Abstract
In this issue, Xu et al. (2008) provide evidence for a new mechanism of T cell receptor regulation. Prior to activation, basic residues in the cytoplasmic domain of the signaling subunits of the T cell receptor associate with the plasma membrane such that the key signaling tyrosines are sequestered in the bilayer.
In order to sense the presence of foreign invaders, many cells of the innate and adaptive immune systems employ activating immune receptors that are comprised of a ligand-binding subunit with no signaling capacity and subunits that couple the receptor to the intracellular signaling machinery (Call and Wucherpfennig, 2007). A key part of the signaling capacity of these receptors resides in immunoreceptor tyrosine-based activation motifs (ITAMs) (Reth, 1989). Of the many receptors that contain ITAMs, the one used by αβT cells to recognize antigen is the most complicated. The α and β chains of the heterodimeric T cell receptor (TCR) typically bind the composite surface of a peptide embedded within a major histocompatibility complex molecule (pMHC). Information about the potency of the pMHC is then transferred to the ITAMs of the associated CD3γε, CD3δε, and CD3ζζ signaling dimers, which are subsequently phosphorylated by the Src kinases, Lck and Fyn. This process is commonly referred to as TCR triggering; it is a complicated process that remains poorly understood despite the efforts of many investigators over more than 20 years. The results of a collaborative effort from the Chou and Wucherpfennig labs, presented in this issue (Xu et al., 2008), represent an important breakthrough in defining a mechanism by which interactions of ITAMs with the plasma membrane act as the “safety” on the TCR trigger.
A fully assembled TCR-CD3 complex contains a total of ten ITAMs: one each for CD3γ, δ, and ε and three in CD3ζ. Experiments in which the ITAMs have been selectively inactivated indicate that the number of ITAMs that are phosphorylated upon TCR engagement determines how thymocytes or mature T cells respond (Holst et al., 2008). Thus, having ten ITAMs is thought to allow for “scalable signaling” to elicit the appropriate thymocyte or T cell response from a range of potential responses that can result from TCR engagement.
This suggests that tight control over ITAM phosphorylation must exist to prevent inappropriate signaling. Aivazian and Stern (2000) showed that a recombinant cytoplasmic domain of CD3ζ, which has a net positive charge due to clusters of basic amino acids, can bind to acidic phospholipid vesicles and that this interaction prevents phosphorylation of the CD3ζ ITAMs by Lck. They proposed a model in which the binding of the CD3ζ cytoplasmic domain to the plasma membrane in some way prevents the spontaneous phosphorylation of the ITAMs, whereas receptor multimerization, upon TCR engagement, results in a high local concentration of the intracellular domains. As a consequence of local competition for lipid binding some ITAMs then fall away from the membrane and become phosphorylated.
The new work by Xu et al. (2008) focused on interactions between the cytoplasmic domain of CD3ε and lipids. Similar to work by Sigalov et al. (2006), the authors demonstrate that the affinity of a soluble CD3ε polypeptide for lipid vesicles depends on the composition of the vesicles. Using mutagenesis, they demonstrate that this interaction is mediated by a cluster of membrane-proximal basic residues, rather than the hydrophobic residues of the ITAM. Importantly, binding prevented the tyrosines from being phosphorylated by Lck. However, one caveat to these and the previous experiments is that the electrostatic interactions observed in these assays may be an in vitro artifact.
To establish that the CD3ε cytoplasmic domain can interact with the electronegative inner leaflet of the plasma membrane of living cells, the authors expressed the extracellular and transmembrane domains of the killer cell immunoglobulin-like receptor KIR2DL3 fused to a teal fluorescent protein (TFP)-tagged CD3ε cytoplasmic domain in Jurkat T cells and labeled the cells with R18 membrane dye. Fluorescence resonance energy transfer (FRET) was observed between TFP and R18 with the wild-type CD3ε reporter construct but was diminished if the membrane-proximal basic residues of the cytoplasmic domain were changed to serines. These data indicate that, in isolation from the full complement of TCR-CD3 cytoplasmic domains, the cytoplasmic domain of CD3ε can reside in close association with the inner leaflet of the membrane of a living cell. The authors then provide a striking picture of how the pieces come together by determining the NMR structure of the CD3ε cytoplasmic domain in association with a lipid bicelle. They show that it is embedded in the polar headgroups of the membrane, with the key tyrosine residues inserting into the interior of the bilayer.
Taking the data together, this study shows that positively charged intracellular domains that contain ITAMs can bind to the inner leaflet of the plasma membrane in living cells such that the crucial tyrosines are completely buried (Figure 1). This would clearly prevent ITAM phosphorylation, and the signaling cascade that follows. Presumably this mechanism is applicable to all of the receptors that utilize this motif.
Figure 1. “On” and “Off” states for ITAM Phosphorylation.
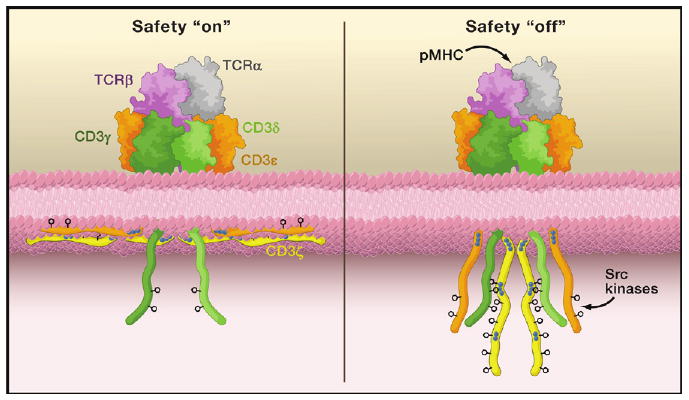
Shown on top are the subunits comprising the extracellular portion of the T cell receptor-CD3 complex: TCRα (gray), TCRβ (purple), CD3γ (dark green), CD3δ (light green), and CD3ε (orange). The cytoplasmic domains, including CD3ζ (yellow), are shown on the bottom along with the inner leaflet of the plasma membrane. The blue dots indicate stretches of basic residues that mediate interactions with the acidic inner leaf of the plasma membrane, while the phenyl rings indicate the critical tyrosines within the ITAM motifs. On the left, the TCR triggering safety is “on” and the ITAM motifs of the CD3ε and CD3ζ cytoplasmic domains are depicted as partially embedded within the plasma membrane with the tyrosines buried in the bilayer. On the right, the safety is “off” due to ligand binding (here as an antigenic peptide embedded in a major histocompatibility complex molecule [pMHC]); this may also occur spontaneously. In this “off” state, the critical tyrosine residues in the ITAM motifs become accessible to cytoplasmic Src kinases and become phosphorylated.
These findings provoke the obvious question of what would dislodge an ITAM motif from the membrane and allow phosphorylation. A mechanical force transmitted across the membrane or a change in the local environment of the complex upon TCR engagement are cited as possibilities. The two are not mutually exclusive, as a change in the local environment presumably requires an initiating signal. Mechanical forces (van der Merwe, 2001), receptor multimerization (Aivazian and Stern, 2000), or both, depending on the density of pMHC, might disrupt the interaction of some CD3ε and/or CD3ζ cytoplasmic domains with the membrane to initiate signaling. Or, they might somehow provoke phosphorylation of the cytoplasmic domains of CD3δ and CD3γ, which lack polybasic clusters and do not associate with acidic lipids (Sigalov et al., 2006). The resultant signaling cascade from either scenario could induce the depletion or accumulation of second messengers, such as phosphatidylinositol bisphosphate (PIP2), inositol triphosphate (IP3), or Ca2+, that could alter the membrane potential and release the associated cytoplasmic domains of CD3ε and CD3ζ similar to mechanisms that regulate phagocytosis (Yeung and Grinstein, 2007). The potency of the pMHC might influence the amount of second messengers that are depleted or released, which could impact the number of ITAMs available for phosphorylation and thus provide a basis for scalable signaling. However, the risk inherent in this scenario is that the change in the environment could nonspecifically release ITAMs from the membrane of nonengaged receptors for access by the Src kinases. This lack of control is not appealing, but other possibilities exist. For example, phosphorylation of serine and threonine residues within polybasic regions can disrupt interactions with the membrane (Yeung and Grinstein, 2007). The cytoplasmic domain of CD3ε has two serines and two threonines within the polybasic region shown to mediate interactions with the membrane (Figure 1 of Xu et al.), whereas the cytoplasmic domain of CD3ζ has two serines within the analogous region (Sigalov et al., 2006). Interestingly, the serine/threonine kinase GRK2 has been shown to constitutively associate with the polybasic region of the CD3ε cytoplasmic domain (DeFord-Watts et al., 2007). A mechanical force or multimerization could be invoked as the causative agent of the serine/threonine phosphorylation. Such a mechanism is appealing because exposing the CD3ε and CD3ζ cytoplasmic domains for phosphorylation would be intrinsic to the TCR-CD3 complex that is engaged. Scalable signaling might also be regulated via serine/threonine phosphorylation within the ITAMs, as has been shown to regulate B cell receptor signaling (Muller et al., 2000). Clearly several more pieces need to be put in place before the TCR signaling puzzle is solved, but the study by Xu and colleagues is an important step forward.
Acknowledgments
We thank E. Newell, L. Klein, B. Lillemeier, A. Girvin, and T. Serwold for thoughtful discussions.
References
- Aivazian D, Stern LJ. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- DeFord-Watts LM, Young JA, Pitcher LA, van Oers NS. J Biol Chem. 2007;282:16126–16134. doi: 10.1074/jbc.M609418200. [DOI] [PubMed] [Google Scholar]
- Holst J, Wang H, Eder KD, Workman CJ, Boyd KL, Baquet Z, Singh H, Forbes K, Chruscinski A, Smeyne R, et al. Nat Immunol. 2008;9:658–666. doi: 10.1038/ni.1611. [DOI] [PubMed] [Google Scholar]
- Muller R, Wienands J, Reth M. Proc Natl Acad Sci USA. 2000;97:8451–8454. doi: 10.1073/pnas.97.15.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reth M. Nature. 1989;338:383–384. [Google Scholar]
- Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Biochemistry. 2006;45:15731–15739. doi: 10.1021/bi061108f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe PA. Immunity. 2001;14:665–668. doi: 10.1016/s1074-7613(01)00155-8. [DOI] [PubMed] [Google Scholar]
- Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, Wucherpfennig KW. Cell. 2008 doi: 10.1016/j.cell.2008.09.044. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Grinstein S. Immunol Rev. 2007;219:17–36. doi: 10.1111/j.1600-065X.2007.00546.x. [DOI] [PubMed] [Google Scholar]


