells in metaphase was counted out of 500 cells. The data were presented from three experiments.
Statistical Analysis
All the values were presented as mean ± standard error of the mean (SEM), and statistically significant differences (p< 0.05) among the GDNF treatment group, the control, and the cells pretreatment with PD98059, were determined using the analysis of variance (ANOVA) and Tukey post-tests.
RESULTS
The C18-4 Cells Express GCNA1, PCNA, Oct-4, GFRα1, and Ret
We first confirmed the identify of the C18-4 cells using various markers for germ cells, proliferating spermatogonia, as well as SSC. RT-PCR analyses showed that the mRNA of PCNA, Oct-4, GFRα1, and Ret was present in the C18-4 cells (Fig. 1A). Immunocytochemical staining revealed that the C18-4 cells expressed GCNA1 (Fig. 1B), PCNA (Fig. 1C), Oct-4 (Fig. 1D), GFRα1 (Fig. 1E), and Ret (Fig. 1F), reflecting that the C18-4 cells are phenotypically SSC.
Figure 1.
The expression of GCNA1, PCNA, Oct-4, GFRα1, and Ret in the C18-4 cells as shown by RT-RCR and immunocytochemistry. (A) RT-PCR analysis shows the mRNA of PCNA, Oct-4, GFRα1, and Ret in the C18-4 cells. All the RNA samples were treated with DNase to remove the potential contamination of genomic DNA before RT, and the RNA sample without RT but with PCR was used as a negative control. (B–F) Immunocytochemical staining shows the protein expression of GCNA1 (red fluorescence, nuclei) (B), PCNA (green fluorescence, nuclei) (C), Oct-4 (green fluorescence, nuclei) (D), GFRα1 (green fluorescence, surface cytoplasm and plasma membrane) (E), Ret (green fluorescence, cytoplasm) (F), in the C18-4 cells. Staining with DAPI (blue fluorescence) was used to identify cell nuclei. Scale bars in B, C, D, E, and F =10 µm.
GDNF Activates the Tyrosine Phosphorylation of Ret and Shc and Induces the Binding of Shc and Grb2 to the Phosphorylated Ret in the C18-4 Cells
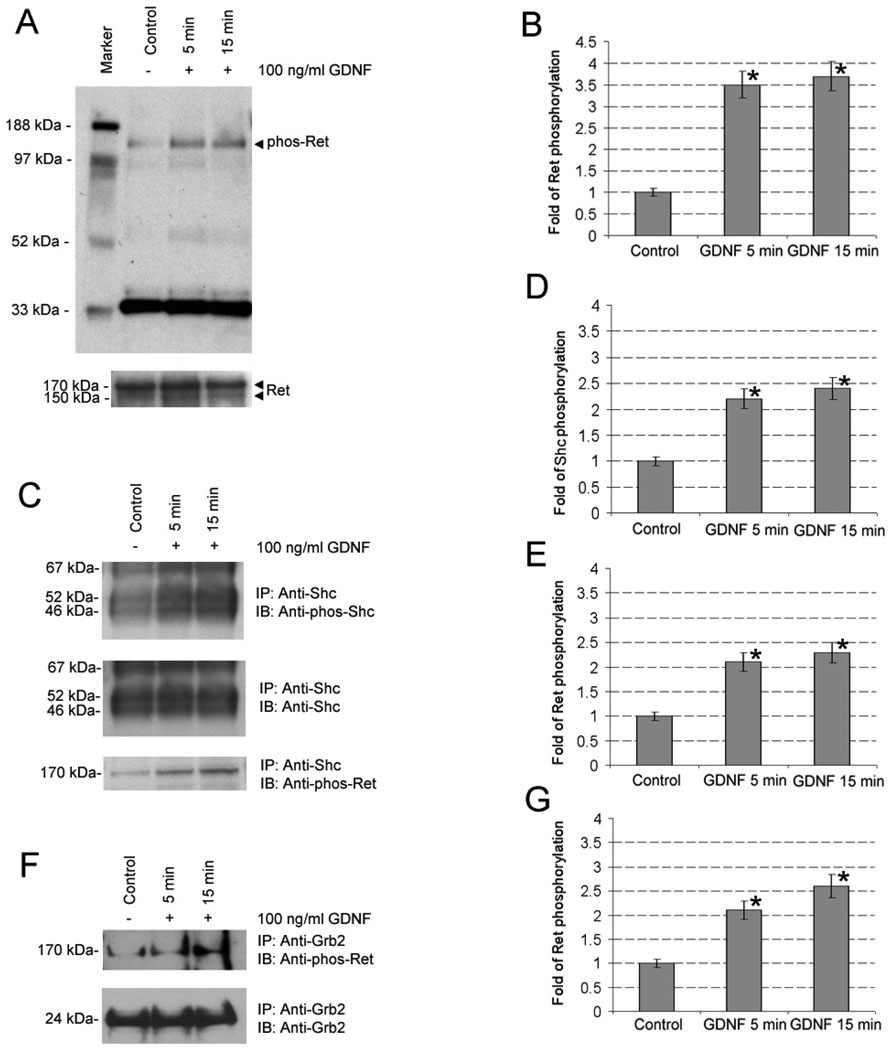
We then examined whether GDNF can activate the tyrosine phosphorylation of Ret and Shc in the C18-4 cells. Western blots clearly showed that the phosphorylation of Ret (170 kDa) at tyrosine 1062 was activated upon GDNF stimulation for 5 min and 15 min (Fig. 2A). Furthermore, immunoprecipitation with anti-Shc and Western blotting using anti-phospho-Shc revealed that all the three isoforms (46, 52, and 67 kDa) of tyrosine phosphorylation of Shc were activated by GDNF treatment for 5 min and 15 min (Fig. 2C). Meanwhile, immunoprecipitation with anti-Shc and Western blotting using anti-phospho-Ret confirmed that the phosphorylation of Ret at tyrosine 1062 was activated by GDNF stimulation in the C18-4 cells and showed that GDNF induced the binding of Shc to the tyrosine phosphorylated Ret (Fig. 2C). Similarly, immunoprecipitation with anti-Grb2 and Western blotting using anti-phospho-Ret further confirmed that the phosphorylation of Ret at tyrosine 1062 was induced upon GDNF stimulation in the C18-4 cells and revealed that GDNF induced the binding of Grb2 to the tyrosine phosphorylated Ret (Fig. 2F, upper panel). Densitometric analyses showed significant differences for each of the experiments upon the addition of GDNF at the indicated time intervals (Fig. 2B, 2D, 2E, 2G).
Figure 2.
The teyrosine phosphorylation of Ret and Shc is activated by GDNF stimulation in the C18-4 cells and both Shc and Grb2 are co-immunoprecipitated with the phosphorylated Ret in the C18-4 cells after GDNF stimulation. (A) The C18-4 cells were serum-starved for 16 h and then treated with 100 ng/ml GDNF for 5 and 15 min or without GDNF treatment. Fifty micrograms of total protein from the GDNF treated or untreated cells were resolved by 4–12% SDS-PAGE and blotted with anti-phospho-Ret (upper panel). The same membrane was re-probed with anti-Ret and showed Ret protein expression in all lanes (lower panel). The molecular mass standards are shown on the left, and the results are representative of three independent experiments. (B) The GDNF-induced phosphorylation of Ret relative to control (1.0) after normalization to the signal obtained with Ret. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (C) Five hundred micrograms of total protein from the GDNF treated or untreated cells were immunoprecipitated with anti-Shc, and half the immunoprecipitates was blotted with anti-phosphor-Shc (upper panel). The same membrane was re-probed with anti-Shc and showed Shc expression in all lanes (middle panel). The same membranes were re-probed with anti-phospho-Ret and they displayed the tyrosine phosphorylation of Ret that were pulled down together with Shc during the immunoprecipitation (lower panel). IP: immunoprecipitation; IB: immunoblotting. The molecular weights are shown on the left, and the results are representative of three independent experiments. (D–E) The GDNF-induced phosphorylation of Shc (52 kDa) and phospho-Ret relative to control (1.0) after normalization to the signal obtained with Shc (52 kDa). Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (F) Three hundred micrograms of total protein from the GDNF treated or untreated cells were immunoprecipitated with anti-Grb2, and half the immunoprecipitates was blotted with anti-phosphor-Ret, showing the tyrosine phosphorylation of Ret that was pulled down together with Grb2 during the immunoprecipitation in the C18-4 cells (upper panel). The same membrane was re-probed with anti-Grb2 and showed Grb2 expression in all lanes (lower panel). IP: immunoprecipitation; IB: immunoblotting. The molecular weights are shown on the left, and the results are representative of three independent experiments. (G) The GDNF-induced phosphorylation of Ret relative to control (1.0) after normalization to the signal obtained with Grb2. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks.
GDNF Activates the Ras/ERK1/2 Pathway in the C18-4 Cells and Type A Spermatogonia
Next, we tested whether GDNF could activate the Ras/ERK1/2 pathway in the C18-4 cells and type A spermatogonia. Immunoprecipitation with GST-Raf1-RBD and immunoblotting using Ras antibody showed that the active Ras (GTP-Ras, 21 kDa) was induced by GDNF as early as 5 min and then decreased at 15 min (Fig. 3A), indicating that GDNF induces a rapid and transient Ras activation. Furthermore, the active Ras induced by GDNF was confirmed in the freshly isolated type A spermatogonia from 6-day-old mice (Fig. 3E and F). Western blots using anti-phospho-ERK1/2 showed that the phosphorylation of ERK1/2 (44 kDa and 42 kDa) was increased by GDNF stimulation for 5 min and 15 min (Fig. 3C). To verify this result, we pretreated the C18-4 cells with PD98059, a specific inhibitor for MEK1, the upstream regulator of ERK1/2. As expected, the phosphorylation of ERK1/2 by GDNF was completely blocked by pretreatment with inhibitor PD98059 (Fig. 3C and 3D). In addition, the phosphorylation of ERK1/2 activated by GDNF was also confirmed in the freshly isolated type A spermatogonia from 6-day old mice (Fig. 3G). Densitometric analyses showed significant differences for each of the experiments upon the addition of GDNF at the indicated time intervals (Fig. 3B, 3D, 3F, 3H).
Figure 3.
The Ras/ERK1/2 pathway is activated by GDNF in the C18-4 cells and type A spermatogonia. (A) Pull-down assay of the activated GTP-loaded Ras with a GST-fusion protein containing the RBD of Raf1 to affinity precipitate active Ras (GTP-Ras) from cell lysates of the C18-4 cells. Five hundred micrograms of cell lysates from the GDNF treated or untreated cells were incubated with GST-Raf1-RBD and an Immobilized Glutathione Disc, and half the pulled-down active Ras was detected by Western blotting using antibody to Ras. IP: immunoprecipitation; IB: immunoblotting. The molecular mass standards are shown on the left, and the results are representative of three independent experiments. (B) The GDNF-induced active Ras relative to the GDNF-untreated control (1.0). Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (C) The C18-4 cells were serum-starved for 16 h and then pretreated with or without PD98059 for 30 min. The cells were treated with 100 ng/ml GDNF for 5 and 15 min or without GDNF treatment. Fifty micrograms of total protein from the GDNF treated or untreated cells were resolved by SDS-PAGE and blotted with anti-phospho-ERK1/2 (upper panel). The same membrane was re-probed with anti-ERK2 and showed ERK2 expression in all lanes (lower panel). The molecular mass standards are shown on the left, and the results are representative of three independent experiments. (D) The GDNF-induced phosphorylation of ERK2 (42 kDa) relative to control (1.0) after normalization to the signal obtained with ERK2. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (E) Pull-down assay of the activated GTP-loaded Ras with a GST-fusion protein containing the RBD to affinity precipitate active Ras from cell lysates of the freshly isolated type A spermatogonia. Three hundred micrograms of cell lysates from the GDNF treated or untreated cells were incubated with GST-Raf1-RBD and an Immobilized Glutathione Disc, and half the pulled-down active Ras was detected by Western blotting using an antibody to Ras. IP: immunoprecipitation; IB: immunoblotting. The molecular weight is shown on the left, and the results are representative of two experiments. (F) The GDNF-induced active Ras relative to the GDNF-untreated control (1.0). Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (G) Type A spermatogonia of 6-day-old mice were serum-starved for 8 h and then treated with 100 ng/ml GDNF for 5 and 15 min or without GDNF treatment. Eighty micrograms of total protein from the GDNF treated or untreated cells were resolved by SDS-PAGE and blotted with anti-phospho-ERK1/2 (upper panel). The same membrane was re-probed with anti-ERK2 and showed ERK2 expression in all lanes (lower panel). The molecular weights are shown on the left, and the results are representative of two experiments. (H) The GDNF-induced phosphorylation of ERK2 (42 kDa) relative to control (1.0) after normalization to the signal obtained with ERK2. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks.
GDNF Activates the Phosphorylation of Transcription Factors CREB-1, ATF-1, and CREM-1, and Enhances the Transcription of the Immediate-Early Gene c-fos in the C18-4 Cells
We next assessed whether the activation of the CREB/ATF-1 family transcription factors was induced by GDNF in the C18-4 cells using anti-phospho-CREB-1. This antibody is able to recognize the phosphorylation of the transcription factor CREB-1 at Ser-133 as well as ATF-1 and CREM-1. We found that GDNF activated the phosphorylation of CREB-1 at Ser-133, ATF-1, and CREM-1, as shown by Western blot analysis (Fig. 4A). We further explored whether GDNF could induce transcription of the immediate-early gene c-fos in the C18-4 cells. The c-fos mRNA was clearly enhanced in the C18-4 cells treated with GDNF for 5 and 15 min and declined at 30 min but was still higher than the control as shown by semi-quantitative RT-PCR analysis (Fig. 4E). To confirm this result, we pretreated the C18-4 cells with the inhibitor PD98059. As anticipated, the up-regulation of c-fos mRNA by GDNF in the C18-4 cells was completely blocked by the pretreatment with PD98059 (Fig. 4G and H). Densitometric analyses showed significant differences for each of the experiments upon the addition of GDNF at the indicated time intervals (Fig. 4B, 4C, 4D, 4F, 4H).
Figure 4.
The phosphorylation of CREB-1, ATF-1, and CREM-1 and c-fos transcription are induced by GDNF in the C18-4 cells and the up-regulation of c-fos transcription by GDNF is blocked by the inhibitor PD98059. (A) The phosphorylation of CREB-1 at Ser-133 and the CREB-related protein ATF-1 and CREM-1 are activated by GDNF in the C18-4 cells as shown by Western blots of 50 µg of total cell lysates with a specific anti-phospho-CREB1 antibody (upper panel). The same membrane was re-probed with anti-β-actin and demonstrated β-actin expression in all lanes (lower panel). The molecular weights are shown on the left, and the results are representative of three independent experiments. (B–D) The GDNF-induced phosphorylation of CREB-1 (B), ATF-1 (C), and CREM-1 (D) relative to control (1.0) after normalization to the signal of β-actin. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (E) RT-PCR analysis shows the time course of increase in c-fos mRNA induced by GDNF in C18-4 cells. The results are representative of three independent experiments. (F) The GDNF-induced c-fos mRNA relative to control (1.0) after normalization to the signal of Gapdh. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks. (G) RT-PCR analysis displays the c-fos mRNA in C18-4 cells pretreated with PD98059 for 30 min and then with GDNF treatment for 5 min and 15 min or without GDNF treatment. The results are representative of three independent experiments. (H) The GDNF-induced c-fos mRNA relative to control (1.0) after normalization to the signal of Gapdh. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks.
The inhibitor PD98059 Blocks GDNF-induced BrdU Incorporation in the C18-4 Cells
Cell proliferation of the C18-4 cells was assessed immunocytochemically using the BrdU incorporation assay, and the nuclei of cells in active DNA synthesis were stained with a BrdU monoclonal antibody. An increase in the BrdU-positive cells was observed in the GDNF-treated cells (Fig. 5B) with respect to the GDNF-untreated control (Fig. 5A), and Fig. 5D showed 2.3-fold induction of the BrdU-positive cells by GDNF. Notably, the increase of the BrdU-positive cells induced by GDNF was completely abolished in the cells pretreated with the inhibitor PD98059 (Fig. 5C and 5D).
Figure 5.
Inhibitor PD98059 blocks GDNF-induced BrdU incorporation in the C18-4 cells. The C18-4 cells were pretreated with or without PD98059 for 30 min, and then GDNF was added to the culture medium as described in the Materials and Methods. After 16 h of culture, the cells were fixed, immunostained with anti-BrdU antibody, followed by incubation with the biotinylated second antibody and streptavidin-peroxidase, and detected by DAB chromogen. Immunocytochemical staining shows the BrdU incorporation in the C18-4 cells without GDNF treatment (A), the cells with GDNF treatment (B), and in the cells pretreated with PD98059 and then with GDNF treatment (C), using anti-BrdU. The BrdU-positive cells are indicated by the arrows, and BrdU negative cells are denoted by asterisks. Scale bars in A, B, and C =10 µm. (D) Percentage of the BrdU-positive cells in the C18-4 cells without or with GDNF treatment, or in the cells pretreated with PD98059 and then with GDNF stimulation. The data were presented as mean ± SEM of BrdU-positive cells out of 500 cells. The percentage of BrdU-positive cells is significantly increased (p <0.05, as indicated by asterisk) in GDNF-treated cells compared to the untreated cells or the cells pretreatment with PD98059.
The inhibitor PD98059 Blocks GDNF-induced Metaphase Counts in the C18-4 Cells
Metaphase counting was also performed to evaluate whether GDNF uses the ERK1/2 pathway to promote cell cycle progression of the C18-4 cells. An increase in the metaphase counts (nuclei with condensed metaphase chromosomes) was observed in the GDNF-treated cells (Fig. 6B) compared to the GDNF-untreated control (Fig. 6A), and Fig. 6D showed 3-fold induction of the metaphase counts by GDNF. Notably, the increase of the GDNF-induced metaphase counts was completely blocked in the cells pretreated with the inhibitor PD98059 (Fig. 6C and 6D).
Figure 6.
Inhibitor PD98059 blocks GDNF-induced metaphase counts in the C18-4 cells and the expression of cell cycle regulators, including cyclin A, cyclin D1, and CDK2 in the C18-4 cells with or without GDNF treatment. The C18-4 cells were cultured in the absence or constant presence of GDNF and/or PD98059 for 24 h. The cells were stained with Giemsa solution, showing morphological characteristics in the C18-4 cells without GDNF treatment (A), the cells with GDNF treatment (B), and in the cells pretreated with PD98059 and then with GDNF treatment (C). The asterisks indicate the condensed metaphase chromosomes typical of mitotic nuclei. Scale bars in A, B, and C =10 µm. (D) Percentage of the cells in metaphse in the C18-4 cells without or with GDNF treatment, or in the cells pretreated with PD98059 and then with GDNF stimulation. The percentage of the cells in metaphase was statistically increased (p <0.05, as indicated by asterisk) in GDNF-treated cells compared to the GDNF-untreated cells or the cells pretreated with PD98059. (E) Western blot analysis reveals an increase in the expression of cyclin A and CDK2 but not cyclin D1 in the C18-4 cells stimulated with GDNF for 6 and 24 h. Inhibitor PD98059 blocked GDNF-induced cyclin A and CDK2 expression in these cells. The membranes were re-probed with anti-β-actin and showed β-actin expression in all lanes. The molecular weights are shown on the left, and the results are representative of three independent experiments. (F) Immunofluorescence analysis shows that an accumulation of cyclin A expression was induced by GDNF treatment for 6 and 24 h in the nuclei of the C18-4 cells when compared to the cells without GDNF stimulation. Staining with DAPI (blue fluorescence) was used to show the cell nuclei. Scale bars =10 µm. (G–H) The GDNF-induced expression of cyclin A (G) and CDK2 (H) relative to control (1.0) after normalization to the signal of β-actin. Satistically significant differences (p< 0.05) between the GDNF-treated and -untreated group were indicated by asterisks.
GDNF Up-regulates Cyclin A and CDK2 But Not Cyclin D1 Expression in the C18-4 Cells
We also tested how GDNF affects cell cycle G1/S transition in the C18-4 cells by examining the expression changes of certain cell cycle regulators, including cyclin A, cyclin D1, and CDK2. As shown by Western blot analysis, the expression of cyclin A and CDK2 was up-regulated in the C18-4 cells stimulated by GDNF for 6 and 24 h (Fig. 6E). Furthermore, the up-regulation of cyclin A and CDK2 expression induced by GDNF in the C18-4 cells was completely blocked by pretreatment with the inhibitor PD98059 (Fig. 6E, 6G, 6H). In contrast, no change of cyclin D1 expression was observed in the C18-4 cells with or without GDNF treatment (Fig. 6E).
The expression change and the subcellular localization of cyclin A in the C18-4 cells with or without GDNF treatment were further determined by immunofluorescence analysis. As shown in Fig. 6F, GDNF induced a marked accumulation of cyclin A in nuclei of the C18-4 cells by GDNF treatment for 6 and 24 h, respectively, compared to the GDNF-untreated control.
DISCUSSION
Spermatogonial stem cells (SSC) have recently assumed great significance in view of their unique characteristics. Firstly, there is no intrinsic barrier to the genetic engineering of SSC using retrovirus, and once inserted, the foreign genes can be transmitted to subsequent generations [28]. Secondly, SSC self-renew throughout life and differentiate into spermatocytes, spermatids, and eventually sperm, and thus they can be used as a good model to elucidate the basic mechanisms of adult stem cell renewal versus differentiation. Thirdly, recent evidence indicates that SSC from immature and adult mouse testis can de-differentiate and acquire embryonic stem (ES) cell properties [29, 30]. These ES-like cells are able to differentiate into derivatives of the three embryonic germ layers [29, 30]. If this is true for human SSC, as proposed by Guan et al [29], this would offer an alternative source of ES-like cells without the ethical issues involved in using discarded embryos. Finally, using SSC transplant [31, 32], patients suffering from cancer could retain their fertility after chemotherapy or irradiation therapy [33].
While there are reports showing that BMP4 induces spermatogonial differentiation via the Smad4/5 nuclear translocation [34] and SCF stimulates spermatogonial proliferation through the activation of phosphoinositide 3-kinase [35–37] and the ERK pathway [22], little is known about the signaling pathways that lead to the self-renewal and differentiation of SSC. GDNF was originally characterized as a survival factor for midbrain dopaminergic neurons [38] and a potent trophic factor for both spinal motoneurons and central noradrenergic neurons [39, 40], and it functions as a morphogen for kidney development [41]. Over the past several years, a growing number of reports demonstrated that GDNF plays a pivotal role in regulating self-renewal of undifferentiated spermatogonia and SSC [4, 8, 9, 17]. We have previously demonstrated that GDNF stimulates the proliferation of C18-4 cells, a murine SSC line [9]. We now report that GDNF/GFRα1/Ret activates the Ras/ERK1/2 pathway to stimulate DNA synthesis and SSC proliferation. Furthermore, the activation of the Ras/ERK1/2 pathway by GDNF in the SSC line was confirmed in freshly isolated type A spermatogonia from immature mice.
We observed, using Western blotting with anti-phospho-Ret, immunoprecipitation with anti-Shc and immunoblotting with anti-phospho-Ret, as well as immunoprecipitation with anti-Grb2 and immunoblotting with anti-phospho-Ret, that GDNF stimulation led to an activation of the Ret phosphorylation at tyrosine 1062 in the C18-4 cells. The tyrosine 1062 in Ret represents a binding site for the docking protein Shc and is crucial for activation of the Ras/ERK signaling pathway [42]. A common route to Ras activation is the recruitment of docking protein Shc by many receptor kinases, including Ret receptor tyrosine kinase [14]. We found, using immunoprecipitation and immunoblotting, that the phosphorylation of all three isoforms of Shc was activated in the C18-4 cells with GDNF stimulation and that GDNF induced the binding of Shc to the tyrosine phosphorylated Ret, indicating a direct association of Shc with Ret receptor. The adaptor protein Grb2 is a major link between the Ret receptor tyrosine kinase, Shc, and the Ras pathway [14, 42]. We discovered that GDNF also induced the binding of Grb2 to the tyrosine phosphorylated Ret in the C18-4 cells, reflecting that Grb2 is directly recruited to the activated Ret in these cells after GDNF stimulation [43].
The activated Ret engages docking protein Shc and adaptor protein Grb2, which results in the activation of the Ras/ERK pathway [44, 45]. We have previously shown using microarray analysis that the mRNA of Ras-related protein was up-regulated in the GFRα1-positive spermatogonia treated with GDNF [9]. The 21 kDa monomeric G protein Ras is recognized as a major regulator for cellular growth and differentiation [19–21]. We thus tested whether Ras could be activated by GDNF/GFRα1/Ret. We detected an obvious and rapid increase of the active Ras in the C18-4 cells stimulated by GDNF for 5 min using the Ras Activation Kit. The activated Ras interacts with a variety of downstream effectors including Raf1. As expected from Ras activation, we also found a significant and rapid increase in the tyrosine phosphorylation of ERK1/2 in the C18-4 cells with GDNF stimulation for 5 min. Moreover, the activated Ras and the phosphorylation of ERK1/2 induced by GDNF were further confirmed in freshly isolated type A spermatogonia from 6-day-old mice. The increase in tyrosine phosphorylation of ERK1/2 by GDNF stimulation can be completely blocked by pretreatment with PD98059, suggesting that GDNF/GFRα1/Ret activates the PD98059-sensitive ERK pathway.
ERK1/2 activation represents a significant signal for the regulation of cellular growth and proliferation. The activation of Ras/ERK1/2 pathway eventually results in the phosphorylation of specific transcription factors and induces transcription of the immediate-early genes in the nuclei [14]. The CREB/ATF-1 family is composed of CREB, CREM, and ATF-1, which constitutes a subfamily of beta-Zip transcription factors, and these factors control eukaryotic gene transcriptional regulation [46, 47]. Knockout studies indicate that the disruption of CREB in mice severely impaired spermatogenesis and resulted in male infertility [48, 49], and CREM deficient mice show a reduction of cell proliferation and DNA synthesis upon partial hepatectomy [50]. Recently, it has been shown that CREB is present in proliferating spermatogonia but not more differentiated germ cells including spermatids and sperm in hydra [51], and a novel splice variant of human testis CREB is chiefly expressed in male germ cells [52]. An important downstream event of the ERK pathway is the phosphorylation of transcription factors CREB, ATF-1, and CREM [14, 53–55], and ERK1/2 regulates the phosphorylation of transcription factor CREB-1 [56]. This prompted us to assess whether GDNF could stimulate the phosphorylation of the members of CREB/ATF-1 family in the C18-4 cells. As shown by Western blotting, the phosphorylation of CREB-1 at Ser-133, ATF-1, and CREM-1, was activated by GDNF treatment in the C18-4 cells for 5 and 15 min. The transcription factors CREB, ATF-1, and CREM are known to be a major regulator for immediate-early gene c-fos transcription [50, 54, 57]. We also found a significant increase in the transcription of c-fos in the C18-4 cells treatment with GDNF for 5 min and 15 min and this remained at a high level up to 30 min. As expected, the increase in c-fos transcription by GDNF stimulation was completely blocked by pretreatment with the inhibitor PD98059, suggesting that GDNF uses the ERK1/2 pathway to motivate c-fos transcription. Considered together, these results demonstrate that GDNF stimulation of the C18-4 cells results in the activation of the Ras/ERK1/2 signaling cascades that stimulates the phosphorylation of transcription factors CREB-1, ATF-1, and CREM-1 and induces c-fos transcription.
GDNF stimulates cell cycle progress in SSC as indicated by our results showing that GDNF induced a marked increase in BrdU-positive cells and metaphase counts in the C18-4 cells. These results also reflect an essential role of GDNF in stimulating DNA synthesis, proliferation, and mitosis of these cells. Significantly, the inhibitor PD98059 completely blocked the GDNF-induced BrdU incorporation and metaphase figure, implicating that the ERK1/2 pathway is required for the GDNF-stimulated cell cycle progression in the C18-4 cells.
The cell cycle through the G1/S checkpoint is regulated by multiple mitogenic signaling pathways, including Ras/ERK pathway. We then addressed how the GDNF/GFRα1/Ret/Shc/Ras/ERK1/2/CREB-1 plus CREM-1/c-fos pathway regulates the G1/S transition in the C18-4 cells. Cyclin A is a key regulatory protein that is involved in control of the S phase of the cell cycle and associates with CDK2 in mammalian cells. The rationale for examining the expression of cyclin A and CDK2 in the C18-4 cells treated with or without GDNF is that cyclin A and CDK2 function primarily in DNA replication at S phase [58–60], and that the transcription factor CREB stimulates cyclin A transcription at G1/S phase [61]. More importantly, a number of reports demonstrate that cyclin A is a target of c-fos in several cell types, including osteoblasts and chondrocytes [62, 63]. There are 2 types of cyclin A, cyclin A1 and A2, which exhibit distinct expression patterns during spermatogenesis. It has been suggested that cyclin A1 is highly expressed in pachytene spermatocytes and required for meiosis, while cyclin A2 is predominantly present in spermatogonia including SSC [22, 64–66]. Using immunohistochemistry, we found that cyclin A is expressed in a subpopulation of spermatogonia along the basement membrane in adult mouse testis (data not shown). Notably, cyclin A is also present in the C18-4 cells further indicating that these cells are SSC. It has been shown that c-fos up-regulates cyclin A but not cyclin D1 and enhances CDK2 activity to accelerate S-phase entry in osteoblasts [62], and co-overexpression of CDK2 plus cyclin A and cyclin E rescued the G1/S block resulting from lack of Myc thus enabling the cells to enter mitosis [67]. We found that GDNF up-regulated cyclin A expression in the nuclei of the C18-4 cells, and that GDNF induced a marked increase of cyclin A-associated CDK2 expression, which would facilitate the G1/S phase transition and DNA synthesis in the C18-4 cells. Importantly, the up-regulation of cyclin A and CDK2 induced by GDNF in these cells was blocked by pretreatment with the inhibitor PD98059, suggesting that ERK1/2 pathway is essential for the GDNF-induced G1/S transition in the C18-4 cells.
We chose to check the expression of cyclin D1, but not cyclin D2 or cyclin D3, in the C18-4 cells treated with or without GDNF because cyclin D1 was suggested to play a role in regulating the cell cycle through G1/S phase progression by the ERK1/2/cyclin D1 pathway [22, 68, 69], and more significantly, cyclin D1 is expressed only in proliferating spermatogonia during spermatogenesis and plays a role in spermatogonial proliferation, in particular during the G1/S phase transition [70]. Conversely, cyclin D2 is expressed at epithelial stage VIII when the Aal differentiate into A1, as well as in spermatocytes and spermatids, but not in SSC, Apr, or Aal spermatogonia and plays a role in spermatogonial differentiation [70, 71], while cyclin D3 is expressed in terminally differentiated Sertoli cells, Leydig cells, and spermatogonia in adult testis [70]. Although it was suggested that cyclin D1 plays a role in regulating the cell cycle through G1/S phase progression [22, 69, 70], we observed that there was no significant change of cyclin D1 expression in the C18-4 cells with or without GDNF stimulation, indicating that cyclin D1 is not involved in regulating the cell cycle progression from G1 to S phase induced by GDNF.
SUMMARY
We have demonstrated that GDNF stimulation of the C18-4 cells results in a series of signaling events in the activation of the Ras/ERK1/2 pathway. These include the phosphorylation of Ret tyrosine kinase, the phosphorylation of docking protein Shc and the recruitment of adaptor protein Grb2, the rapid activation of the Ras/ERK1/2 pathway, the phosphorylation of transcription factors CREB-1, ATF-1, and CREM-1, the induction in c-fos transcription, and the enhancement of cyclin A and CDK2 expression, which leads to accelerate S-phase entry in the C18-4 cells and promotes DNA synthesis and SSC proliferation (see Fig. 7). This study thus offers a novel insight into the dynamics of GDNF signaling in SSC and new molecular signatures for possible therapy of male infertility and testicular tumors.
Figure 7.
The schematic diagram demonstrates intracellular signaling events in the Ras/ERK1/2 pathway as well as the upstream and downstream cascades activated by GDNF in the C18-4 cells. “P” indicates “phosphorylate”, and “A” denotes “activate”.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. George C. Enders, Department of Anatomy and Cell Biology, University of Kansas Medical Center, for providing an antibody to GCNA1. This study was supported by NIH grants RO1-HD033728 and R01-HD044543.
Footnotes
DISCLOSURES: The authors indicate no potential conflicts of interest.
References
- 1.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776–798. [PubMed] [Google Scholar]
- 2.Brinster RL. Germline stem cell transplantation and transgenesis. Science. 2002;296:2174–2176. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dym M. Spermatogonial stem cells of the testis. Proc Natl Acad Sci U S A. 1994;91:11287–11289. doi: 10.1073/pnas.91.24.11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng X, Lindahl M, Hyvonen ME, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 5.Sariola H, Saarma M. Novel functions and signalling pathways for GDNF. J Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- 6.Tadokoro Y, Yomogida K, Ohta H, et al. Homeostatic regulation of germinal stem cell proliferation by the GDNF/FSH pathway. Mech Dev. 2002;113:29–39. doi: 10.1016/s0925-4773(02)00004-7. [DOI] [PubMed] [Google Scholar]
- 7.Viglietto G, Dolci S, Bruni P, et al. Glial cell line-derived neutrotrophic factor and neurturin can act as paracrine growth factors stimulating DNA synthesis of Ret-expressing spermatogonia. Int J Oncol. 2000;16:689–694. doi: 10.3892/ijo.16.4.689. [DOI] [PubMed] [Google Scholar]
- 8.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann MC, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Dev Biol. 2005;279:114–124. doi: 10.1016/j.ydbio.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braydich-Stolle L, Kostereva N, Dym M, et al. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol. 2007;304:34–45. doi: 10.1016/j.ydbio.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jing S, Wen D, Yu Y, et al. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell. 1996;85:1113–1124. doi: 10.1016/s0092-8674(00)81311-2. [DOI] [PubMed] [Google Scholar]
- 12.Treanor JJ, Goodman L, de Sauvage F, et al. Characterization of a multicomponent receptor for GDNF. Nature. 1996;382:80–83. doi: 10.1038/382080a0. [DOI] [PubMed] [Google Scholar]
- 13.Poteryaev D, Titievsky A, Sun YF, et al. GDNF triggers a novel ret-independent Src kinase family-coupled signaling via a GPI-linked GDNF receptor alpha1. FEBS Lett. 1999;463:63–66. doi: 10.1016/s0014-5793(99)01590-2. [DOI] [PubMed] [Google Scholar]
- 14.Trupp M, Scott R, Whittemore SR, et al. Ret-dependent and -independent mechanisms of glial cell line-derived neurotrophic factor signaling in neuronal cells. J Biol Chem. 1999;274:20885–20894. doi: 10.1074/jbc.274.30.20885. [DOI] [PubMed] [Google Scholar]
- 15.Trupp M, Raynoschek C, Belluardo N, et al. Multiple GPI-anchored receptors control GDNF-dependent and independent activation of the c-Ret receptor tyrosine kinase. Mol Cell Neurosci. 1998;11:47–63. doi: 10.1006/mcne.1998.0667. [DOI] [PubMed] [Google Scholar]
- 16.Golden JP, DeMaro JA, Osborne PA, et al. Expression of neurturin, GDNF, and GDNF family-receptor mRNA in the developing and mature mouse. Exp Neurol. 1999;158:504–528. doi: 10.1006/exnr.1999.7127. [DOI] [PubMed] [Google Scholar]
- 17.Naughton CK, Jain S, Strickland AM, et al. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod. 2006;74:314–321. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- 18.Buageaw A, Sukhwani M, Ben-Yehudah A, et al. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol Reprod. 2005;73:1011–1016. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- 19.Wittinghofer A. Signal transduction via Ras. Biol Chem. 1998;379:933–937. [PubMed] [Google Scholar]
- 20.Yamamoto T, Taya S, Kaibuchi K. Ras-induced transformation and signaling pathway. J Biochem (Tokyo) 1999;126:799–803. doi: 10.1093/oxfordjournals.jbchem.a022519. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Sala D, Rebollo A. Novel aspects of Ras proteins biology: regulation and implications. Cell Death Differ. 1999;6:722–728. doi: 10.1038/sj.cdd.4400557. [DOI] [PubMed] [Google Scholar]
- 22.Dolci S, Pellegrini M, Di Agostino S, et al. Signaling through extracellular signal-regulated kinase is required for spermatogonial proliferative response to stem cell factor. J Biol Chem. 2001;276:40225–40233. doi: 10.1074/jbc.M105143200. [DOI] [PubMed] [Google Scholar]
- 23.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann MC, Braydich-Stolle L, Dettin L, et al. Immortalization of mouse germ line stem cells. Stem Cells. 2005;23:200–210. doi: 10.1634/stemcells.2003-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Z, Feng L, Zhang X, et al. Expression of Col1a1, Col1a2 and procollagen I in germ cells of immature and adult mouse testis. Reproduction. 2005;130:333–341. doi: 10.1530/rep.1.00694. [DOI] [PubMed] [Google Scholar]
- 26.Bellve AR, Cavicchia JC, Millette CF, et al. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dym M, Jia MC, Dirami G, et al. Expression of c-kit receptor and its autophosphorylation in immature rat type A spermatogonia. Biol Reprod. 1995;52:8–19. doi: 10.1095/biolreprod52.1.8. [DOI] [PubMed] [Google Scholar]
- 28.Nagano M, Brinster CJ, Orwig KE, et al. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proc Natl Acad Sci U S A. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan K, Nayernia K, Maier LS, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- 30.Baba S, Heike T, Umeda K, et al. Generation of Cardiac and Endothelial Cells from Neonatal Mouse Testis-derived Multipotent Germline Stem Cells. Stem Cells. 2007 doi: 10.1634/stemcells.2006-0574. [DOI] [PubMed] [Google Scholar]
- 31.Ryu BY, Orwig KE, Oatley JM, et al. Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells. 2006;24:1505–1511. doi: 10.1634/stemcells.2005-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brinster RL. Male germline stem cells: from mice to men. Science. 2007;316:404–405. doi: 10.1126/science.1137741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanatsu-Shinohara M, Ogonuki N, Iwano T, et al. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development. 2005;132:4155–4163. doi: 10.1242/dev.02004. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini M, Grimaldi P, Rossi P, et al. Developmental expression of BMP4/ALK3/SMAD5 signaling pathway in the mouse testis: a potential role of BMP4 in spermatogonia differentiation. J Cell Sci. 2003;116:3363–3372. doi: 10.1242/jcs.00650. [DOI] [PubMed] [Google Scholar]
- 35.Blume-Jensen P, Jiang G, Hyman R, et al. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3'-kinase is essential for male fertility. Nat Genet. 2000;24:157–162. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- 36.Kissel H, Timokhina I, Hardy MP, et al. Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. The Embo Journal. 2000;19:1312–1326. doi: 10.1093/emboj/19.6.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng LX, Ravindranath N, Dym M. Stem cell factor/c-kit up-regulates cyclin D3 and promotes cell cycle progression via the phosphoinositide 3-kinase/p70 S6 kinase pathway in spermatogonia. Journal of Biological Chemistry. 2000;275:25572–25576. doi: 10.1074/jbc.M002218200. [DOI] [PubMed] [Google Scholar]
- 38.Lin LF, Doherty DH, Lile JD, et al. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 39.Henderson CE, Phillips HS, Pollock RA, et al. GDNF: a potent survival factor for motoneurons present in peripheral nerve and muscle. Science. 1994;266:1062–1064. doi: 10.1126/science.7973664. [DOI] [PubMed] [Google Scholar]
- 40.Arenas E, Trupp M, Akerud P, et al. GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron. 1995;15:1465–1473. doi: 10.1016/0896-6273(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 41.Vega QC, Worby CA, Lechner MS, et al. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc Natl Acad Sci U S A. 1996;93:10657–10661. doi: 10.1073/pnas.93.20.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Besset V, Scott RP, Ibanez CF. Signaling complexes and protein-protein interactions involved in the activation of the Ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J Biol Chem. 2000;275:39159–39166. doi: 10.1074/jbc.M006908200. [DOI] [PubMed] [Google Scholar]
- 43.Alberti L, Borrello MG, Ghizzoni S, et al. Grb2 binding to the different isoforms of Ret tyrosine kinase. Oncogene. 1998;17:1079–1087. doi: 10.1038/sj.onc.1202046. [DOI] [PubMed] [Google Scholar]
- 44.Arighi E, Alberti L, Torriti F, et al. Identification of Shc docking site on Ret tyrosine kinase. Oncogene. 1997;14:773–782. doi: 10.1038/sj.onc.1200896. [DOI] [PubMed] [Google Scholar]
- 45.Lorenzo MJ, Gish GD, Houghton C, et al. RET alternate splicing influences the interaction of activated RET with the SH2 and PTB domains of Shc, and the SH2 domain of Grb2. Oncogene. 1997;14:763–771. doi: 10.1038/sj.onc.1200894. [DOI] [PubMed] [Google Scholar]
- 46.De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Prog Nucleic Acid Res Mol Biol. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. [DOI] [PubMed] [Google Scholar]
- 47.Fimia GM, De Cesare D, Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- 48.Hummler E, Cole TJ, Blendy JA, et al. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci U S A. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blendy JA, Kaestner KH, Schmid W, et al. Targeting of the CREB gene leads to up-regulation of a novel CREB mRNA isoform. Embo J. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- 50.Servillo G, Della Fazia MA, Sassone-Corsi P. Transcription factor CREM coordinates the timing of hepatocyte proliferation in the regenerating liver. Genes Dev. 1998;12:3639–3643. doi: 10.1101/gad.12.23.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chera S, Kaloulis K, Galliot B. The cAMP response element binding protein (CREB) as an integrative HUB selector in metazoans: clues from the hydra model system. Biosystems. 2007;87:191–203. doi: 10.1016/j.biosystems.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Huang X, Zhang J, Lu L, et al. Cloning and expression of a novel CREB mRNA splice variant in human testis. Reproduction. 2004;128:775–782. doi: 10.1530/rep.1.00036. [DOI] [PubMed] [Google Scholar]
- 53.Impey S, Obrietan K, Wong ST, et al. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 54.Cui H, Cai F, Belsham DD. Leptin signaling in neurotensin neurons involves STAT, MAP kinases ERK1/2, and p38 through c-Fos and ATF1. Faseb J. 2006;20:2654–2656. doi: 10.1096/fj.06-5989fje. [DOI] [PubMed] [Google Scholar]
- 55.Verploegen S, Lammers JW, Koenderman L, et al. Identification and characterization of CKLiK, a novel granulocyte Ca(++)/calmodulin-dependent kinase. Blood. 2000;96:3215–3223. [PubMed] [Google Scholar]
- 56.Gee K, Angel JB, Ma W, et al. Intracellular HIV-Tat expression induces IL-10 synthesis by the CREB-1 transcription factor through Ser133 phosphorylation and its regulation by the ERK1/2 MAPK in human monocytic cells. J Biol Chem. 2006;281:31647–31658. doi: 10.1074/jbc.M512109200. [DOI] [PubMed] [Google Scholar]
- 57.Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 58.Girard F, Strausfeld U, Fernandez A, et al. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 59.Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 60.Pagano M, Pepperkok R, Lukas J, et al. Regulation of the cell cycle by the cdk2 protein kinase in cultured human fibroblasts. J Cell Biol. 1993;121:101–111. doi: 10.1083/jcb.121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Desdouets C, Matesic G, Molina CA, et al. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol Cell Biol. 1995;15:3301–3309. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sunters A, Thomas DP, Yeudall WA, et al. Accelerated cell cycle progression in osteoblasts overexpressing the c-fos proto-oncogene: induction of cyclin A and enhanced CDK2 activity. J Biol Chem. 2004;279:9882–9891. doi: 10.1074/jbc.M310184200. [DOI] [PubMed] [Google Scholar]
- 63.Beier F, Taylor AC, LuValle P. Activating transcription factor 2 is necessary for maximal activity and serum induction of the cyclin A promoter in chondrocytes. J Biol Chem. 2000;275:12948–12953. doi: 10.1074/jbc.275.17.12948. [DOI] [PubMed] [Google Scholar]
- 64.Ravnik SE, Wolgemuth DJ. Regulation of meiosis during mammalian spermatogenesis: the A-type cyclins and their associated cyclin-dependent kinases are differentially expressed in the germ-cell lineage. Dev Biol. 1999;207:408–418. doi: 10.1006/dbio.1998.9156. [DOI] [PubMed] [Google Scholar]
- 65.Giuili G, Tomljenovic A, Labrecque N, et al. Murine spermatogonial stem cells: targeted transgene expression and purification in an active state. EMBO Rep. 2002;3:753–759. doi: 10.1093/embo-reports/kvf149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ravnik SE, Wolgemuth DJ. The developmentally restricted pattern of expression in the male germ line of a murine cyclin A, cyclin A2, suggests roles in both mitotic and meiotic cell cycles. Dev Biol. 1996;173:69–78. doi: 10.1006/dbio.1996.0007. [DOI] [PubMed] [Google Scholar]
- 67.Prathapam T, Tegen S, Oskarsson T, et al. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci U S A. 2006;103:2695–2700. doi: 10.1073/pnas.0511186103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Du HJ, Tang N, Liu BC, et al. Benzo[a]pyrene-induced cell cycle progression is through ERKs/cyclin D1 pathway and requires the activation of JNKs and p38 mapk in human diploid lung fibroblasts. Mol Cell Biochem. 2006;287:79–89. doi: 10.1007/s11010-005-9073-7. [DOI] [PubMed] [Google Scholar]
- 69.Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 70.Beumer TL, Roepers-Gajadien HL, Gademan IS, et al. Involvement of the D-type cyclins in germ cell proliferation and differentiation in the mouse. Biol Reprod. 2000;63:1893–1898. doi: 10.1095/biolreprod63.6.1893. [DOI] [PubMed] [Google Scholar]
- 71.de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.