Abstract
An acyl-CoA dehydrogenase has been identified as part of the mitochondrial β-oxidation pathway in the ascomycete fungus Aspergillus nidulans. Disruption of the scdA gene prevented use of butyric acid (C4) and hexanoic acid (C6) as carbon sources and reduced cellular butyryl-CoA dehydrogenase activity by 7.5-fold. While the mutant strain exhibited wild-type levels of growth on erucic acid (C22:1) and oleic acid (C18:1), some reduction in growth was observed with myristic acid (C14). The ΔscdA mutation was found to be epistatic to a mutation downstream in the β-oxidation pathway (disruption of enoyl-CoA hydratase). The ΔscdA mutant was also unable to use isoleucine or valine as a carbon source. Transcription of scdA was observed in the presence of either fatty acids or amino acids. When the mutant was grown in medium containing either isoleucine or valine, organic acid analysis of culture supernatants showed accumulation of 2-oxo acid intermediates of branched chain amino acid catabolism, suggesting feedback inhibition of the upstream branched-chain α-keto acid dehydrogenase.
Keywords: Aspergillus nidulans, fatty acid, beta-oxidation, valine, isoleucine, acyl-CoA dehydrogenase, isobutyryl-CoA, 2-methylbutyryl-CoA, butyryl-CoA
Introduction
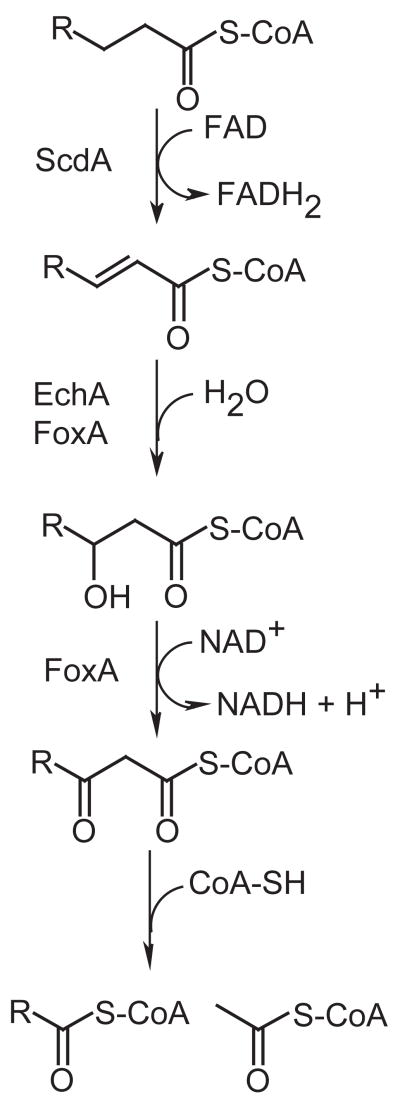
Fatty acids are metabolized to acetyl-CoA units via β-oxidation (Fig. 1). In mammals multiple distinct β-oxidation pathways are known, differing primarily in substrate specificity, stereochemistry and subcellular localization (pathways are located in both mitochondria and peroxisomes). Extensive study of yeast fungi (primarily Saccharomyces cerevisiae and Candida tropicalis) has supported the existence of only one, peroxisomal β-oxidation pathway (Kunau et al., 1988; Hiltunen et al., 1992; Kurihara et al., 1992; Smith et al., 2000). An additional mitochondrial β-oxidation pathway has been described in the filamentous ascomycete Aspergillus nidulans (Maggio-Hall and Keller, 2004), a pathway that now appears to be conserved in all non-yeast fungi based on available genome sequences.
Figure 1.
Reactions of fatty acid β-oxidation. Fatty acyl-CoAs are first oxidized to enoyl-CoAs; depending on subcellular localization the electrons are passed either to ubiquinone (via electron transfer flavoproteins in the mitochondria) or to oxygen (producing hydrogen peroxide in peroxisomes). Enoyl-CoAs are hydrated to hydroxyacyl-CoAs, which are in turn oxidized to ketoacyl-CoAs (this time electrons are passed to NAD). The acetyl group is then transferred to a free CoA molecule (release of acetyl-CoA). The remaining acyl-CoA, now shorter by two carbon units, may undergo additional rounds of β-oxidation. Gene products whose functions have been previously described (Maggio-Hall and Keller, 2004) or described in this study are indicated.
Study of the two β-oxidation pathways in A. nidulans has suggested possibly overlapping substrate specificity (Maggio-Hall and Keller, 2004). Analysis of the mitochondrial pathway included disruption of the enoyl-CoA hydratase (2nd step of the β-oxidation cycle). The echA mutant was completely unable to grow on short-chain fatty acids (butyric and hexanoic acids) as sole carbon source and growth was severely restricted on long- (myristic and oleic acids) and very-long (erucic acid) chain fatty acids. Disruption of the foxA gene, encoding the bifunctional protein (catalyzing both 2nd and 3rd steps) of peroxisomal β-oxidation, eliminated growth on erucic acid, had an intermediate effect on oleic acid and had no effect on growth on short-chain fatty acids.
Here we describe the identification and disruption of the gene encoding short-chain acyl-CoA dehydrogenase (scdA), the first step of the mitochondrial β-oxidation pathway. Analysis of this mutant has provided new insight into the division of labor between the peroxisomal and mitochondrial pathways. Similar to the ΔechA phenotype, disruption of scdA eliminated growth on short-chain fatty acids. However, unlike disruption of echA, there was little to no effect on growth on long- or very long-chain fatty acids. Furthermore, the scdA mutation was genetically epistatic to the echA mutation on these fatty acids, suggesting that the ΔechA phenotype with these substrates was primarily due to toxicity of accumulated enoyl-CoAs or their derivatives. Consistent with the genetic analysis, disruption of scdA resulted in a 7.5-fold reduction in butyryl-CoA dehydrogenase activity in cell extracts. The scdA mutant was also unable to grow on isoleucine or valine as sole carbon source, a phenotype that had also been found for the echA mutant (Maggio-Hall and Keller, 2004). Metabolism of these compounds in A. nidulans appears to be via pathways that are similar to those known in mammals (Robinson et al. 1956; Bachhawat et al., 1957; Robinson and Coon, 1957). GC/MS analysis of culture supernatants of the ΔscdA strain showed increased accumulation of 2-oxo acid intermediates of all three branched-chain amino acids (Val, Ile and Leu) when either Ile or Val was provided in the medium. This finding suggests feedback inhibition of the upstream branched-chain α-keto acid dehydrogenase when a downstream pathway is blocked.
Materials and Methods
Strains and culture media
A. nidulans strains used in this study are listed in Table 1. Biotin was supplemented at 0.1 μM. Culture media were based on the glucose minimal medium as previously described (Shimizu and Keller, 2001). Where indicated, glucose was replaced with 1% lactose, 4.9 mM erucic acid, 6 mM oleic acid, 8.5 mM myristic acid, 20 mM hexanoic acid, or 0.1 M acetate, L-isoleucine, L-valine, L-leucine or L-methionine. When testing for growth inhibition by fatty acids in the presence of an alternate carbon source, 6 mM erucic acid, 6 mM myristic acid or 20 mM hexanoic acid was used in combination with 1% lactose. Erucic, oleic and myristic acids were solubilized with 0.5% (v/v) Tergitol NP40, a surfactant that is not metabolized by A. nidulans as a carbon source (16). Where indicated, nitrate (the sole nitrogen source in the glucose minimal medium) was replaced with 10 mM L-isoleucine, L-valine, L-leucine or L-methionine. Solid media were made by adding 1.5% agar prior to autoclaving. Cultures were grown at 37°C. The ΔechA; ΔscdA double mutant strain (RLMH63) was generated by a sexual cross between RLMH41 and RLMH62 using standard methods (Pontecorvo et al., 1953). Progeny were screened by phenotype and by PCR to confirm inheritance of both gene disruptions.
Table 1.
Strain List
| Strain Name | Genotype | Reference |
|---|---|---|
| A26 | biA1; veA1 | FGSCa |
| A89 | biA1; argB2; veA1 | FGSC |
| RLMH41 | biA1; pyrG89; ΔechA::pyrG; veA1 | (Maggio-Hall and Keller, 2004) |
| RLMH62 | pyrG89; argB2; ΔscdA::argB; veA1 | This Study |
| RLMH63 | biA1; pyrG89; ΔechA::pyrG; ΔscdA::argB; veA1 | This Study |
| TLMH17 | biA1; argB2; ΔscdA::argB; veA1 | This Study |
Fungal Genetic Stock Center, Kansas City, KS
Disruption of scdA
The scdA gene appears in GenBank as AN0824.2 (Accession No. EAA65654). scdA was amplified from genomic DNA, prepared as previously described (Yang and Griffiths, 1993), and cloned into the blunt-ended BamHI site of pBluescript SK− (Stratagene) to generate pLMH23. PCR primers were designed using the A. nidulans genome sequence (Broad Institute; http://www.broad.mit.edu/) and were as follows: TAATCTGGAGCAGGCTACACT and TTCGCAAAATACACAATGTGG. Identity of the cloned 3.5 kb PCR product was confirmed by sequencing. A 817 bp BamHI fragment was excised from the scdA gene (removing 709 bp of coding region plus 108 bp upstream) and replaced with the A. nidulans argB gene (Upshall et al., 1986). This plasmid (pLMH24) was linearized with NotI and transformed into A. nidulans strain A89 (biA1; argB2; veA1) using standard protoplast transformation techniques (Yelton et al., 1984). Thirty six tranformants were tested for growth on hexanoic acid. Hexanoic acid non-utilizing strains (6/36) were then analyzed by Southern hybridization to confirm the disruption of scdA.
Complementation of the scdA mutant strain
The pyrG gene from A. fumigatus (Weidner et al., 1998) was cloned into the NotI site of the scdA clone (pLMH23). This plasmid (pLMH27) was used to transform strain RLMH62 (pyrG89; ΔscdA::argB; argB2; veA1) to pyrimidine prototrophy. The phenotypes of 36 transformants were tested, and 33 had regained the ability to grow on hexanoic acid. Two complementing and two non-complementing transformants were analyzed by Southern hybridization to confirm the presence and absence, respectively, of the transformed scdA gene.
Dry cell weight analysis
Liquid culture medium (50 ml in 125 ml flasks) was inoculated with 106 spores. Cultures were shaken at 300 rpm for 72 h. Mycelium was harvested by vacuum filtration through Miracloth (Calbiochem). Mycelium was lyophilized and weighed. Each strain was inoculated in triplicate or quintuplicate, where indicated.
Butyryl-CoA dehydrogenase assay—
Wild-type and ΔscdA cultures were cultivated in glucose medium 24 h and then transferred to medium containing 1% Tween 80, 25 mM isoleucine and 25 mM valine as carbon sources. Sixteen hours after transfer, mycelium was ground using quartz sand as described by Valenciano (Valenciano et al., 1996), except that 10 mM Tris-HCl, pH 7.5 + 1 mM EDTA was used as the isolation buffer. The resulting homogenate was filtered through Whatman paper and centrifuged 5 min. at 900 x g. The cleared extract was then sonicated 2 x 1 min. with an Ultrasonics W375 sonicator (microtip set at output level 3, 30% duty cycle). Butyryl-CoA dehydrogenase activity was measured by following the reduction of ferricenium ion (at 300 nm, ε=4.3 mM−1) as previously described (Lehman and Thorpe, 1990), except that the buffer used was 42 mM Tris-HCl, pH 7.5, and 0.2 mM butyryl-CoA was used as substrate. Low residual reduction of ferricenium ion by cell extracts in the absence of substrate (corresponding to ca. 0.007 μmole ion min−1mg−1 for both wild type and mutant extracts) was subtracted from specific activities reported in the results section.
GC/MS analysis of culture supernatants—
Strains were grown 24 h in liquid glucose minimal medium and transferred to carbon-free minimal medium. After 1.5 h, mycelium was transferred to minimal medium containing 5 mM of the indicated carbon source. Supernatants were isolated by filtration after 24 h incubation and were stored at −80°C prior to GC/MS analysis. Ten milliliters of culture supernatant was lyophilized, extracted, derivatized and analyzed by GC/MS as described by Hoffmann et al. (Hoffmann et al., 1989). 4-Nitrophenol and 2-oxocaproic acid were included as internal standards. The oxo acids were treated with O-(2,3,4,5,6-pentafluorobenzyl)hydroxylamine hydrochloride prior to extraction to form oximes, which both stabilizes the compounds and aids in chromatography. Peaks were identified by comparison to a mass spectra library of known compounds. 2-oxo-3-methylvalerate and 2-oxoisocaproate were distinguished from each other based on GCMS peak of 382 (mass/charge) for 2-oxoisocaproate and differences in the 255/280 peak ratio between the two compounds.
mRNA analysis
Strains were grown 24 hours on glucose minimal medium and then transferred to minimal media containing a variety of carbon sources (described above). Six hours after transfer, mycelium was collected by filtration and lyophilized. Total RNA was isolated from powdered mycelium using Trizol reagent (Invitrogen). Northern blots were prepared as described (Sambrook and Russell, 2001). The scdA probe was generated using a random primer method (Sambrook and Russell, 2001) using the scdA clone (the 3.5 kb fragment from pLMH23, above). Radioactivity was visualized with a phosphorimager (Molecular Dynamics).
Results
scdA is required for growth on hexanoic acid, butyric acid, isoleucine and valine
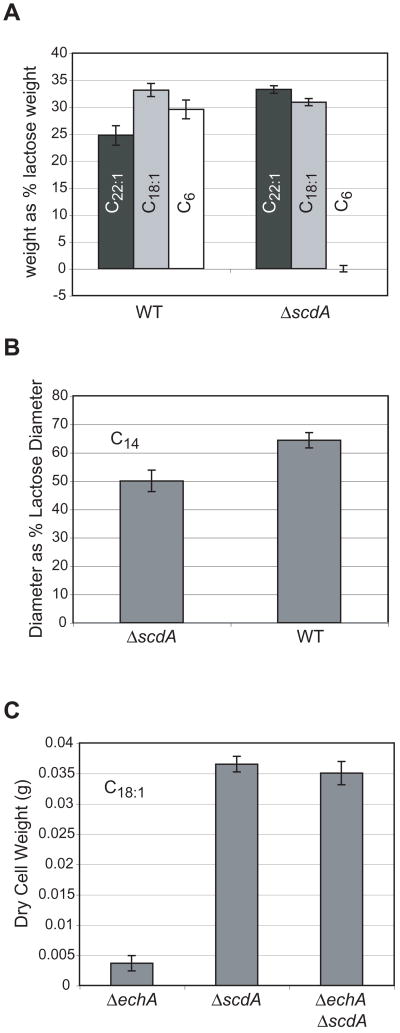
Based on the homology of its gene product to characterized mammalian enzymes, the scdA gene had been previously proposed to encode the first enzyme of a mitochondrial short-chain β-oxidation pathway in A. nidulans (Maggio-Hall and Keller, 2004). We now report that disruption of this gene eliminated growth on the short-chain fatty acids butyric (C4, data not shown) and hexanoic (C6, Fig. 2) acid, similar to what was reported for disruption of the gene encoding the second enzyme of the pathway (echA). The ΔscdA mutant showed no phenotype on erucic acid (C22:1, Fig. 2) and oleic acid (C18:1, Fig. 2), and this was markedly different than the phenotype observed for ΔechA (reduced to 4.7% and 9.3% of wild-type growth on erucic and oleic acids, respectively, Maggio-Hall and Keller, 2004). Growth of the ΔscdA mutant on myristic acid (C14) was approximately 80% of the wild type level (Fig. 2). The ΔechA and ΔscdA mutations were combined by sexually crossing the two individual mutants, and the ΔscdA mutation was found to be epistatic, i.e. the double mutant showed wild type-growth on oleic acid (Fig. 2). Like the echA mutant, growth of the scdA mutant was completely inhibited by short-chain fatty acids in the presence of another usable carbon source (lactose, data not shown). No inhibition was observed on erucic acid, and the ΔscdA mutation was found to be epistatic to the ΔechA mutation in terms of inhibition by this fatty acid (Figure S1 in the supplement). Myristic acid produced a very small but statistically significant growth inhibition of the ΔscdA mutant compared to wild type. Again, the ΔscdA mutation was found to be epistatic to ΔechA with respect to inhibition by myristic acid (Figure S1 in the supplement).
Figure 2.
Growth of A. nidulans strains on fatty acids as sole carbon source. A. Dry cell weight of mycelium was measured after 72 h incubation in liquid minimal media containing lactose, erucic acid (C22:1, black bar), oleic acid (C18:1, grey bar) or hexanoic acid (C6, white bar). Cultures for each strain were set up in triplicate. Yields for the fatty acids are presented as a percent of the yield for lactose for the indicated strain (WT, A26; ΔscdA, TLMH17). Error bars represent standard error and include error from measurement of the lactose cultures. B. Colony diameter after 96 h incubation on solid minimal medium containing myristic acid, as a percent of the diameter on medium containing lactose (WT, A36; ΔscdA, TLMH17). Spores (105) were inoculated at the center of a standard Petri plate containing 25 ml solid medium. Plates were inoculated in triplicate. Error bars represent standard deviation and include error from measurement of diameters with both lactose and myristic acid. C. Dry cell weight of mycelium was measured after 72 h incubation in minimal medium containing oleic acid. Cultures for each strain were set up in quintuplicate (ΔechA, RLMH41; ΔscdA, TLMH17; ΔechA ΔscdA, RLMH63). Error bars represent standard error.
The echA gene was previously shown to be required for metabolism of isoleucine and valine (Maggio-Hall and Keller, 2004), and the ΔscdA mutant was similarly unable to grow on either of these amino acids as carbon source (Fig. 3). Growth on leucine—a branched-chain fatty acid that requires a different dehydrogenase (isovaleryl-CoA dehydrogenase, IvdA) for catabolism in A. nidulans (Rodríguez et al., 2004a)—was unaffected (Fig. 3). The ΔscdA mutation did not prevent utilization of either Ile or Val as sources of nitrogen. While growth diameter was similar to wild type when point inoculated on solid medium, asexual sporulation appeared to be inhibited at the center of the ΔscdA colony (Figure S2 in the supplement).
Figure 3.
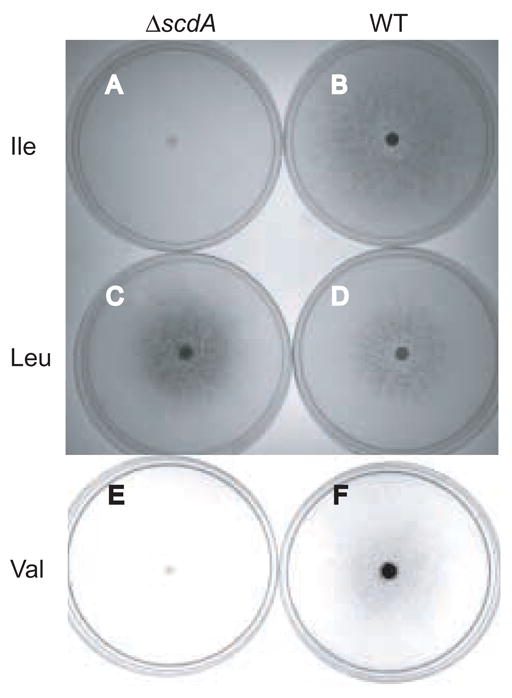
Growth of wild-type and ΔscdA strains on branched-chain amino acids as sole carbon source. Plates containing 30 ml solid medium were inoculated at the center with 5 x 106 spores and incubated at 37°C for 9 days (Ile and Leu media) or 10 days (Val medium). After two days, plates were sealed with one layer of parafilm to prevent dessication. Shown are: (A) TLMH17 (ΔscdA) and (B) A26 (wild type) inoculated on isoleucine medium, (C) TLMH17 and (D) A26 grown on leucine medium, and (E) TLMH17 and (F) A26 on valine medium. The dark color indicates successful formation of green conidia, the result of asexual development in A. nidulans.
Disruption of scdA resulted in a significant reduction (7.5-fold) in butyryl-CoA dehydrogenase activity measured in mycelial extracts. Sixteen hours after transfer to medium containing isoleucine, valine and Tween 80 (polysorbate monooleate), wild type specific activity was 0.081 ±0.007 μmole (crotonyl-CoA formed) min−1 mg−1 compared to 0.011 ±0.003 μmole min−1mg−1 in the mutant strain.
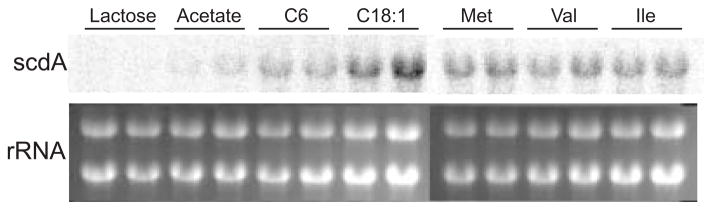
Transcription of the scdA gene was found to respond to fatty acids and amino acids (Fig. 4). Oleic acid was found to be a stronger inducer of scdA than hexanoic acid, despite our finding that scdA was not required for growth on the long-chain fatty acids. Fatty acid catabolism regulatory proteins have been recently discovered in A. nidulans (Hynes et al., 2006). The FarA regulator was found to be necessary for induction of gene expression by both long- and short-chain fatty acids and was shown to bind DNA upstream of echA. It is likely FarA also regulates scdA, explaining, at least mechanistically, the induction by oleic acid. Putative binding sites for FarA and the short-chain responsive FarB are present upstream of the scdA gene (CCGAGG or CCTCGG present at −1239, −1192, −704, −188, and −155 relative to the start codon). Expression of scdA may also be induced generally by the presence of a number of amino acids, as methionine—which does not require scdA for its use as a carbon source (data not shown)—was found to induce expression of scdA at levels similar to isoleucine and valine (Fig. 3). Similar results were reported for Northern analyses of genes required for the metabolism of leucine in A. nidulans (Rodríguez et al., 2004b). Rodríguez et al. (2004b) found expression of mccA and mccB responded to phenylalanine and methionine in addition to isoleucine, valine, and leucine.
Figure 4.
Transcriptional control of scdA by fatty acids and amino acids. Shown is a Northern analysis of scdA transcript levels in the wild-type strain (A26) after 6 h incubation in minimal media containing the following carbon sources: lactose, acetate, hexanoic acid (C6), oleic acid (C18:1), Met, Val, and Ile. Total RNA was isolated from two biological replicates for each carbon source in this analysis. Ethidium bromide-stained rRNA is shown as a control for total RNA loading.
Organic acids accumulate in the culture supernatants of ΔscdA strains
Wild type and mutant strains were grown on glucose minimal medium in submerged culture and then transferred to media containing valine, isoleucine or short-chain fatty acids as sole carbon source. After 24 h supernatants were subjected to GC/MS analysis. Supernatants were lyophilized, extracted and converted to trimethylsilyl derivatives. Peaks were identified by comparison of the corresponding mass spectrum to a library of known mass spectra derived from the analysis of commercially available standards. Mutant and wild-type peak areas were normalized to the internal standard 2-oxocaproic acid to allow for quantitative comparisons. No major cellular metabolites were observed to accumulate in the mutant strain in the presence of short-chain fatty acids (data not shown). Metabolites observed in the presence of the two amino acids are described below.
Valine
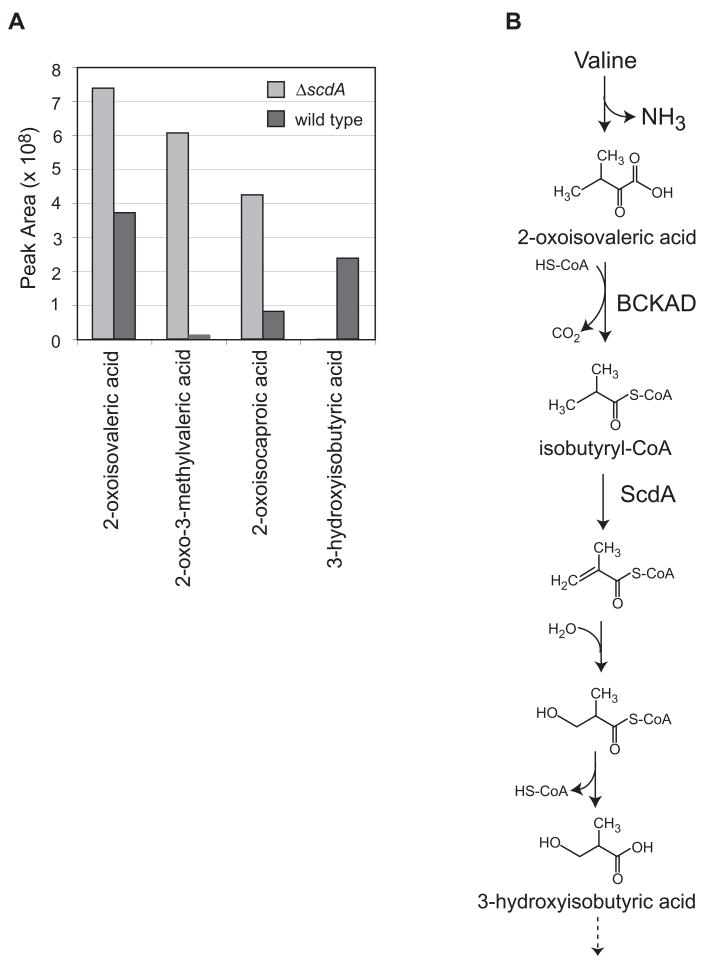
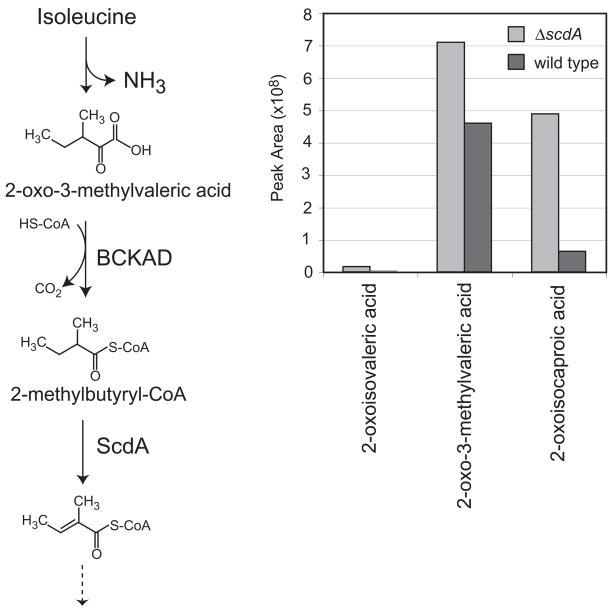
Proposed intermediates upstream of the acyl-CoA dehydrogenase in the valine catabolic pathway are 2-oxoisovaleric acid (the product of transamination) and isobutyryl-CoA (Fig. 5). Isobutyrate is too volatile for the organic acid analysis procedure employed, and isobutyryl-glycine, which has been observed in urine of humans with mutations in the valine-specific dehydrogenase (Sass et al., 2004), was not observed. Glycine conjugation may not be observed in fungi, since similar organic acid analyses of metabolites from leucine catabolism also did not indicate the accumulation of glycine derivatives (Rodríguez et al., 2004b). However, the upstream intermediate 2-oxoisovaleric acid was observed to accumulate to higher levels in the ΔscdA mutant in the presence of valine (Fig. 5 and Fig. S3 in the supplement). Additionally, 2-oxo-3-methylvaleric acid and 2-oxo-isocaproic acid, the 2-oxo acid intermediates of the isoleucine and leucine pathways, respectively, were observed to accumulate in the mutant strain. 3-Hydroxyisobutyric acid, an intermediate downstream of the acyl-CoA dehydrogenase step, was found to accumulate only in wild-type supernatants. Valine is a relatively poor carbon source for A. nidulans (Fig. 3), which may explain the build-up of detectable levels of valine pathway intermediates in culture supernatants of the wild-type strain.
Figure 5.
Accumulation of intermediates of branched-chain amino acid catabolism in the ΔscdA mutant, TLMH17, incubated with Val. A. Detection of intermediates of Val, Ile, and Leu catabolism in culture supernatants of wild type and mutant (ΔscdA) strains of A. nidulans: 2-oxoisovaleric acid, 2-oxo-3-methylvaleric acid, 2-oxoisocaproic acid, and 3-hydroxyisobutyric acid. 2-oxocaproic acid was added to culture supernatants as an internal standard prior to sample lyophilization and derivatization. Shown are normalized GC/MS peak area measurements for wild type (A26, dark bars) and ΔscdA mutant (TLMH17, light bars). The 2-oxo acids were normalized using the peak areas of the 2-oxocaproic acid internal standard. The 3-hydroxyisobutyric acid peak areas were normalized using the 4-nitrophenol internal standard. B. Shown is the proposed Val catabolic pathway in A. nidulans, based on studies in other eukaryotes (Bachhawat et al., 1957; Robinson and Coon, 1957) and genetic and genomic data for A. nidulans (Maggio-Hall and Keller, 2004). Valine is converted to 2-oxoisovaleric acid by the action of an unidentified transaminase. 2-Oxoisovaleric acid is activated by the branched-chain α-keto acid dehydrogenase (BCKAD, Pettit et al., 1978) to yield a branched-chain acyl-CoA, isobutyryl-CoA. This is converted to 3-hydroxyisobutyryl-CoA by two enzymes of β-oxidation, acyl-CoA dehydrogenase and enoyl-CoA hydratase. After release of CoA, 3-hydroxyisobutyrate is expected to be converted to methylmalonate semialdehyde and then to propionyl-CoA and CO2. Propionyl-CoA is metabolized for energy via the methylcitric acid cycle in A. nidulans (Brock et al., 2000).
Isoleucine
Partial catabolism of isoleucine by the ΔscdA mutant was expected to produce 2-oxo-3-methylvaleric acid (the product of transamination) and 2-methyl-butyryl-CoA (Fig. 6). Here 2-oxo-3-methylvaleric acid was found to accumulate in the ΔscdA mutant in excess of wild type by over two-fold (Fig. 6 and Fig. S3 in the supplement). Additionally accumulation of 2-oxo-isocaproic acid (Leu pathway) and a minor accumulation of 2-oxo-isovaleric acid (Val pathway) were observed.
Figure 6.
Accumulation of intermediates of branched-chain amino acid catabolism in the ΔscdA mutant incubated with Ile. A. Detection of intermediates of Val, Ile, and Leu catabolism in culture supernatants of wild type and mutant (ΔscdA) strains of A. nidulans: 2-oxoisovaleric acid, 2-oxo-3-methylvaleric acid, and 2-oxoisocaproic acid. 2-oxocaproic acid was added to culture supernatants as an internal standard prior to sample lyophilization and derivatization. Shown are normalized GC/MS peak area measurements for wild type (A26, dark bars) and ΔscdA mutant (TLMH17, light bars). B. Shown is the proposed Ile catabolic pathway in A. nidulans, based on studies in other eukaryotes (Robinson et al., 1956) and genetic and genomic data for A. nidulans (Maggio-Hall and Keller, 2004). Ile is converted to 2-oxo-3-methylvaleric acid by the action of an unidentified transaminase. Branched-chain α-keto acid dehydrogenase (BCKAD) then generates 2-methylbutyryl-CoA, a substrate for β-oxidation enzymes, including ScdA (as indicated). One complete oxidative cycle yields acetyl-CoA and propionyl-CoA.
Discussion
Disruption of the short-chain acyl-CoA dehydrogenase homolog scdA eliminated growth on short-chain fatty acids and significantly reduced butyryl-CoA dehydrogenase activity in cell extracts. The mutant exhibited a small growth defect on myristic acid but essentially no defect on longer-chain fatty acids tested (oleic and erucic acids). Although the subcellular localization of ScdA was not explored in this work, mitochondrial localization has been demonstrated for the downstream enoyl-CoA hydratase using a GFP fusion (Maggio-Hall and Keller, 2004), and mitochondrial localization is predicted for ScdA using the PSORTII algorithm (http://psort.nibb.ac.jp/form2.html). Mitochondrial degradation of the branched-chain amino acids isoleucine and valine has long been recognized (Johnson et al., 1972), and we have now presented genetic and biochemical evidence for the participation of scdA in both of these pathways in A. nidulans. Interestingly, mammals have distinct dehydrogenases for both of these pathways in addition to those involved in metabolism of straight-chain fatty acids (Andresen et al., 2000; Ikeda et al., 1983; Nguyen et al., 2002).
The crystal structures of a number of acyl-CoA dehydrogenases have been solved, and the position of the catalytic glutamate residue is thought to play a role in substrate specificity (Battaile et al. 2002 and references therein). The spacing of the glutamate residue in ScdA is conserved with those found in the medium-, short- and short/branched-chain specific enzymes (Battaile et al. 1996, sequence alignment provided in Figure S4 in the supplement). The combined substrate range of these enzymes in terms of chain length is C4 to C12 (Ikeda et al., 1985; Rozen et al., 1994). Of these three acyl-CoA dehydrogenases ScdA shares the highest amino acid identity with the short/branched enzymes (SBCAD). SBCADs exhibit fairly wide substrate specificity, giving the highest activity with 2-methylbutyryl-CoA in the Ile degradation pathway and somewhat lower activities with butyryl-CoA, hexanoyl-CoA and isobutyryl-CoA (Rozen et al., 1994; He et al., 2003). Essentially no in vitro activity has been reported for chain lengths longer than C8. A number active site residues that affect substrate specificity have been identified in the human and rat SBCAD enzymes (He et al., 2003) and all of these are conserved—matching either the human or rat residue at each position—in ScdA (Figure S4 in the supplement). As assessed by growth rate, Ile and Val are degraded far more slowly than fatty acids in A. nidulans, but this is not necessarily a reflection on the substrate specificity of ScdA alone. Our results show that intermediates of Ile and Val metabolism were excreted into the culture medium even by the wild type. Yeast fungi appear to lack enzymes necessary for mitochondrial fatty acid β-oxidation, and, consistently, they are also unable to degrade isoleucine and valine for use as a source of carbon. Rather these amino acids are converted to so-called “fusel” alcohols after deamination in yeast (Dickinson et al., 1998; Dickinson et al., 2000).
The intermediates observed to accumulate in isoleucine and valine cultures of the ΔscdA strain are among those that are indicators of Maple Syrup Urine Disease in humans. MSUD is caused by a variety of mutations in the branched-chain keto acid dehydrogenase (BCKAD) that is shared by the leucine, isoleucine, and valine catabolic pathways, leading to the accumulation of 2-oxo acids and their corresponding branched-chain amino acids (Chuang and Shih, 2001). We have found that addition of either isoleucine or valine leads to significant build-up of 2-oxoisocaproic acid from the leucine pathway. Since scdA is not required for leucine catabolism (based on the observed genetic phenotype, reported here, and identification of a separate isovaleryl-CoA dehydrogenase reported previously by Rodríguez et al., 2004a), it is most likely that build-up of branched-chain acyl-CoAs from the isoleucine or valine pathway eventually inhibits the upstream BCKAD. Both isobutyryl-CoA and isovaleryl-CoA have been reported to inhibit bovine BCKAD (Parker and Randle, 1978; Pettit and Reed, 1988). Despite the addition of just one of the three branched-chain amino acids to the culture medium, all three were apparently being metabolized by the fungus at the time of sampling (24h after transfer from glucose-containing medium). The other two were likely mobilized from cellular storage. Both isoleucine and valine have been found to induce leucine catabolic genes (Rodríguez et al., 2004b). Another possible explanation is that accumulation of one 2-oxo acid may lead to synthesis of the others via biosynthetic pathways. For example, 2-oxoisovalerate (Val pathway) may be converted to 2-oxoisocaproate via the leucine biosynthetic pathway. This apparent mixing of both anabolic and catabolic pathways has been suggested from labeling studies of fusel alcohol synthesis in yeast (Dickinson et al., 1998; Webb and Ingraham, 1963). However, 2-oxo-3-methylvalerate (Ile intermediate) would not be easily converted to either 2-oxoisovalerate or 2-oxoisocaproate along known biosynthetic routes—this would require backing up the Ile biosynthetic pathway all the way to pyruvate—and therefore this does not likely account for the observed accumulation of these compounds in the isoleucine cultures.
The oleic acid phenotypes of the ΔscdA mutant and the ΔscdA; ΔechA double mutant support a division of labor between mitochondrial and peroxisomal β-oxidation pathways—generally short-chain fatty acids are processed by the former and long-chain fatty acids by the latter. A determination, based on growth phenotypes, of whether the substrate specificities of the pathways overlap in the middle has been hampered by lack of growth or poor growth with medium-chain length fatty acids (discussed further below). The oleic acid phenotype of the ΔechA mutant was previously attributed at least in part to toxicity, since the strain was somewhat sensitive to oleic acid in the presence of lactose (Maggio-Hall and Keller, 2004). The phenotype of the ΔscdA; ΔechA mutant strengthens this conclusion and implicates accumulated enoyl-CoAs—the products of ScdA—or their derivatives as the source of the toxicity in the ΔechA mutant. Enolates are believed to act as alkylating agents (Speir and Barnsley, 1971). Short-chain enoyl-CoA hydratase deficiency has never been identified in humans, which may indicate that animals inheriting mutant alleles are not viable. Because toxicity is observed on long-chain fatty acids when the mitochondrial pathway is blocked, we infer that at least some carbon is flowing through that pathway. It remains to be determined if this is due to a very small amount of activity with long-chain substrates (discussed further below) or if some butyryl-CoA, produced by metabolism of long-chain fatty acids in the peroxisome, is transferred to the mitochondrion. Transfer of butyryl-CoA via the carnitine shuttle system that delivers peroxisomal β-oxidation-derived acetyl-CoA to the TCA cycle is known to occur in mammals (Vanhove et al., 1993). The growth defect and small amount of inhibition of the ΔscdA mutant observed with myristic acid (C14)—assuming it was not due to minor amounts of shorter-chain fatty acids (which are much more inhibitory) present in the commercial preparation—may indicate an upper limit on chain length accommodated in the mitochondrial pathway. A single acyl-CoA dehydrogenase with a substrate range from C4 to C14 would be unprecedented. Fatty acids of length C8 to C12 fail to support growth of even wild-type A. nidulans as sole carbon source but they are still metabolized to some extent (inferred from increased inhibition of the ΔechA strain compared to wild type when octanoic acid was added to lactose minimal medium, Maggio-Hall and Keller, 2004). The phenotype data reported here and the lack of an obvious medium chain-specific dehydrogenase homolog in the A. nidulans genome call for a more thorough, kinetic analysis of purified ScdA. Additionally a more thorough understanding of organelle-specific fatty acid transport and activation in A. nidulans could help us determine which acyl-CoAs are actually available for metabolism by a given pathway.
Supplementary Material
Growth inhibition by fatty acids in the presence of an alternate carbon source. Erucic acid or myristic acid (6 mM) was added to solid minimal medium containing 1% lactose. Spores (105) of the indicated strains were inoculated at the center of individual plates. Shown are growth diameters after 5 days incubation (37ºC) of strains A26 (wild type), RLMH41 (ΔechA), RLMH63 (ΔechA ΔscdA), and TLMH17 (ΔscdA). Bar heights are averages of 3 replicates. Error bars represent standard deviation.
Growth of wild type and ΔscdA strains with individual amino acids as sole nitrogen source. The sodium nitrate in glucose or lactose minimal medium was replaced with 10 mM of the indicated amino acid. Plates were incubated for two days at 37ºC. Shown are representative results (from duplicate trials) with glucose minimal medium. Similar results were obtained with lactose minimal medium (data not shown).
Mass spectra of observed 2-oxo acid intermediates of branched-chain amino acid metabolism. Oxo acids were converted to their corresponding oximes after treatment with O-(2,3,4,5,6-pentafluorobenzyl)-hydroxylamine hydrochloride, which increases their stability and generates the signature peak at 181 amu. Shown are spectra for intermediates observed in culture supernatants of the ΔscdA strain incubated with isoleucine or valine: (A) 2-oxoisovaleric acid, (B) 2-oxo-3-methylvaleric acid, and (C) 2-oxo-isocaproic acid.
Amino acid alignment of select acyl-CoA dehydrogenase enzymes. The alignment was generated by Clustal W method using DNAstar software. Catalytic residues (inferred for IvdA and ScdA by homology, but based on crystal structures for the mammalian homologs) are indicated in cyan. Residues of mammalian SBCAD enzymes (and the corresponding ScdA residues) that are known to control substrate specificity (for 2-methylbutyryl-, hexanoyl- and isobutyryl-CoA) are indicated in pink. Sequences were obtained from GenBank: A. nidulans IvdA (AAR23112), human IVD (P26440), human LCAD (AAH39063), porcine MCAD (AAW30430), porcine SCAD (BAA13964), A. nidulans ScdA (EAA65654), rat SBCAD (P70584), and human SBCAD (AAH13756). For references regarding active site residues, see Battaile et al., 1996 and He et al., 2003. ScdA is 48.1% and 46.9% identical to human and rat SBCAD, respectively. A. nidulans IvdA (whose gene is located in the leucine catabolic cluster, Rodríguez et al., 2004a) is 55.8% identical to human IVD (isovaleryl-CoA dehydrogenase).
Acknowledgments
L.M.-H. was supported by an NIH Postdoctoral Fellowship (F32 AI052654). We thank Lane Milde and Marisa Trapp for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andresen BS, Christensen E, Corydon TJ, Bross P, Pilgaard B, Wanders RJ, Ruiter JP, Simonsen H, Winter V, Knudsen I, Schroeder LD, Gregersen N, Skovby F. Isolated 2-methylbutyrylglycinuria caused by short/branched-chain acyl-CoA dehydrogenase deficiency: identification of a new enzyme defect, resolution of its molecular basis, and evidence for distinct acyl-CoA dehydrogenases in isoleucine and valine metabolism. Am J Hum Genet. 2000;67:1095–1103. doi: 10.1086/303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat BK, Coon MJ, Kupiecki FP, Nagle R, Robinson WG. Coenzyme A thiol esters of isobutyric, methacrylic, and beta-hydroxyisobutyric acids as intermediates in the enzymatic degradation of valine. J Biol Chem. 1957;224:1–11. [PubMed] [Google Scholar]
- Battaile KP, Mohsen AA, Vockley J. Functional role of the active site glutamate-368 in rat short chain acyl-CoA dehydrogenase. Biochemistry. 1996;35:15356–15363. doi: 10.1021/bi961113r. [DOI] [PubMed] [Google Scholar]
- Battaile KP, Molin-Case J, Paschke R, Wang M, Bennett D, Vockley J, Kim JJP. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases. J Biol Chem. 2002;277:12200–12207. doi: 10.1074/jbc.M111296200. [DOI] [PubMed] [Google Scholar]
- Brock M, Fischer R, Linder D, Buckel W. Methylcitrate synthase from Aspergillus nidulans: implications for proprionate as an antifungal agent. Mol Microbiol. 2000;35:961–973. doi: 10.1046/j.1365-2958.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- Chuang DT, Shih VE. Maple syrup urine disease (branched-chain ketoaciduria) In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. McGraw-Hill; New York: 2001. pp. 1971–2006. [Google Scholar]
- Dickinson JR, Harrison SJ, Hewlins MJE. An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 1998;273:25751–25756. doi: 10.1074/jbc.273.40.25751. [DOI] [PubMed] [Google Scholar]
- Dickinson JR, Harrison SJ, Dickinson JA, Hewlins MJE. An investigation of the metabolism of isoleucine to active amyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 2000;275:10937–10942. doi: 10.1074/jbc.275.15.10937. [DOI] [PubMed] [Google Scholar]
- He M, Burghardt TP, Vockley J. A novel approach to the characterization of substrate specificity in short/branched chain acyl-CoA dehydrogenase. J Biol Chem. 2003;278:37974–37986. doi: 10.1074/jbc.M306882200. [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Wenzel B, Beyer A, Erdmann R, Fosså A, Kunau WH. Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J Biol Chem. 1992;267:6646–6653. [PubMed] [Google Scholar]
- Hoffmann G, Aramaki S, Blum-Hoffmann E, Nyhan WL, Sweetman L. Quantitative analysis of organic acids in biological samples: batch isolation followed by gas chromatographic-mass spectrometric analysis. Clin Chem. 1989;35:587–595. [PubMed] [Google Scholar]
- Hynes MJ, Murray SL, Duncan A, Khew GS, Davis MA. Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans. Eukaryot Cell. 2006;5:794–805. doi: 10.1128/EC.5.5.794-805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Dabrowski C, Tanaka K. Separation and properties of five distinct acyl-coA dehydrogenases from rat liver mitochondria. J Biol Chem. 1983;258:1066–1076. [PubMed] [Google Scholar]
- Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. J Biol Chem. 1985;260:1311–1325. [PubMed] [Google Scholar]
- Johnson WA, Connelly JL, Glynn MT. Cellular localization and characterization of bovine liver branched-chain α-keto acid dehydrogenases. Biochemistry. 1972;11:1967–1973. doi: 10.1021/bi00760a036. [DOI] [PubMed] [Google Scholar]
- Kunau WH, Buhne S, de la Garza M, Kionka C, Mateblowski M, Schultz-Borchard U, Thieringer R. Comparative enzymology of β-oxidation. Biochem Soc Trans. 1988;16:418–420. doi: 10.1042/bst0160418. [DOI] [PubMed] [Google Scholar]
- Kurihara T, Ueda M, Okada H, Kamasawa N, Naito N, Osumi M, Tanaka A. β-Oxidation of butyrate, the short-chain-length fatty acid, occurs in peroxisomes in the yeast Candida tropicalis. J Biochem. 1992;111:783–787. doi: 10.1093/oxfordjournals.jbchem.a123836. [DOI] [PubMed] [Google Scholar]
- Lehman TC, Thorpe C. Alternate electron acceptors for medium-chain acyl-CoA dehydrogenase: use of ferricenium salts. Biochemistry. 1990;29:10594–10602. doi: 10.1021/bi00499a004. [DOI] [PubMed] [Google Scholar]
- Maggio-Hall LA, Keller NP. Mitochondrial β-oxidation in Aspergillus nidulans. Mol Microbiol. 2004;54:1173–1185. doi: 10.1111/j.1365-2958.2004.04340.x. [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Andresen BS, Corydon TJ, Ghisla S, Abd-El Razik N, Mohsen AW, Cederbaum SD, Roe DS, Roe CR, Lench NJ, Vockley J. Identification of isobutyryl-CoA dehydrogenase and its deficiency in humans. Mol Genet Metab. 2002;77:68–79. doi: 10.1016/s1096-7192(02)00152-x. [DOI] [PubMed] [Google Scholar]
- Parker PJ, Randle PJ. Partial purification and properties of branched-chain 2-oxo acid dehydrogenase of ox liver. Biochem J. 1978;171:751–757. doi: 10.1042/bj1710751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit FH, Yeaman SJ, Reed LJ. Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci USA. 1978;75:4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit FH, Reed LJ. Branched-chain alpha-keto acid dehydrogenase complex from bovine kidney. Methods Enzymol. 1988;166:309–312. doi: 10.1016/s0076-6879(88)66042-3. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, MacDonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Robinson WG, Bachhawat BK, Coon MJ. Tiglyl coenzyme A and α-methylacetoacetyl coenzyme A, intermediates in the enzymatic degradation of isoleucine. J Biol Chem. 1956;218:391–400. [PubMed] [Google Scholar]
- Robinson WG, Coon MJ. The purification and properties of beta-hydroxyisobutyric dehydrogenase. J Biol Chem. 1957;225:511–521. [PubMed] [Google Scholar]
- Rodríguez JM, Ruíz-Sala P, Ugarte M, Peñalva MA. Fungal metabolic model for type I 3-methylglutaconic aciduria. J Biol Chem. 2004a;279:32385–32392. doi: 10.1074/jbc.M313044200. [DOI] [PubMed] [Google Scholar]
- Rodríguez JM, Ruíz-Sala P, Ugarte M, Peñalva MA. Fungal metabolic model for 3-methycrotony-CoA carboxylase deficiency. J Biol Chem. 2004b;279:4578–4587. doi: 10.1074/jbc.M310055200. [DOI] [PubMed] [Google Scholar]
- Rozen R, Vockley J, Zhou L, Milos R, Willard J, Fu K, Vicanek C, Low-Nang L, Torban E, Fournier B. Isolation and expression of a cDNA encoding the precursor for a novel member (ACADSB) of the acyl-CoA dehydrogenase gene family. Genomics. 1994;24:280–287. doi: 10.1006/geno.1994.1617. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sass JO, Sander S, Zschocke J. Isobutyryl-CoA dehydrogenase deficiency: isobutyrylglycinuria and ACAD8 gene mutations in two infants. J Inherit Metab Dis. 2004;27:741–745. doi: 10.1023/B:BOLI.0000045798.12425.1b. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Brown TW, Eitzen GA, Rachubinski RA. Regulation of peroxisome size and number by fatty acid β-oxidation in the yeast Yarrowia lipolytica. J Biol Chem. 2000;275:20168–20178. doi: 10.1074/jbc.M909285199. [DOI] [PubMed] [Google Scholar]
- Speir TW, Barnsley EA. The conjugation of glutathione with unsaturated acyl thiol esters and the metabolic formation of S-carboxyalkylcysteines. Biochem J. 1971;125:267–273. doi: 10.1042/bj1250267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upshall A, Gilbert T, Saari G, O'Hara PJ, Weglenski P, Berse B, Miller K, Timberlake WE. Molecular analysis of the argB gene of Aspergillus nidulans. Mol Gen Genet. 1986;204:349–354. doi: 10.1007/BF00425521. [DOI] [PubMed] [Google Scholar]
- Valenciano S, De Lucas JR, Pedregosa A, Monistrol IF, Laborda F. Induction of β-oxidation enzymes and microbody proliferation in Aspergillus nidulans. Arch Microbiol. 1996;166:336–341. doi: 10.1007/s002030050392. [DOI] [PubMed] [Google Scholar]
- Vanhove GF, Van Veldhoven PP, Fransen M, Denis S, Eyssen HJ, Wanders RJ, Mannaerts GP. The CoA esters of 2-methyl-branched chain fatty acids and of the bile acid intermediates di-and trihydroxycoprostanic acids are oxidized by one single peroxisomal branched chain acyl-CoA oxidase in human liver and kidney. J Biol Chem. 1993;268:10335–10344. [PubMed] [Google Scholar]
- Webb AD, Ingraham JL. Fusel oil. Adv Appl Microbiol. 1963;5:317–353. [Google Scholar]
- Weidner G, d'Enfert C, Koch A, Mol PC, Brakhage AA. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5'-monophosphate decarboxylase. Curr Genet. 1998;33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- Yang X, Griffiths AJ. Plasmid diversity in senescent and nonsenescent strains of Neurospora. Mol Gen Genet. 1993;237:177–186. doi: 10.1007/BF00282799. [DOI] [PubMed] [Google Scholar]
- Yelton MM, Hamer JE, Timberlake WE. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth inhibition by fatty acids in the presence of an alternate carbon source. Erucic acid or myristic acid (6 mM) was added to solid minimal medium containing 1% lactose. Spores (105) of the indicated strains were inoculated at the center of individual plates. Shown are growth diameters after 5 days incubation (37ºC) of strains A26 (wild type), RLMH41 (ΔechA), RLMH63 (ΔechA ΔscdA), and TLMH17 (ΔscdA). Bar heights are averages of 3 replicates. Error bars represent standard deviation.
Growth of wild type and ΔscdA strains with individual amino acids as sole nitrogen source. The sodium nitrate in glucose or lactose minimal medium was replaced with 10 mM of the indicated amino acid. Plates were incubated for two days at 37ºC. Shown are representative results (from duplicate trials) with glucose minimal medium. Similar results were obtained with lactose minimal medium (data not shown).
Mass spectra of observed 2-oxo acid intermediates of branched-chain amino acid metabolism. Oxo acids were converted to their corresponding oximes after treatment with O-(2,3,4,5,6-pentafluorobenzyl)-hydroxylamine hydrochloride, which increases their stability and generates the signature peak at 181 amu. Shown are spectra for intermediates observed in culture supernatants of the ΔscdA strain incubated with isoleucine or valine: (A) 2-oxoisovaleric acid, (B) 2-oxo-3-methylvaleric acid, and (C) 2-oxo-isocaproic acid.
Amino acid alignment of select acyl-CoA dehydrogenase enzymes. The alignment was generated by Clustal W method using DNAstar software. Catalytic residues (inferred for IvdA and ScdA by homology, but based on crystal structures for the mammalian homologs) are indicated in cyan. Residues of mammalian SBCAD enzymes (and the corresponding ScdA residues) that are known to control substrate specificity (for 2-methylbutyryl-, hexanoyl- and isobutyryl-CoA) are indicated in pink. Sequences were obtained from GenBank: A. nidulans IvdA (AAR23112), human IVD (P26440), human LCAD (AAH39063), porcine MCAD (AAW30430), porcine SCAD (BAA13964), A. nidulans ScdA (EAA65654), rat SBCAD (P70584), and human SBCAD (AAH13756). For references regarding active site residues, see Battaile et al., 1996 and He et al., 2003. ScdA is 48.1% and 46.9% identical to human and rat SBCAD, respectively. A. nidulans IvdA (whose gene is located in the leucine catabolic cluster, Rodríguez et al., 2004a) is 55.8% identical to human IVD (isovaleryl-CoA dehydrogenase).