Abstract
The highly polymorphic major histocompatibility complex class Ia (MHC-Ia) molecules present a broad array of peptides to the clonotypically diverse αβ T-cell receptors. In contrast, MHC-Ib molecules exhibit limited polymorphism and bind a more restricted peptide repertoire, in keeping with their major role in innate immunity. Nevertheless, some MHC-Ib molecules do play a role in adaptive immunity. While human leukocyte antigen E (HLA-E), the MHC-Ib molecule, binds a very restricted repertoire of peptides, the peptide binding preferences of HLA-G, the class Ib molecule, are less stringent, although the basis by which HLA-G can bind various peptides is unclear. To investigate how HLA-G can accommodate different peptides, we compared the structure of HLA-G bound to three naturally abundant self-peptides (RIIPRHLQL, KGPPAALTL and KLPQAFYIL) and their thermal stabilities. The conformation of HLA-GKGPPAALTL was very similar to that of the HLA-GRIIPRHLQL structure. However, the structure of HLA-GKLPQAFYIL not only differed in the conformation of the bound peptide but also caused a small shift in the α2 helix of HLA-G. Furthermore, the relative stability of HLA-G was observed to be dependent on the nature of the bound peptide. These peptide-dependent effects on the substructure of the monomorphic HLA-G are likely to impact on its recognition by receptors of both innate and adaptive immune systems.
Keywords: human leukocyte antigen G, HLA-G; structural immunology; innate immunity; antigen presentation; adaptive immunity
Introduction
Polymorphism is a hallmark of major histocompatibility complex class Ia (MHC-Ia) molecules, enabling them to present a wide spectrum of peptides and providing specificity in recognition by the highly variable αβ T-cell receptor (TCR) expressed on the surface of cytotoxic T lymphocytes. Maintenance of HLA (human leukocyte antigen) polymorphism reflects natural selection for enhanced protective immunity, whereby MHC molecules can differ from one another by many or only a few amino acids. MHC-Ia polymorphism is generally concentrated in the antigen (Ag)-binding cleft, where it can control the size and diversity of the peptide repertoire and the role of tapasin in peptide loading. Moreover, HLA polymorphism can result in altered conformation of the bound peptide or subtly altered juxtapositioning of the α-helices within the Ag-binding cleft, thereby influencing cognate TCR recognition and TCR allorecognition.1–3
In contrast, MHC-Ib molecules are much less polymorphic and often exhibit limited tissue distribution. The MHC-Ib family members include HLA-E, HLA-F, HLA-G and Hfe (HLA-H) in humans and gene products from the H2-M, H2-Q and H2-T regions in mice. The limited polymorphism present in MHC-Ib genes results in a marked reduction in the diversity of peptides that can be presented by these molecules. For instance, HLA-E has 8 alleles that produce only 3 proteins,4 while HLA-G has 36 alleles that make 14 proteins plus additional isoforms from alternate splicing events.5 Moreover, class Ib molecules often possess a larger number of primary anchor sites when compared with most class Ia molecules, which further constrains their intrinsic peptide repertoire. For example, the peptide binding groove of HLA-E has evolved to bind peptides derived from the leader sequences of other class I molecules in which residues 2, 3, 6, 7 and 9 of the peptide act as anchor sites.6–8 The differences in the repertoire of peptides bound between MHC-Ia and MHC-Ib molecules reflect disparate roles for MHC-Ib in immunity; MHC-Ia molecules are typically involved in adaptive immunity, while MHC-Ib molecules are often considered to act as ligands in innate immunity. For example, HLA-E in humans and Qa-1b in mice regulate the activation of natural killer (NK) cells by acting as a ligand for the CD94–NKG2 receptors.9
The primary site of expression of HLA-G is at the fetal–maternal interface,10–13 where it appears to play a role in promoting maternal tolerance of the fetus.14 The peptide repertoire of HLA-G has been studied both in vitro15,16 and in vivo,17 leading to the identification of 15 distinct HLA-G-restricted self-peptides.18 Although HLA-G possesses a large number of anchor sites, the repertoire of peptides that HLA-G can bind appears somewhat less stringent for that observed in HLA-E. HLA-G-restricted peptides display a preference for a basic residue at P1, a proline or small hydrophobic residue at P3 and a leucine at P9.16 Recently, the high-resolution crystal structure of HLA-G bound to RIIPRHLQL, a peptide derived from histone H2A and one of the most abundant peptides eluted from HLA-G in transfected cells,15,16 has been determined.19 Furthermore, one study reported the low-resolution structure of a disulfide-linked HLA-GRIIPRHLQL dimer,20 consistent with a report that HLA-G exists as a disulfide-linked dimer on the cell surface.21 The HLA-GRIIPRHLQL structure revealed an extensive network of bonds between the peptide and HLA-G, providing a molecular basis for the restricted peptide repertoire of HLA-G. The structure also revealed that the mode of peptide binding between HLA-E and HLA-G was remarkably well conserved. Nevertheless, it was unclear how HLA-G could bind disparate self-peptides.
HLA-G has been reported to regulate NK cell activation both directly acting as ligand for the KIR2DL4 receptor and indirectly through the provision of peptides that promote the cell surface expression of HLA-E expression in the trophoblast. HLA-G also binds the receptors LILRB1 (leukocyte immunoglobulin-like receptor B1 or immunoglobulin-like transcript 2) and LILRB2 (leukocyte immunoglobulin-like receptor B2 or immunoglobulin-like transcript 4),22–25 where the recognition site is in the α3 domain,26 distal to the Ag-binding cleft, and therefore unlikely to be affected by the identity of the peptide. The docking mode between HLA-G and KIR2DL4 is unknown; however, HLA-G residues Met76 and Gln79 of the α1 helix are involved,27 suggesting that KIR2DL4 sits atop the Ag-binding cleft—a mode consistent with the binding of other KIRs (killer cell immunoglobulin-like receptors) to MHC-Ia molecules.28,29 Thus, the surface formed by the peptide and Ag-binding cleft is likely to influence KIR2DL4 recognition of HLA-G.
To investigate how different peptides are accommodated by HLA-G and how these peptides differentially impact on the conformation on the Ag-binding cleft of HLA-G, we solved the structure of HLA-G presenting two unrelated self-peptides: KGPPAALTL is the most abundant single peptide derived from placenta,17 whereas KLPQAFYIL is the major species eluted from HLA-G-transfected cells.15 The observed conformational malleability in the binding modes of these HLA-G-restricted epitopes has implications for the recognition of HLA-G by ligands of adaptive and innate immune systems.
Results
Structure of HLA-GKGPPAALTL
The structure of the HLA-G presenting the peptide KGPPAALTL was determined to 2.4-Å resolution to Rcryst and Rfree values of 21.6% and 29.9%, respectively. The electron density for the peptide, and the contacting residues, was unambiguous and free from crystal contacts. The KGPPAALTL peptide is bound in a linear and extended manner, with a small bulge centered at P4-Pro (Fig. 1a). The peptide is anchored in the Ag-binding cleft at the N- and C-termini via a series of interactions that are conserved throughout MHC-I. Namely, P1-Lys forms H-bonds with Tyr7, Tyr171 and Tyr159 and salt bridges with Glu62 and Glu63 (Fig. 2a). P9-Leu is involved in a network of H-bonds with Asn77, Tyr84, Ser143 (a Thr in MHC-Ia) and Lys146 and a water-mediated H-bond with Thr80 (Fig. 2b).
Fig. 1.
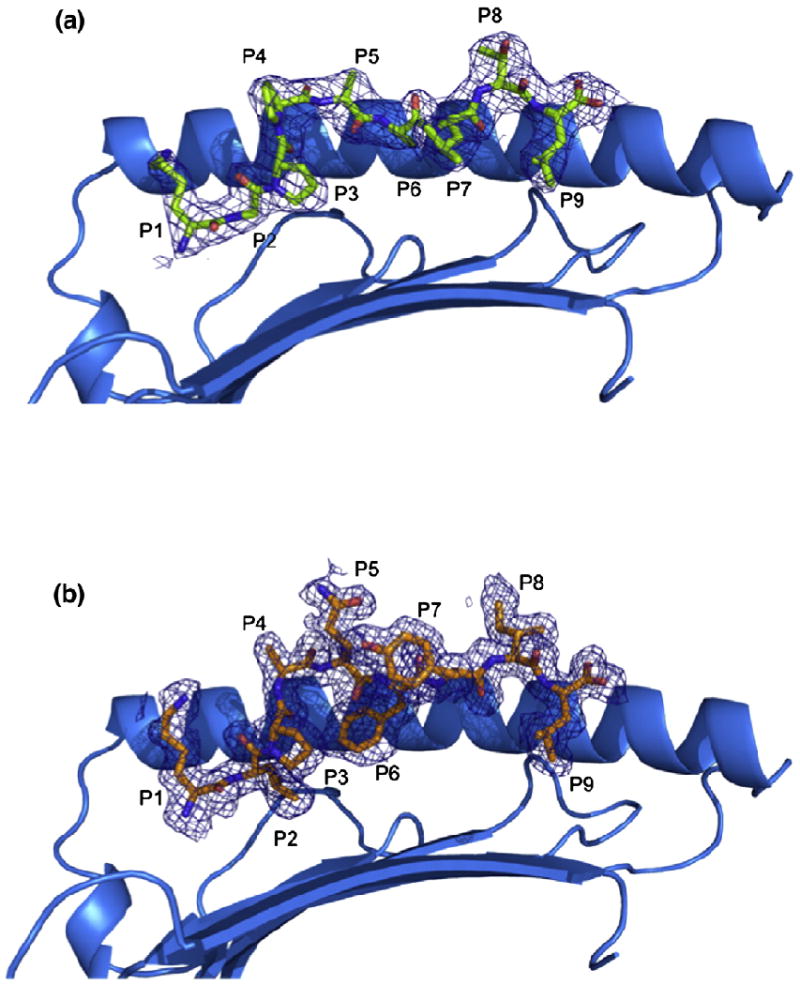
Structure of pHLA-G complexes. (a and b) Side views of the HLA-GKGPPAALTL complex (a) and the HLA-GKLPAQFYIL complex (b) showing 2.4- and 1.7-Å omit maps, respectively (contoured at 1σ). The α2 helix has been removed for clarity.
Fig. 2.
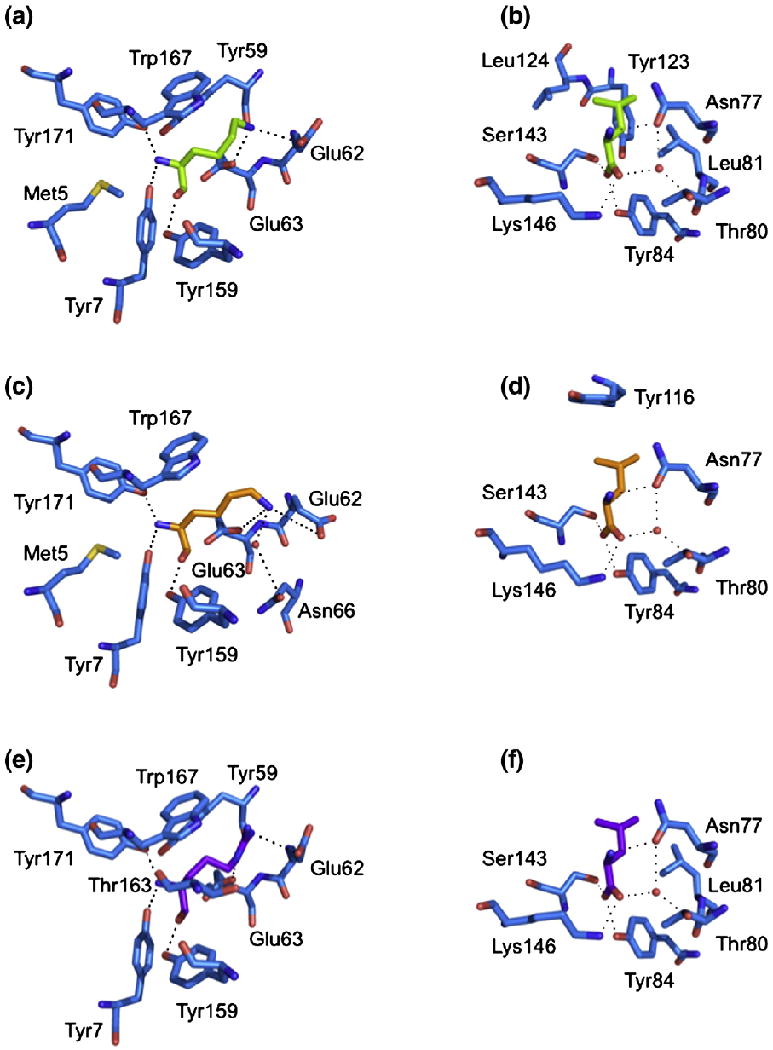
Conserved interactions between peptide and HLA-G. The polar and non-polar contacts of the P1 residue (a, c, e) and the P9 residue (b, d, f) of each pHLA-G complex are shown. (a and b) HLA-GKGPPAALTL. (c and d) HLA-GKLPAQFYIL. (e and f) HLA-GRIIPRHLQL. H-bonds and salt bridges are represented by dashed lines, and water molecules are represented by red spheres.
Collectively, the peptide makes extensive polar and non-polar contacts with HLA-G (Table 1), totaling one salt bridge, 12 H-bonds, eight water-mediated H-bonds and numerous van der Waals (vdW) interactions. Accordingly, the peptide sits lower in the Ag-binding cleft than in MHC-Ia and most closely resembles HLA-E-bound peptides. Indeed, the KGPPAALTL epitope provides a relatively featureless surface in which only the side chain of P8-Thr protrudes from the Ag-binding cleft (Fig. 1a).
Table 1.
HLA-GKGPPAALTL contacts
| Peptide residue | HLA-G residue | Interaction |
|---|---|---|
| Lys1 | ||
| Lys | Met5, Tyr7, Tyr59, Glu62, Glu63, Tyr159, Trp167, Tyr171 | vdW |
| LysN | Tyr7OH | H-bond |
| Tyr171OH | H-bond | |
| LysNz | (Glu62Oε2), Glu62Oε1 | Salt bridge |
| (Glu63Oε2, Glu63Oε1) | Salt bridge | |
| LysO | Tyr159OH | H-bond |
| Gly2 | ||
| Gly | Tyr7, Glu63, Tyr159 | vdW |
| GlyN | Tyr7OH | H-bond |
| Glu63Oε2 | H-bond | |
| GlyO | Glu63Oε1 | Water-mediated H-bond |
| Asn66Nδ2 | Water-mediated H-bond | |
| Pro3 | ||
| Pro | Asn66, Trp97, Tyr159 | vdW |
| Pro4 | ||
| Pro | Asn66 | vdW |
| Ala5 | ||
| Ala | Arg156 | vdW |
| AlaO | Arg156NH1 | H-bond |
| Arg156NH2 | H-bond | |
| Ala6 | ||
| Ala | His70 | vdW |
| AlaN | Ala69O | Water-mediated H-bond |
| Thr73Oγ1 | Water-mediated H-bond | |
| Leu7 | ||
| Leu | Thr73, Asn77, Glu114, Tyr116, Trp133, Arg156 | vdW |
| LeuN | Trp97Nε1 | Water-mediated H-bond |
| Tyr116OH | Water-mediated H-bond | |
| LeuO | Asn77Nδ2 | H-bond |
| Thr8 | ||
| Thr | Asn77, Lys146 | vdW |
| Leu9 | ||
| Leu | Asn77, Thr80, Leu81, Tyr84, Tyr123, Leu124, Ser143, Lys146 | vdW |
| LeuN | Asn77Oδ1 | H-bond |
| LeuO | Tyr84OH | H-bond |
| Ser143Oγ | H-bond | |
| LeuOXT | Lys146Nζ | H-bond |
| Asn77Oδ1 | Water-mediated H-bond | |
| Thr80Oγ1 | Water-mediated H-bond |
The presence of a glycine, two alanines and two prolines within the KGPPAALTL epitope provides little opportunity for side-chain interactions with HLA-G. The two proline residues do not form any polar contacts and only participate in vdW interactions with three residues of HLA-G, suggesting that the prolines are important for maintaining the conformation of the peptide. The main chain of P2-Gly forms H-bonds with Tyr7 and Glu63 and water-mediated H-bonds with Glu63 and Asn66. The O of P5-Ala forms two H-bonds with Arg156, and the N of Ala6 forms water-mediated H-bonds with Ala69 and Thr73. The P7-Leu main chain forms a H-bond with Asn77 and water-mediated H-bonds with Trp97 and Tyr116 (Table 1).
The overall structure of HLA-GKGPPAALTL is similar to HLA-GRIIPRHLQL with an r.m.s.d. of 0.40 Å over the whole molecule and that of 0.26 Å within the Ag-binding cleft. Furthermore, the KGPPAALTL peptide adopts a conformation remarkably similar to that of the RIIPRHLQL peptide (r.m.s.d. of 0.24 Å between the peptides) despite the former not possessing the buried P6-His residue (Fig. 3a). Additional notable differences between the KGPPAALTL and RIIPRHLQL peptides are the smaller solvent-exposed side chains in the former peptide, specifically Ala and Thr in place of Arg and Gln in P5 and P8, respectively. Accordingly, the solvent-accessible area of the peptide is only ∼300 Å2 compared with that of ∼400 Å2 for the RIIPRHLQL peptide.
Fig. 3.
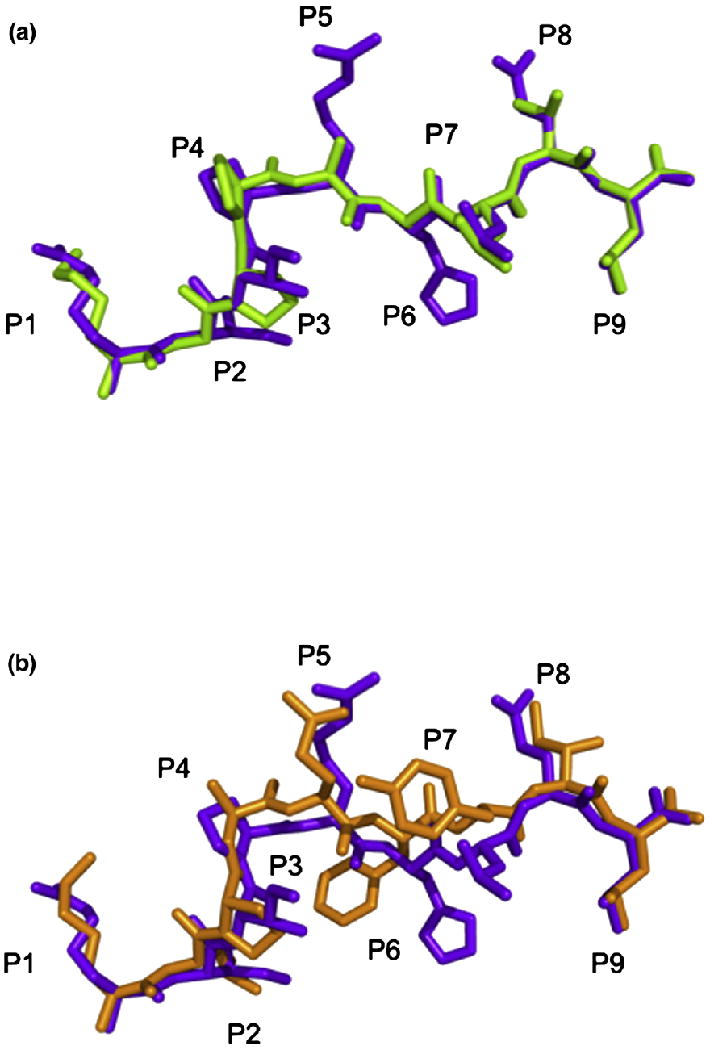
Peptide conformation. (a) Comparison of the conformations of the KGPPAALTL peptide (green) and the RIIPRHLQL peptide (purple) when presented by HLA-G highlighting the similarity between the two peptides. (b) Comparison of the conformations of the KLPAQFYIL peptide (orange) and the RIIPRHLQL peptide (purple) when presented by HLA-G highlighting the central bulge in KLPAQFYIL.
The contacts formed between HLA-G and the peptide backbone are well conserved between the two peptides, with only two H-bonds and one water-mediated H-bond absent in the HLA-GKGPPAALTL complex when compared with HLA-GRIIPRHLQL. The two lost H-bonds, from P3 and P4, are due to the different orientation of His70 (Fig. 4), while the lost water-mediated bond between P7-Leu and Arg156 is due to a slight shift in the water molecule. Furthermore, the conserved side-chain interactions are seen at P1, which is occupied by a conserved basic residue, and at P9-Leu (Fig. 2). The remaining polar contacts that are absent in the HLA-GKGPPAALTL complex (three H-bonds and four water-mediated H-bonds) are due to the substitution of P5-Arg and P6-His for two Ala residues in the KGPPAALTL epitope.
Fig. 4.
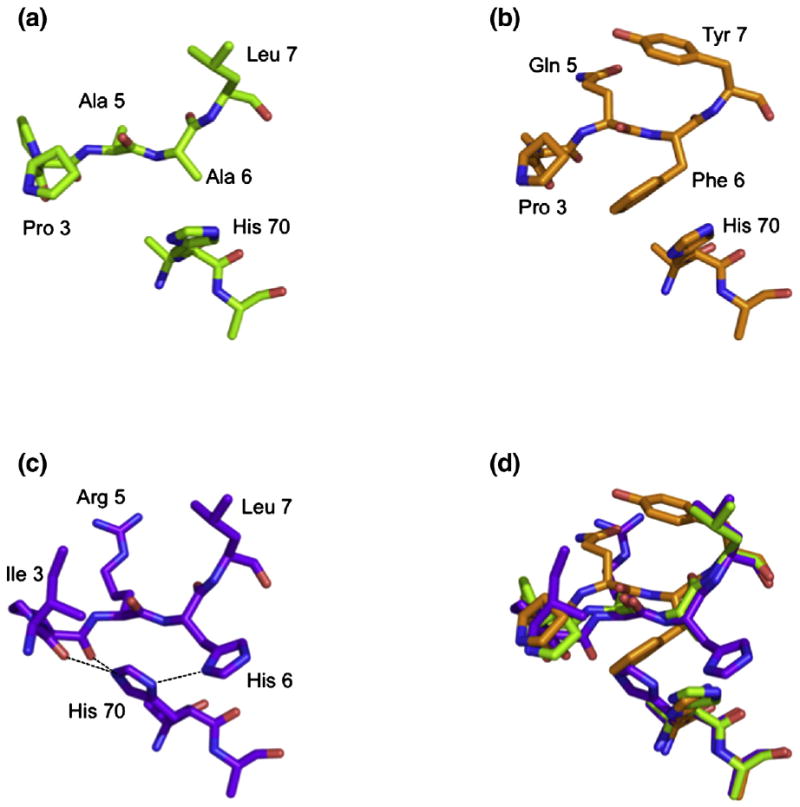
Orientation of His70. The orientation of His70 side chain is altered by the presence of a His at P6 of RIIPRHLQL (c) compared with that seen in the presence of KGPPAALTL (a) and KLPAQFYIL (b). An overlay of the three peptides is shown in (d).
The most significant difference observed between HLA-GRIIPRHLQL and HLA-GKGPPAALTL resides within the altered conformation of the side chain of His70, which moves away from P4-Pro and toward the Cβ of P6-Ala in the HLA-GKGPPAALTL complex (Fig. 4a). This reorientation of His70 could not be accommodated in the HLA-GRIIPRHLQL complex as it would result in a steric clash with the side chain of P6-His. In HLA-GRIIPRHLQL, His70 makes contacts with P3-Ile, P4-Pro and P5-Arg of the peptide, whereas in HLA-GKGPPAALTL, His70 of HLA-G sits farther from the N-terminal region of the peptide; the cavity created by the movement of His70 is occupied by a water molecule. Notably, as mentioned above, the shift in the conformation of His70 results in a loss of two H-bonds and several vdW interactions between the KGPPAALTL peptide and HLA-G when compared with the HLA-GRIIPRHLQL complex.
Accordingly, the structures of HLA-GRIIPRHLQL and HLA-GKGPPAALTL are very similar in that malleability of the His70 side chain of HLA-G permits a similar mode of binding.
Structure of HLA-GKLPAQFYIL
The structure of HLA-GKLPAQFYIL was determined to 1.7-Å resolution to Rcryst and Rfree values of 18.2% and 22.6%, respectively. There were two molecules in the asymmetric unit, exhibiting an r.m.s.d. of 0.61 Å—with the difference between the two monomers primarily due to differing juxtapositioning of the α3 domain. The Ag-binding clefts have an r.m.s.d. of 0.34 Å, and the peptides have an r.m.s.d. of 0.30 Å with respect to the binding cleft. Unless stated otherwise, the interactions made with the KLPAQFYIL peptide are confined to one HLA-G molecule. The electron density for the peptide, and the contacting residues, was unambiguous and free from crystal contacts (Fig. 1b).
The peptide termini are anchored through a series of interactions that are conserved throughout MHC-I. These involve H-bonds between P1-Lys and Tyr7, between Tyr171 and Tyr159 (Fig. 2c), between P9-Leu and Asn77 and between Tyr84 and Ser143 (Fig. 2d). The side chain of P1-Lys also forms salt bridges with Glu62 and Glu63 and a water-mediated H-bond with Asn66, while P9-Leu forms an additional water-mediated H-bond with Thr80.
The backbone of the KLPAQFYIL peptide forms a total of 12 H-bonds and seven water-mediated H-bonds with HLA-G. P2-Leu forms both a H-bond and a water-mediated H-bond with Glu65 and a water-mediated H-bond with Asn66. Due to a difference in the orientation of the Asn66 side chain, a H-bond between P3-Pro and Asn66 is seen in one molecule in the asymmetric unit, whereas both molecules have H-bonds between P5-Gln and Arg156 and between P7-Tyr and Asn77 and water-mediated H-bonds between P7-Tyr and Tyr116 and between Arg156 and Asp74 (Table 2).
Table 2.
HLA-GKLPAQFYIL contacts
| Peptide residue | HLA-G residue | Interaction |
|---|---|---|
| Lys1 | ||
| Lys | Met5, Tyr7, Glu62, Glu63, Tyr159, Trp167, Tyr171 | vdW |
| LysN | Tyr7OH | H-bond |
| Tyr171OH | H-bond | |
| LysNζ | Glu62Oε2, Glu62Oε1a | Salt bridge |
| Glu63Oε2, Glu63Oε1 | Salt bridge | |
| Asn66Nδ2a | Water-mediated H-bond | |
| Asn66Oδ1b | Water-mediated H-bond | |
| LysO | Tyr159OH | H-bond |
| Leu2 | ||
| Leu | Tyr7, Met45, Glu63, Asn66b, Thr67, Tyr159 | vdW |
| LeuN | Glu63Oε2 | H-bond |
| LeuO | Glu63Oε1 | Water-mediated H-bond |
| Asn66Nδ2a | Water-mediated H-bond | |
| Pro3 | ||
| Pro | Asn66b, Ile99a, Arg156, Tyr159 | vdW |
| ProO | Asn66Oδ1b | H-bond |
| Ala4 | ||
| Ala | Asn66 | vdW |
| Gln5 | ||
| Gln | Gln155, Arg156 | vdW |
| GlnNε2 | Gln155Oε1a | H-bond |
| GlnO | Arg156NH1 | H-bond |
| Arg156NH2 | H-bond | |
| Phe6 | ||
| Phe | Asn66, His70, Trp97, Arg156 | vdW |
| Tyr7 | ||
| Tyr | Thr73, Asn77, Val152, Gln155, Arg156 | vdW |
| TyrN | Tyr116OH | Water-mediated H-bond |
| Arg156NH2 | Water-mediated H-bond | |
| TyrOH | Gln155Oε1 | H-bond |
| Arg156NH1 | Water-mediated H-bond | |
| TyrO | Asp74Oδ1 | Water-mediated H-bond |
| Asn77Nδ2 | H-bond | |
| Ile8 | ||
| Ile | Asn77, Lys146 | vdW |
| Leu9 | ||
| Leu | Asn77, Thr80, Tyr84, Tyr116, Ser143, Lys146 | vdW |
| LeuN | Asn77Oδ1 | H-bond |
| LeuO | Tyr84OH | H-bond |
| Ser143Oγ | H-bond | |
| LeuOXT | Lys146Nζ | H-bond |
| Asn77Oδ1 | Water-mediated H-bond | |
| Thr80Oγ1 | Water-mediated H-bond |
In molecule 1, not in molecule 2.
In molecule 2, not in molecule 1.
Given the limited side chains available for polar interactions in the KLPAQFYIL peptide, it is not surprising that there are only two salt bridges, two H-bonds and two water-mediated H-bonds between the peptide side chains and HLA-G. Aside from the P1 interactions described above, the only side-chain interactions are with P5-Gln and P7-Tyr. A H-bond is seen in one molecule between P5-Gln and Gln155, and P7-Tyr forms a H-bond with Gln155 and a water-mediated H-bond with Arg156.
The conformation of the termini of the KLPAQFYIL peptide and the nature of the interacting residues with HLA-G are similar to those observed in the HLA-GRIIPRHLQL and HLA-GKGPPAALTL complexes (Fig. 2). However, major deviations, up to 2.0 Å, were observed in the central region of the peptide, spanning residues P4-Ala to P8-Ile. As a consequence, the KLPAQFYIL peptide sits slightly higher in the Ag-binding cleft and closer to the α2 helix than the other two peptides when bound to HLA-G (Fig. 5a and b).
Fig. 5.
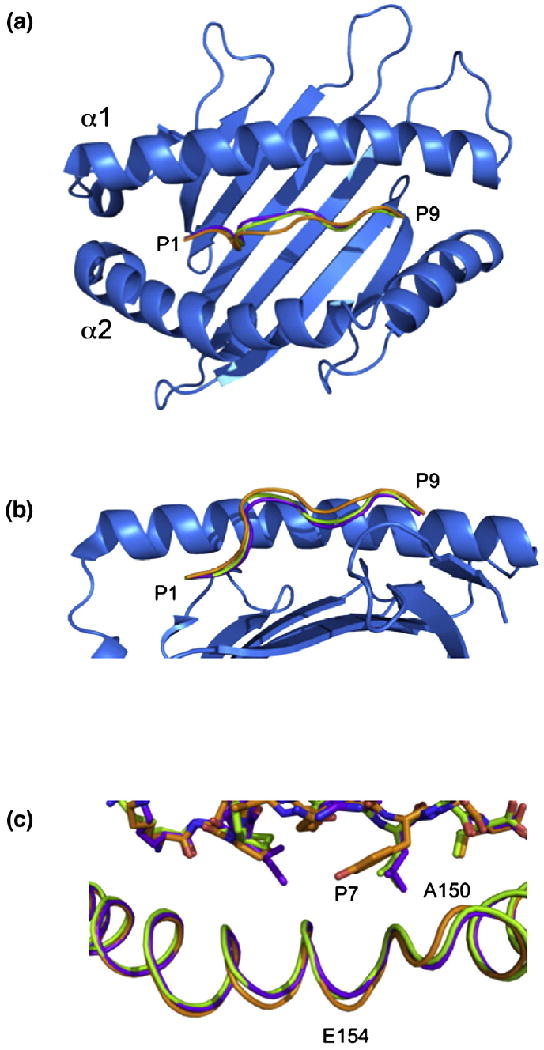
Comparison of the three pHLA-G complexes. Top view (a) and side view (b) of HLA-G showing the backbones of RIIPRHLQL (purple), KGPPAALTL (green) and KLPAQFYIL (orange). The α2 helix has been removed from (b) for clarity. (c) A 0.9-Å shift in the α2 helix centered at V152 is seen in the HLA-GKLPAQFYIL complex (orange) when compared with HLA-GRIIPRHLQL (purple) and HLA-GKGPPAALTL (green).
This shift may be due to the presence of a Phe at P6 of the peptide (Fig. 3b). P6-Phe cannot sit as low in the cleft as P6-Ala of KGPPAALTL due to its size, whereas P6-His of RIIPRHLQL, although being similarly bulky, forms multiple H-bonds with HLA-G, which anchors the peptide in the cleft. Between residues P5 and P7, the KLPAQFYIL backbone deviates by 1.5–2.0 Å from that of the RIIPRHLQL and KGPPAALTL peptides.
This movement of the P6 residue alters the H-bonding network of the KLPAQFYIL peptide backbone with respect to RIIPRHLQL. In total, two H-bonds and three water-mediated H-bonds are lost at P3, P4, P6 and P7, with another water-mediated H-bond gained at P7-Tyr. As with the HLA-GKGPPAALTL complex, the absence of P6-His eliminates two H-bonds and four water-mediated H-bonds. The substitution of Gln for Arg at P5 allows the conservation of the H-bond with Gln155 that is lost in the HLA-GKGPPAALTL complex. In the HLA-GKLPAQFYIL complex, His70 adopts the same orientation as observed in the HLA-GKGPPAALTL complex, thereby eliminating the two H-bonds with P3 and P4 seen in HLA-GRIIPRHLQL (Fig. 4).
The Ag-binding cleft of HLA-GKLPQAFYIL has r.m.s.d. values of 0.53 and 0.54 Å when superposed to HLA-GRIIPRHLQL and HLA-GKGPPAALTL, respectively. The greatest deviation was observed in residues 150–154, which are shifted by up to 0.9 Å, creating a slightly wider Ag-binding cleft in HLA-GKLPQAFYIL (Fig. 5c). This region is adjacent to the center of the peptide (residues 4–7), which deviates up to 2.0 Å from the other two peptides toward the α2 helix (Fig. 5a). The side chain of the P7 residue, a Tyr in KLPPAALTL and a Leu in the other two peptides, points toward this region of the α2 helix (Fig. 5c). This suggests that the α2 helix shifts to accommodate the movement in the peptide and the bulky P7-Tyr side chain.
Across all three pHLA-G complexes, one salt bridge, 10 H-bonds and five water-mediated H-bonds are conserved in total (Fig. 6). The major sites of conservation are P1, P2 and P9, where all polar interactions are conserved. The P1 residue forms H-bonds with Tyr7, Tyr171 and Tyr159, a salt bridge with Glu62 and a H-bond (for P1-Arg) or a salt bridge (for P1-Lys) with Glu63. The P2 residue forms a H-bond with Glu63 and water-mediated H-bonds with Glu63 and Asn66. The P9 residues form a H-bond network with Asn77, Tyr84, Ser143 and Lys146, as well as water-mediated H-bonds with Asn77 and Thr80. The other conserved interactions are H-bonds between P5 and Arg156 and a water-mediated H-bond between P7 and Tyr116.
Fig. 6.
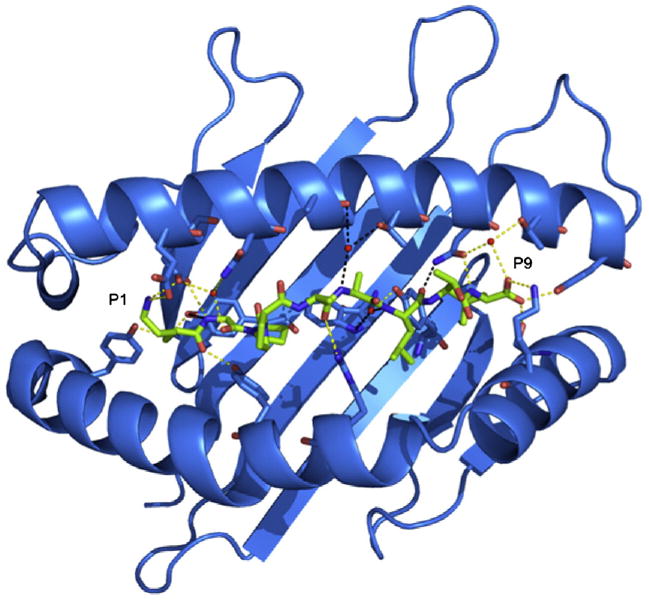
Conserved peptide contacts. Polar contacts between the KGPPAALTL peptide and HLA-G are represented by dashed lines shown in yellow for conserved contacts and in black for non-conserved contacts. Water molecules are represented by red spheres.
Thermal stability of pHLA-G complexes
In order to evaluate the contribution of the bound peptide to the stability of HLA-G, we assessed the thermal stability of HLA-G in complex with four peptides (RIIPRHLQL, KGPPAALTL, KLPAQFYIL and RLPKDFRIL) using circular dichroism (CD). The ellipticity at 218 nm was measured as temperature was increased from 20 to 90 °C. The relative thermal stability of each pHLA-G complex was determined from the temperature at which the protein was 50% unfolded (Tm) (Fig. 7). The fourth peptide, RLPKDFRIL, was identified after elution of peptides from HLA-G-transfected cells.15 We found that the yield of HLA-GRLPKDFRIL following in vitro refolding was too low for structural studies but of sufficient quantity for thermal denaturation experiments. HLA-GRLPKDFRIL was also by far the least thermally stable of the four complexes, with a Tm of 64.3 ± 1.9 °C. The other three peptides stabilised HLA-G to similar degrees, with HLA-GRIIPRHLQL marginally showing the greatest stability with Tm = 73.5 ± 0.5 °C, followed by HLA-GKGPPAALTL and then HLA-GKLPAQFYIL with Tm values of 71.8 ± 1.8 and 69.0 ± 1.7 °C, respectively.
Fig. 7.
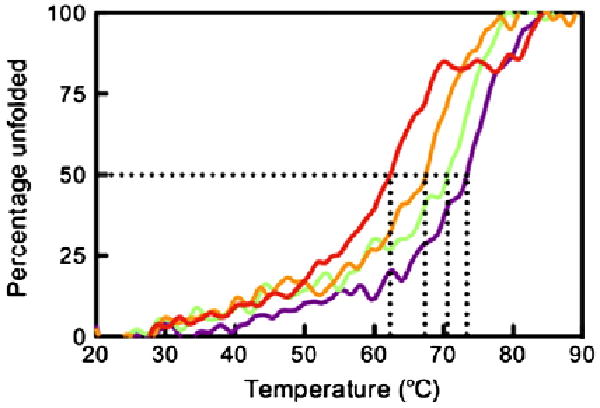
Thermal stability of pHLA-G complexes. The ellipticity at 218 nm was measured as temperature increased from 20 to 90 °C. The data were normalised to determine the midpoint of thermal denaturation (Tm). Samples were diluted to 5 and 10 μM in 10 mM Tris, pH 8.0, containing 150 mM NaCl. The complexes were measured twice at both 5 and 10 μM, and the Tm was averaged over the four experiments. The figure is representative of denaturation at 5 μM. Data for HLA-GRLPKDFRIL, HLA-GKLPAQFYIL, HLA-GKGPPAALTL and HLA-GRIIPRHLQL are shown in red, orange, green and purple, respectively.
Discussion
Typical of MHC-Ib molecules, and in contrast to MHC-Ia molecules, HLA-G binds a restricted repertoire of peptides. The structure of HLA-GRIIPRHLQL revealed an extensive network of bonds with the epitope along the Ag-binding cleft, thereby providing a snapshot for understanding the peptide binding properties of HLA-G. The mode of HLA-G peptide binding was similar to that of HLA-E, although the peptide repertoire of HLA-G is not as restricted as that of HLA-E.19 We determined the structure of HLA-G in complex with two unrelated self-peptides, KGPPAALTL and KLPAQFYIL, to simultaneously evaluate the conserved features of peptide binding by HLA-G and provide a basis for the adaptability of HLA-G in being able to accommodate disparate peptide sequences. We demonstrated that an extensive network of contacts along the length of the peptide is present in all pHLA-G complexes. However, superposition of the three pHLA-G complexes showed that while there was little difference at the N- and C-termini, the central region of the peptide demonstrated a greater degree of conformational flexibility. This malleability in the peptide also resulted in a degree of variability in the HLA-G contacting residues, which ultimately impinges on a region of the α2 helix, whereby widening of the Ag-binding cleft in the HLA-GKLPAQFYIL complex was observed.
The nature of the peptide also affected the thermal stability of pHLA-G. The RIIPRHLQL peptide had the greatest stabilising effect, possibly due to the large number of polar interactions it makes with HLA-G, whereas HLA-GRLPKDFRIL was the least stable. We investigated whether the peptide–HLA-G interface surface area (SA) and shape complementarity (SC) influenced the stability of the complexes. The values were as follows: HLA-GRIIPRHLQL, SA ≈ 750 Å2 and SC = 0.76; HLA-GKLPAQFYIL, SA ≈ 750 Å2 and SC = 0.76; and HLA-GKGPPAALTL, SA ≈ 625 Å2 and SC = 0.70. Despite the differences between HLA-GKGPPAALTL and HLA-GRIIPRHLQL/HLA-GKLPAQFYIL, the three pHLA-G complexes exhibit similar Tm values, suggesting that the SA and the SC of the interface have little bearing on complex stability. The results of the thermal stability studies are consistent with cell surface expression studies on HLA-G, where the RLPKDFRIL peptide was the least effective at stabilising HLA-G on the cell surface, while the RIIPRHLQL and KIPAQFYIL peptides (KLPAQFYIL with a substitution at P2 to improve binding) were more efficient.30 These data suggest that the peptide influences the stability of HLA-G and in turn its cell surface expression. The bound peptide is therefore likely to affect recognition of HLA-G both directly through interaction with cognate receptors and indirectly through the regulation of the levels of HLA-G on the cell surface.
HLA-G is involved in cancer, inflammation and pregnancy,31 all processes involving cells of the innate immune response. HLA-G is recognised by two families of innate immune receptors: LILRB1 and LILRB2 and KIR2DL4, with the latter binding to the Ag-binding cleft of HLA-G.27 The influence of the bound peptide on recognition of MHC-Ia by KIR is well established. The identity of the peptide presented by HLA-B27 affects recognition by KIR3DL1, with the P8 position of primary importance.32 Structural and peptide substitution studies have also demonstrated that the P8 position of the peptide presented by HLA-Cw3 is critical for KIR2DL2 binding.28 Similarly, KIR2DL1 recognition of HLA-Cw4 is influenced by the P7 and P8 residues of the bound peptide.29,33 KIR2DL1 and KIR2DL3 both bind HLA-Cw7 but display differing peptide preferences.34 Recognition of MHC-Ib by NK receptors is also influenced by the bound peptide. Subtle alterations in the HLA-E-bound peptide can have dramatic effects on recognition by CD94–NKG2 receptors.35–39 We therefore suggest that variations in the peptide and the Ag-binding cleft of HLA-G are likely to impact on KIR2DL4 binding.
Although MHC-Ib molecules play a primary role in innate immunity, MHC-Ib molecules can play a critical role in adaptive immunity.40,41 For example, Qa-1b-restricted T cells specific for Salmonella typhimurium42 and HLA-E-restricted T cells for a range of pathogens, including cytomegalovirus,43 S. typhimurium44 and Mycobacterium tuberculosis,45 have been described. As yet, a direct role for HLA-G in adaptive immunity has not been described, although HLA-G-specific cytotoxic T lymphocytes have been raised in vivo in transgenic mice.46 There is currently only one published TCR–pMHC-Ib structure, that of HLA-E presenting a cytomegalovirus-derived peptide to the KK50.4 TCR.6 This TCR displays exquisite specificity for a single peptide residue, with substitution of the P8 residue abrogating binding. With only one TCR–pMHC-Ib structure, the influence of the conformational variability of MHC on TCR recognition cannot be assessed. However, TCR–pMHC-Ia structures have demonstrated that small changes in peptide sequence47 and subtle alterations in the juxtapositioning of the α-helices can dramatically influence TCR binding and dictate the nature of the MHC-restricted response.1–3,48,49 Accordingly, we speculate that that conformational variability observed in the pHLA-G complexes will not only influence innate recognition but also be of sufficient magnitude to allow self-discrimination from non-self-discrimination by receptors of the adaptive immune system.
Materials and Methods
Cloning, expression and crystallization of HLA-G
Details pertaining to the cloning, expression and crystallization of HLA-G have been published previously.50 Briefly, the gene encoding HLA-G*0101 was cloned from the human choriocarcinoma cell line JEG-3 using standard protocols. The codon for cysteine at position 42 was changed to encode serine using Quik-Change site-directed mutagenesis (Stratagene, La Jolla, CA). The Cys-to-Ser mutation improved the yield of correctly folded HLA-G and was previously shown not to affect the mode of peptide binding.50 The HLA-G1 was expressed in BL21 Escherichia coli, and inclusion body protein was prepared, refolded and purified essentially as described previously.19,51
Data collection, structure determination and refinement
Diffracting crystals of HLA-GKGPPAALTL were obtained using the vapour diffusion technique at 4 °C with 16% polyethylene glycol, pH 7.2, plus 10 mM CoCl2 as the precipitant and streak seeding from crystals of HLA-GRIIPRHLQL.50 The crystals belonged to space group P3221 with unit cell dimensions a = b = 76.9 Å and c = 151.9 Å (Table 3). Diffracting crystals of HLA-GKLPQAFYIL were obtained using the vapour diffusion technique at 4 °C with 16% polyethylene glycol, pH 7.2, plus 10 mM CoCl2 as the precipitant. Although these crystals were grown in similar conditions to HLA-GRIIPRHLQL and HLA-GKGPPAALTL, they were of a different morphology and belonged to space group P21 with unit cell dimensions a = 58.6 b = 86.0 Å, c = 111.6 Å and β = 95.6° (Table 3), with two molecules per asymmetric unit.
Table 3.
Data collection and refinement statistics
| HLA-GKGPPAALTL | HLA-GKLPAQFYIL | |
|---|---|---|
| Data collection statistics | ||
| Peptide | KGPPAALTL | KLPAQFYIL |
| Temperature (K) | 100 | 100 |
| X-ray source | BioCARS, APS | GM/CA-CAT, APS |
| Detector | Quantum 4 CCD | MARmosaic 300 CCD |
| Space group | P3221 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 76.9, 76.9, 151.9 | 58.6, 86.0, 111.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 95.6, 90 |
| Resolution (Å) | 37.3–2.4 | 43.0–1.7 |
| Total no. of observations | 65,643 | 435,436 |
| No. of unique observations | 20,503 | 119,833 |
| Multiplicity | 3.20 | 3.63 |
| Data completeness (%) | 97.7 (93.4) | 99.4 (98.7) |
| I/σI | 24.4 (2.8) | 33.4 (2.9) |
| Rmerge (%)a | 6.1 (41.6) | 8.2 (58.9) |
| Refinement statistics | ||
| No. of reflections used | 19,333 | 113,595 |
| No. of reflections used for Rfree | 1072 | 6044 |
| Non-hydrogen atoms | ||
| Protein | 3138 | 6288 |
| Water | 113 | 993 |
| Cobalt | 1 | 2 |
| Chloride | 2 | — |
| Rcryst (%)b | 21.6 | 18.2 |
| Rfree (%)b | 29.9 | 22.6 |
| r.m.s.d from ideality | ||
| Bond lengths (Å) | 0.012 | 0.022 |
| Bond angles (°) | 1.7 | 1.9 |
| Ramachandran plot (%)c | ||
| Favoured regions | 95.8 | 98.1 |
| Allowed regions | 4.2 | 1.9 |
| B-factors (Å2) | ||
| Average main chain | 51.1 | 28.9 |
| Average side chain | 53.2 | 34.1 |
| Average water molecule | 48.1 | 45.6 |
| Cobalt | 42.4 | 84.7 |
| Chloride | 66.1 | — |
Values in parentheses are for the highest-resolution shell.
Rmerge = 100 Σ |Ihkl − 〈Ihkl〉| / ΣIhkl.
Rcryst = 100 Σhkl | |Fo| − |Fc| | / Σhkl |Fo| for all data except for 5% that was used for the Rfree calculation.
MolProbity.52
Data sets for the HLA-GKGPPAALTL and HLA-GKLPQAFYIL complexes were collected from flash-frozen crystals at the BioCARS and GM/CA-CAT beamlines using CCD detectors. The data were processed and scaled using the HKL package.53 The crystal structures of the complexes were solved by the molecular replacement method using Phaser,54 with the unliganded HLA-G structure (Protein Data Bank ID 1YDP) as the search model. For the search model, all water molecules and the bound peptide were removed. Unbiased features in the initial electron density map confirmed the correctness of the molecular replacement solution. The progress of refinement was monitored by the Rfree value (5% of the data) with neither a sigma nor a low-resolution cutoff being applied to the data. The structure was manually built in the program Coot interspersed with rounds of refinement using programs from the CCP4 package.55 The HLA-GKGPPAALTL structure was also subjected to simulated annealing using PHENIX56 to eliminate bias from the search model. Tightly restrained individual B-factor refinement was employed, and bulk solvent corrections were applied to the data set. H-bonds were located using programs from the CCP4 package (contacts). Data collection and refinement statistics are shown in Table 3.
Thermostability assay
The thermal stability of HLA-GRIIPRHLQL, HLA-GKGPPAALTL, HLA-GKLPAQFYIL and HLA-GRLPKDFRIL was investigated using CD. CD spectra were measured on a Jasco 810 spectropolarimeter using a temperature-controlled cuvette with a 0.1-cm path length. Initially, a CD scan of each protein was performed to determine the wavelength minimum, at which unfolding was measured. The ellipticity (θ) of each pHLA-G complex was measured at 218 nm as temperature was increased from 20 to 90 °C over 70 min. The θ218 at 80–90 °C was averaged to provide a value for 100% unfolded protein, and the average θ218 at 20–30 °C was taken to represent 0% unfolded protein. The midpoint (Tm) of these two values corresponded to the temperature at which the protein was 50% unfolded. Each pHLA-G complex was measured in duplicate at two concentrations, 5 and 10 μM.
Protein Data Bank accession numbers
Coordinates have been deposited in the Protein Data Bank with codes 3KYN and 3KYO.
Acknowledgments
The National Health and Medical Research Council and the Australian Research Council (ARC) supported this work. J.R. is supported by an ARC Federation Fellowship, and C.S.C. is supported by an ARC Queen Elizabeth II Fellowship. We thank the staff at the BioCARS and GM/CA-CAT for their assistance with data collection.
Abbreviations used
- MHC
major histocompatibility complex
- HLA
human leukocyte antigen
- TCR
T-cell receptor
- Ag
antigen
- NK
natural killer
- LILR
leukocyte immunoglobulin-like receptor
- KIR
killer cell immunoglobulin-like receptor
- vdW
van der Waals
- SA
surface area
- SC
shape complementarity
References
- 1.Archbold JK, Macdonald WA, Gras S, Ely LK, Miles JJ, Bell MJ, et al. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med. 2009;206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macdonald WA, Purcell AW, Mifsud NA, Ely LK, Williams DS, Chang L, et al. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J Exp Med. 2003;198:679–691. doi: 10.1084/jem.20030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tynan FE, Borg NA, Miles JJ, Beddoe T, El-Hassen D, Silins SL, et al. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I: implications for T-cell receptor engagement and T-cell immunodominance. J Biol Chem. 2005;280:23900–23909. doi: 10.1074/jbc.M503060200. [DOI] [PubMed] [Google Scholar]
- 4.Grimsley C, Kawasaki A, Gassner C, Sageshima N, Nose Y, Hatake K, et al. Definitive high resolution typing of HLA-E allelic polymorphisms: identifying potential errors in existing allele data. Tissue Antigens. 2002;60:206–212. doi: 10.1034/j.1399-0039.2002.600302.x. [DOI] [PubMed] [Google Scholar]
- 5.Lajoie J, Boivin AA, Jeanneau A, Faucher MC, Roger M. Identification of six new HLA-G alleles with non-coding DNA base changes. Tissue Antigens. 2009;73:379–380. doi: 10.1111/j.1399-0039.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- 6.Hoare HL, Sullivan LC, Pietra G, Clements CS, Lee EJ, Ely LK, et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat Immunol. 2006;7:256. doi: 10.1038/ni1312. [DOI] [PubMed] [Google Scholar]
- 7.O'Callaghan CA, Tormo J, Willcox BE, Braud VM, Jakobsen BK, Stuart DI, et al. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol Cell. 1998;1:531–541. doi: 10.1016/s1097-2765(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 8.Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278:5082–5090. doi: 10.1074/jbc.M208268200. [DOI] [PubMed] [Google Scholar]
- 9.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaschitz A, Lenfant F, Mallet V, Hartmann M, Bensussan A, Geraghty D, et al. Endothelial cells in chorionic fetal vessels of first trimester placenta express HLA-G. Eur J Immunol. 1997;27:3380–3388. doi: 10.1002/eji.1830271237. [DOI] [PubMed] [Google Scholar]
- 11.Chu W, Fant ME, Geraghty DE, Hunt JS. Soluble HLA-G in human placentas: synthesis in trophoblasts and interferon-γ-activated macrophages but not placental fibroblasts. Hum Immunol. 1998;59:435–442. doi: 10.1016/s0198-8859(98)00045-7. [DOI] [PubMed] [Google Scholar]
- 12.Kovats S, Main E, Librach C, Stubblebine M, Fisher S, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 13.McMaster MT, Librach CL, Zhou Y, Lim KH, Janatpour MJ, DeMars R, et al. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 14.Le Bouteiller P, Solier C, Proll J, Aguerre-Girr M, Fournel S, Lenfant F. Mini symposium. The major histocompatibility complex in pregnancy: Part II. Placental HLA-G protein expression in vivo: where and what for? Hum Reprod Update. 1999;5:223–233. doi: 10.1093/humupd/5.3.223. [DOI] [PubMed] [Google Scholar]
- 15.Diehl M, Munz C, Keilholz W, Stevanovic S, Holmes N, Loke YW, Rammensee HG. Nonclassical HLA-G molecules are classical peptide presenters. Curr Biol. 1996;6:305–314. doi: 10.1016/s0960-9822(02)00481-5. [DOI] [PubMed] [Google Scholar]
- 16.Lee N, Malacko A, Ishitani A, Chen M, Bajorath J, Marquardt H, Geraghty DE. The membrane-bound and soluble forms of HLA-G bind identical sets of endogenous peptides but differ with respect to TAP association. Immunity. 1995;3:591–600. doi: 10.1016/1074-7613(95)90130-2. [DOI] [PubMed] [Google Scholar]
- 17.Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, Geraghty DE. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal–placental immune recognition. J Immunol. 2003;171:1376–1384. doi: 10.4049/jimmunol.171.3.1376. [DOI] [PubMed] [Google Scholar]
- 18.Clements CS, Kjer-Nielsen L, McCluskey J, Rossjohn J. Structural studies on HLA-G: implications for ligand and receptor binding. Hum Immunol. 2007;68:220–226. doi: 10.1016/j.humimm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Clements CS, Kjer-Nielsen L, Kostenko L, Hoare HL, Dunstone MA, Moses E, et al. Crystal structure of HLA-G: a nonclassical MHC class I molecule expressed at the fetal–maternal interface. Proc Natl Acad Sci USA. 2005;102:3360–3365. doi: 10.1073/pnas.0409676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiroishi M, Kuroki K, Ose T, Rasubala L, Shiratori I, Arase H, et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J Biol Chem. 2006;281:10439–10447. doi: 10.1074/jbc.M512305200. [DOI] [PubMed] [Google Scholar]
- 21.Boyson JE, Erskine R, Whitman MC, Chiu M, Lau JM, Koopman LA, et al. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc Natl Acad Sci USA. 2002;99:16180–16185. doi: 10.1073/pnas.212643199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–225. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 23.Chapman TL, Heikema AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 24.Gonen-Gross T, Achdout H, Arnon TI, Gazit R, Stern N, Horejsi V, et al. The CD85J/leukocyte inhibitory receptor-1 distinguishes between conformed and β2-microglobulin-free HLA-G molecules. J Immunol. 2005;175:4866–4874. doi: 10.4049/jimmunol.175.8.4866. [DOI] [PubMed] [Google Scholar]
- 25.Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G α1 domain involved in KIR2DL4 recognition. Cell Res. 2005;15:176. doi: 10.1038/sj.cr.7290283. [DOI] [PubMed] [Google Scholar]
- 28.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 29.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1–HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 30.Munz C, Stevanovic S, Rammensee HG. Peptide presentation and NK inhibition by HLA-G. J Reprod Immunol. 1999;43:139. doi: 10.1016/s0165-0378(99)00029-7. [DOI] [PubMed] [Google Scholar]
- 31.Favier B, LeMaoult J, Rouas-Freiss N, Moreau P, Menier C, Carosella ED. Research on HLA-G: an update. Tissue Antigens. 2007;69:207–211. doi: 10.1111/j.1399-0039.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 32.Stewart-Jones GBE, di Gleria K, Kollnberger S, McMichael AJ, Jones EY, Bowness P. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur J Immunol. 2005;35:341–351. doi: 10.1002/eji.200425724. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maenaka K, Juji T, Nakayama T, Wyer JR, Gao GF, Maenaka T, et al. Killer cell immunoglobulin receptors and T cell receptors bind peptide–major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem. 1999;274:28329–28334. doi: 10.1074/jbc.274.40.28329. [DOI] [PubMed] [Google Scholar]
- 35.Brooks AG, Borrego F, Posch PE, Patamawenu A, Scorzelli CJ, Ulbrecht M, et al. Specific recognition of HLA-E, but not classical, HLA class I molecules by soluble CD94/NKG2A and NK cells. J Immunol. 1999;162:305–313. [PubMed] [Google Scholar]
- 36.Kaiser BK, Barahmand-pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174:2878–2884. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan LC, Clements CS, Rossjohn J, Brooks AG. The major histocompatibility complex class Ib molecule HLA-E at the interface between innate and adaptive immunity. Tissue Antigens. 2008;72:415–424. doi: 10.1111/j.1399-0039.2008.01138.x. [DOI] [PubMed] [Google Scholar]
- 38.Petrie EJ, Clements CS, Lin J, Sullivan LC, Johnson D, Huyton T, et al. CD94–NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J Exp Med. 2008;205:725–735. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan LC, Clements CS, Beddoe T, Johnson D, Hoare HL, Lin J, et al. The heterodimeric assembly of the CD94–NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 2007;27:900–911. doi: 10.1016/j.immuni.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan LC, Hoare HL, McCluskey J, Rossjohn J, Brooks AG. A structural perspective on MHC class Ib molecules in adaptive immunity. Trends Immunol. 2006;27:413. doi: 10.1016/j.it.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Lo WF, Ong H, Metcalf ES, Soloski MJ. T cell responses to Gram-negative intracellular bacterial pathogens: a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. J Immunol. 1999;162:5398–5406. [PubMed] [Google Scholar]
- 43.Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci USA. 2003;100:10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salerno-Goncalves R, Fernandez-Vina M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. 2004;173:5852–5862. doi: 10.4049/jimmunol.173.9.5852. [DOI] [PubMed] [Google Scholar]
- 45.Heinzel AS, Grotzke JE, Lines RA, Lewinsohn DA, McNabb AL, Streblow DN, et al. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J Exp Med. 2002;196:1473–1481. doi: 10.1084/jem.20020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lenfant F, Pizzato N, Liang S, Davrinche C, Le Bouteiller P, Horuzsko A. Induction of HLA-G-restricted human cytomegalovirus pp65 (UL83)-specific cytotoxic T lymphocytes in HLA-G transgenic mice. J Gen Virol. 2003;84:307–317. doi: 10.1099/vir.0.18735-0. [DOI] [PubMed] [Google Scholar]
- 47.Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- 48.Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 50.Clements CS, Kjer-Nielsen L, Kostenko L, McCluskey J, Rossjohn J. The production, purification and crystallization of a soluble form of the nonclassical MHC HLA-G: the essential role of cobalt. Acta Crystallogr, Sect F, Struct Biol Cryst Commun. 2006;62:70–73. doi: 10.1107/S1744309105041473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clements CS, Kjer-Nielsen L, MacDonald WA, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr, Sect D: Biol Crystallogr. 2002;58:2131–2134. doi: 10.1107/s0907444902015482. [DOI] [PubMed] [Google Scholar]
- 52.Lovell SC, Davis IW, Arendall BWI, de Bakker PIW, Word JM, Prisant MG, et al. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins: Struct, Funct, Genet. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 54.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr, Sect D: Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 55.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr, Sect D: Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]


