Abstract
Background
Alternative DNA conformations are of particular interest as potential signals to mark important sites on the genome. The structural variability of CA microsatellites is particularly pronounced; these are repetitive poly(CA) · poly(TG) DNA sequences spread in all eukaryotic genomes as tracts of up to 60 base pairs long. Many in vitro studies have shown that the structure of poly(CA) · poly(TG) can vary markedly from the classical right handed DNA double helix and adopt diverse alternative conformations. Here we have studied the mechanism of formation and the structure of an alternative DNA structure, named Form X, which was observed previously by polyacrylamide gel electrophoresis of DNA fragments containing a tract of the CA microsatellite poly(CA) · poly(TG) but had not yet been characterized.
Results
Formation of Form X was found to occur upon reassociation of the strands of a DNA fragment containing a tract of poly(CA) · poly(TG), in a process strongly stimulated by the nuclear proteins HMG1 and HMG2. By inserting Form X into DNA minicircles, we show that the DNA strands do not run fully side by side but instead form a DNA knot. When present in a closed DNA molecule, Form X becomes resistant to heating to 100°C and to alkaline pH.
Conclusions
Our data strongly support a model of Form X consisting in a DNA loop at the base of which the two DNA duplexes cross, with one of the strands of one duplex passing between the strands of the other duplex, and reciprocally, to form a semicatenated DNA junction also called a DNA hemicatenane.
Background
Alternative DNA conformations are of particular interest as potential signals to mark important sites on the genome, contrasting with the rather uniform structure of the classical B-form DNA double helix which shows little variation as a function of its nucleotide sequence. The structural variability of CA microsatellites is particularly pronounced. These are repetitive poly (CA) · poly (TG) DNA sequences spread in all eukaryotic genomes as tracts of up to 60 base pairs long, sometimes longer, which are particularly abundant in the human genome where they are present in about 105 copies [1]. Because of this abundance, combined with their frequent length variability between different individuals, they have served as a very useful source of markers in human genetics. Many in vitro studies have shown that the structure of poly (CA) · poly (TG) can vary markedly from the classical right-handed DNA double helix and adopt diverse conformations (for a review see e.g. [2] and references therein), the best known of which being left-handed Z-DNA [3,4]. In the course of our work with DNA fragments containing this repetitive sequence we have observed the formation of several alternative structures which appeared as retarded bands upon gel electrophoresis. While some of them have been shown to correspond to multistranded complexes [5,6,7], a series of closely spaced bands initially named 'bands X' [5] were drawn to our attention for two reasons. First, they migrated near the regular double-stranded form of the fragment, suggesting that they might correspond to double-stranded, not multi-stranded, structures. Second, they were bound with high affinity by proteins HMG1 and HMG2, two abundant non-histone nuclear proteins for which no double-stranded DNA substrate with such a high affinity was known. Here we describe a mechanism of formation of these structures, and their characterization as DNA loops at the base of which the DNA duplexes form a unique knot in which one of the strands of one duplex passes between the strands of the other duplex, and reciprocally, to form a semicatenated DNA junction, also called a DNA hemicatenane.
Results
A 120 base pair (bp) DNA fragment containing a 60 bp tract of poly(CA) · poly(TG), when incubated under specific conditions and analyzed by polyacrylamide gel electrophoresis, can give rise to a series of bands. While some of these bands have been shown to correspond to multistranded complexes [5,6,7], a series of closely spaced bands initially named 'Form X' [5] were drawn to our attention for two reasons. First, they migrated near the regular double-stranded form of the fragment, suggesting that they might correspond to double-stranded, not multi-stranded, structures. Second, they were bound with high affinity by proteins HMG1 and HMG2, two abundant non-histone nuclear proteins for which no double-stranded DNA substrate with such a high affinity was known.
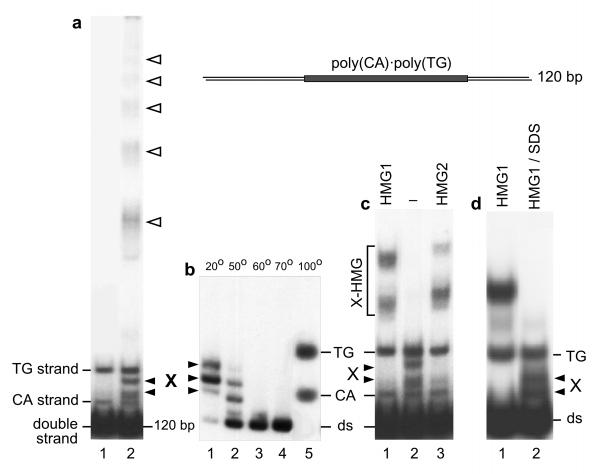
These bands are shown in Figure 1a, where they are noted X and migrate on this polyacrylamide gel between the two single strands of the DNA fragment. These structures are stable and can be purified by electroelution from a polyacrylamide gel ([5]; Fig. 1b lane 1). The study of their thermal stability (Fig. 1b) shows that they dissociate upon moderate heating to give the regular double-stranded fragment, which dissociates in turn at higher temperature to give its two single strands. Form X, therefore, contains both strands of the fragment in equimolar amount. Figure 1c shows the affinity of proteins HMG1 and HMG2 for Form X DNA. Under conditions where neither double-stranded nor single-stranded DNA is bound, Form X is entirely complexed with HMG1/2 even at a ratio of competitor DNA to Form X higher than 106. A detailed analysis of the relative affinities of HMG1/2 to different DNA substrates (double-stranded or single-stranded DNA, loops, minicircles, cruciform, Form X) will be presented elsewhere (C.G. and F.S., in preparation).
Figure 1.
Formation of alternative structures by a DNA fragment containing a poly (CA) · poly (TG) tract. a. A 120 bp DNA fragment containing a 60 bp tract of poly (CA) · poly (TG) was 32P end labelled and analyzed on a 4% polyacrylamide gel (lane 1). Under specific conditions of incubation [5,6,7,8] this fragment can give rise to a series of bands (lane 2). Two bands, labelled CA and TG, correspond to the single strands of the fragment. The upper ladder of bands (empty arrowheads) corresponds to multistranded forms [5]. Bands labelled X have not been studied previously and are the subject of the present paper, b. Form X is stable and can be electroeluted (lane 1, see Methods). After elution, Form X was incubated in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5, for 10 min. at the indicated temperatures and analyzed on a polyacrylamide gel, showing that it dissociates at ~ 50-60°C to reform the regular double-stranded fragment, c. Specific interaction of proteins HMG1 and HMG2 with Form X. The starting DNA material (lane 2) contains, in addition to the regular double-stranded fragment, small amounts of single strands and of Form X. In the presence of E. coli competitor DNA, purified HMG1 and HMG2 proteins [5] (lanes 1 and 3 respectively) bind exclusively to Form X. Increasing the amount of competitor DNA up to 4 μg per sample does not modify the result (not shown). d. Formation of Form X by strand reassociation in the presence of HMG1/2. The DNA fragment, labelled on its TG strand, was heat-denatured and allowed to reassociate in the presence of protein HMG1. By electrophoresis on a polyacrylamide gel, complexes between Form X and HMG1 are obtained (lane 1), and can be dissociated by SDS to yield free Form X (lane 2).
To study the detailed structure of Form X it was necessary to purify it in sufficient amounts. We have never been able to induce its formation directly, even by incubating the double-stranded fragment in the presence of a wide variety of agents or by varying the pH between 6.0 and 9.0. But, after it was shown that the formation of multistranded structures by fragments containing a tract of poly(CA) · poly(TG) was correlated to some extent with the opening of the double helix [6,7,8,9], we found that the most efficient way of producing Form X was to dissociate the DNA strands by thermal denaturation and to let them reassociate in the presence of protein HMG1 or HMG2. In this manner, we obtain the complexes of Form X with HMG1/2, which can then be dissociated by SDS and Form X purified (Fig. 1d, lane 2). In the absence of HMG1/2, this same process gives no Form X or hardly detectable amounts of Form X.
To study the structure of Form X, experiments were performed to study its sensitivity to single-strand specific nucleases (S1 and P1 nucleases), and to chemicals which react specifically with non-B regions of DNA (diethylpyrocarbonate, hydroxylamine, permanganate). Such experiments (not shown) clearly showed a change of conformation limited to the repetitive region of the fragments, but did not allow us to determine its exact structure. For example, the hypothesis that Form X might correspond to four-stranded structures could not be ruled out at that stage. In addition, structures containing staggered single-stranded loops resulting from shifted reassociation in the repetitive region had to be considered [7, 8]. Starting with this hypothesis, in an attempt to measure the length of such loops, we set out to determine the linking number of DNA in Form X, i.e. the number of times one strand turns around the other strand in such a structure. Indeed, if Form X contains unpaired regions, the linking number is expected to decrease by one unit for each unpaired turn of double helix (10.5 bp).
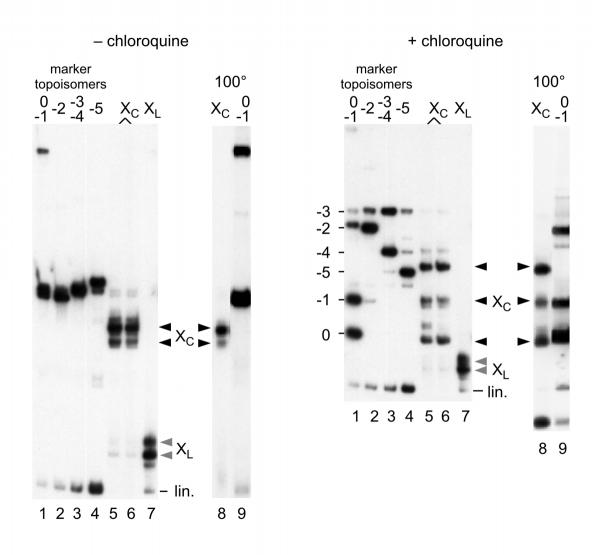
To test this hypothesis, Form X obtained with a 258 bp fragment containing the same 60 bp tract of poly(CA) · poly(TG) as above was circularized, and the products obtained were compared to a series of marker topoisomers obtained by circularization of the regular linear fragment in the presence of increasing amounts of ethidium bromide [10]. To clearly resolve all the topoisomers, the analysis was performed on polyacrylamide gels in the absence or in the presence of chloroquine [11] and is shown in Figure 2. It is observed that circularized Form X (noted Xc) does not migrate like any of the marker topoisomers. It can also be seen that, unlike linear Form X (XL), circular Form X is extremely stable and is not modified by incubation at 100°C (nor by alkaline pH, result not shown). Neither is it modified by incubation with calf thymus topoisomerase I or by human topoisomerase II, suggesting that it contains no superhelical stress. Since circular Form X does not correspond to any band in the series of topoisomer markers, we then considered that Form X might contain a large number of negative supercoils, larger than in the most supercoiled of the marker topoisomers, and that the corresponding torsional stress was absorbed and constrained by a change of conformation strictly limited to the poly (CA) · poly (TG) region and stabilized by supercoiling of the small circles. For example, a change of conformation from B-DNA to Z-DNA might have corresponded to this hypothesis.
Figure 2.
Circularization of Form X. In these experiments a 258 bp linear fragment containing the same 60 bp tract of poly(CA) · poly(TG) as above was used. Linear Form X (bands labelled XL lane 7, greyed arrowheads) was incubated in the presence of DNA ligase, and the circular forms obtained (bands labelled Xc lanes 5 and 6, black arrowheads) were analyzed by electrophoresis on polyacrylamide gels in the absence (left panel) or in the presence (right panel) of 20 μM chloroquine. A series of marker topoisomers was prepared by ligation of the regular 258 bp linear fragment in the presence of variable amounts of ethidium bromide [10], yielding a series of topoisomers containing increasing numbers of negative supercoils (lanes 1-4), with up to 5 negative superturns for the most supercoiled topoisomer. In lanes 8 and 9, circularized Form X and topoisomers 0 and -1 were analyzed after incubation for 5 min. at 100°C. Note that circularized Form X does not migrate like any of the topoisomer markers.
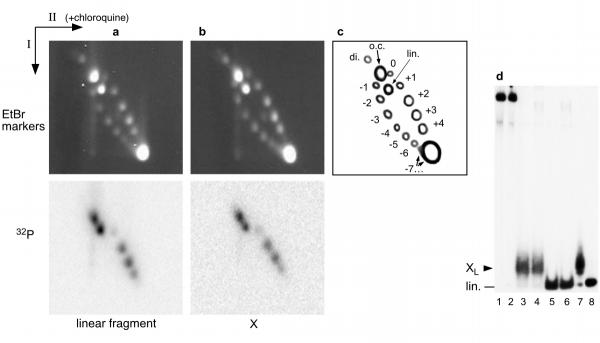
This was shown not to be the case when Form X was inserted in a large DNA fragment, leading to the formation of 2384 bp circles in which the linking number was measured by two-dimensional agarose gel electrophoresis, the first dimension in the absence of chloroquine and the second dimension in the presence of chloroquine [11] (Figure 3). No difference of migration is visible between circles containing Form X and circles containing the regular form of the fragment. If the circles containing Form X are preincubated at 100°C before electrophoresis, exactly the same distribution of topoisomers is obtained (not shown). Therefore the presence of Form X on a DNA circle of that size does not modify its electrophoretic mobility, and it is impossible to show any change of linking number in Form X relative to regular DNA. In addition, as observed with 258 bp minicircles (Fig. 2), recutting the 2384 bp circles containing Form X shows that Form X is completely stable and resistant to heating at 100°C when contained in a covalently closed circle (Fig. 3d, lanes 3 and 4). Therefore the hypothesis of a global change of conformation of poly(CA) · poly(TG) induced or stabilized by supercoiling had to be ruled out. This experiment also shows with no ambiguity that Form X contains two ends only, since ligation of Form X with the vector yields almost exclusively monomeric circles, even in the presence of an excess of vector. Therefore Form X can only be a two-stranded structure.
Figure 3.
Insertion of Form X in a full length plasmid. Plasmid pE10 was cut so as to obtain two fragments, a short 120 bp fragment containing the poly (CA) · poly (TG) tract, and a large 2264 bp fragment. The small fragment was 32P end labelled and part of it was converted to Form X. After reinsertion of the short fragment by ligation into the large 2264 bp fragment, the original plasmid pE10 was reconstituted either in its regular form, or as a Form X containing plasmid. To compare the linking numbers of both plasmids, they were analyzed by two dimension agarose gel electrophoresis with the first dimension without chloroquine and the second dimension in the presence of 1.3 μM chloroquine. The markers consisted of a series of topoisomers of plasmid pE10 obtained by recircularization of the linear plasmid in the presence of variable amounts of ethidium bromide. After electrophoresis, the gels were first stained with ethidium bromide and photographed to detect the markers, then dried and exposed to detect the radioactivity of the reformed plasmid. a. The experiment was performed with the 120 bp fragment in its regular linear form. b. Same experiment with Form X of the 120 bp fragment, c. Scheme of the different species present on the gels: o.c. open circles; lin. linear fragment; di. dimeric circles; +4 to -7: number of supercoils, positive or negative, in the marker topoisomers which were separated on the gel. Beyond 7 negative supercoils, all topoisomers migrate together under the conditions used. d. Analysis of the products on a 4% polyacrylamide gel, to show that Form X has remained stable after insertion in plasmid pE10. Lanes 1 and 2: pE10 containing Form X or the regular fragment, respectively. Lane 3: pE10 containing Form X was redigested with EcoRI + ClaI, the 120 bp fragment is recovered as Form X. Lane 4: pE10 containing Form X was incubated 2 min at 100°C and redigested with EcoRI + ClaI, Form X is recovered, showing that it is resistant to 100°C when inserted in a circular molecule. Lanes 5 and 6: redigestion of pE10 containing the regular linear form of the 120 bp fragment, without or with previous incubation at 100°C, respectively. The regular form of the 120 bp fragment is recovered, as expected. Lanes 7 and 8: controls showing respectively Form X and the regular 120 bp linear fragment used in these experiments.
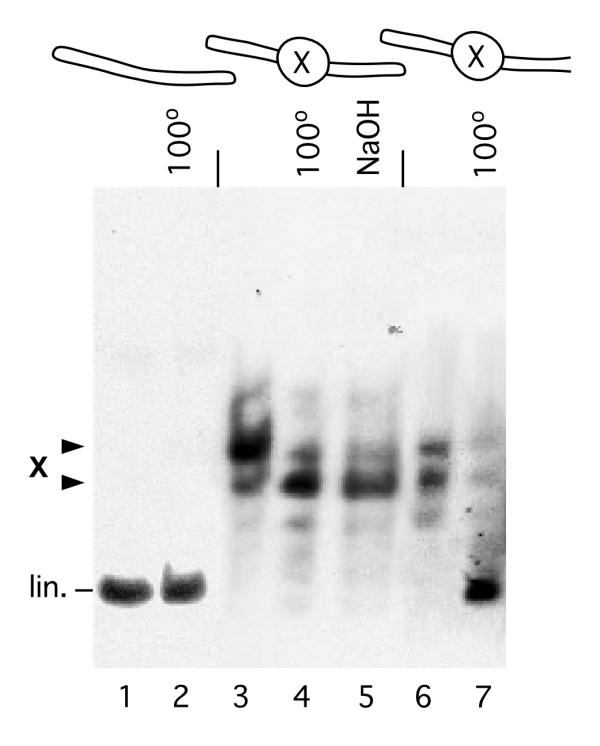
At this stage of the work, Form X looked as a paradox: a stable non-B double stranded DNA structure, with no visible change of DNA linking number. This puzzle was resolved by ligating hairpin oligonucleotides at the ends of linear Form X. Figure 4 shows that Form X remains stable in molecules with closed ends, and resists heating to 100°C and treatment by alkaline pH (lanes 4 and 5), as do circular molecules, but unlike open linear molecules. It should also be noted that upon adding a hairpin oligonucleotide at one end only, Form X is not as stable and can be dissociated by heat treatment (lane 7), although not as easily as when it is contained in an open linear fragment.
Figure 4.
Form X on a linear fragment with closed ends. Hairpin oligonucleotides were added by ligation to the ends of purified Form X. The ligation products were gel-purified and analyzed on a polyacrylamide gel with or without preincubation at 100°C or in 0.1N NaOH. Lanes 1 and 2: linear fragment with closed ends, unincubated (lane 1) or incubated at 100°C (lane 2). Lanes 3-5: Form X with closed ends, unincubated (lane 3), incubated at 100°C (lane 4), or incubated in 0.1N NaOH (lane 5). Lanes 6 and 7: Form X with one end closed and the other end open, unincubated (lane 6) or incubated at 100°C (lane 7). It is observed that Form X with both ends closed is completely resistant to denaturation (lanes 4 and 5).
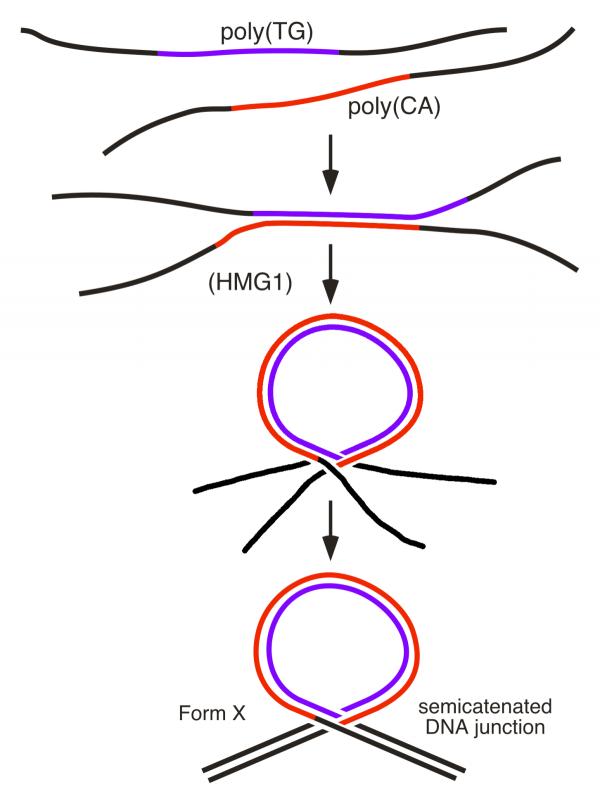
These results show that both DNA strands in Form X do not simply run side by side as in regular DNA, but that they are somehow associated in a knot. A simple model appears if one considers the process used for producing Form X. During their reassociation, the two repetitive DNA strands do not necessarily pair in perfect register, but can also pair with a shift in the repetitive poly (CA) · poly (TG) region. In such a case, one should expect a pause when the reassociation process reaches the sides of the repetitive region. We suggest that during this pause one of the single strands of one end can insert in the fork formed by the two single strands at the opposite end, possibly through interactions between the short single-stranded repetitive sequences remaining on both sides of the central double-stranded region (Fig. 5). Then, complete pairing of the non repetitive sequences at both ends yields the formation of a loop at the base of which two duplexes cross, with one of the strands of one duplex passing between the two strands of the other duplex, and reciprocally, to form a structure which is schematically represented in Figure 5. Several parameters of such a structure can vary, including the location of the junction within the repetitive nucleotide sequence, the size of the loop, and the DNA linking number inside the loop. This is probably the explanation for the number of different bands shown by Form X on a polyacrylamide gel, where up to seven bands can be seen depending on the gel concentration.
Figure 5.
Model of Form X: a DNA loop with a semicatenated DNA junction. After denaturation, the reassociation of the strands of a DNA fragment containing the sequence poly (CA) · poly (TG) can occur with a shift in the repetitive sequence. In such a case, the reassociation is expected to pause when it reaches the sides of the repetitive region, allowing one of the single-stranded ends to insert into the fork formed by the two single strands at the opposite end. This process is facilitated by HMG1 or HMG2, and possibly also by the presence of complementary sequences left on both sides of the double-stranded region. The final result is a loop at the base of which two DNA duplexes cross, forming a knot in which one of the strands of one duplex passes between the two strands of the other duplex, and reciprocally, to form a semicatenated DNA junction.
The role of HMG1/2 in the process of formation of Form X can be envisioned at two stages. On one hand, HMG1/2 increases the flexibility of DNA [12, 13] which should facilitate the formation of a loop in the central region. On the other hand, the known affinity of HMG1/2 for DNA junctions should help stabilize the transient junction before complete pairing of the non-repetitive terminal regions.
The possibility that two DNA duplexes might associate to form such junctions has been proposed previously [14,15,16,17,18,19], and such structures have been termed 'hemicatenanes' to reflect the fact that each duplex is linked to the other duplex by only one of its strands, although from an etymological point of view the term 'semicatenane' formed with two Latin roots would seem more correct. We have continued to use the term 'Form X' which we used before we began to determine its structure. Semicatenanes have been considered to explain the association of meiotic chromosomes [16, 17], although a double Holliday junction structure has so far been preferred. Semicatenanes have also been proposed as intermediates in the replication of the genome of SV40 virus, to explain some intermediates which seem not to correspond to fully catenated structures [15, 18]. The possibility for producing such structures in vitro and their remarkable stability should now allow the study of their characteristics, which should, in turn, facilitate the investigation of their significance in vivo. It should be noted that the repetitive sequence is required only because of the particular process used to prepare Form X, but that there should be no theoretical objection to the existence of such structures with non-repetitive sequences.
The remarkable stability of this structure makes it possible to consider several experiments. For example, it should be possible to insert such structures into vectors and to introduce them into living cells, allowing to study their evolution and their effect on the biological activity of the DNA molecule in which they are inserted. Equally interesting would be the study of the enzymes and more generally of the proteins able to interact with these structures. The fact that proteins HMG1 and HMG2, two of the most abundant non-histone proteins, bind to Form X with very high affinity (C.G. and F.S., in preparation) already suggests that this structure might play a role in the function of the genome. In addition, the formation of DNA loops has often been proposed to explain several chromosomal structures or many regulation processes (see e.g. [20,21]), and the semicatenated loop described here is certainly among the most stable of all DNA loops observed so far.
Conclusions
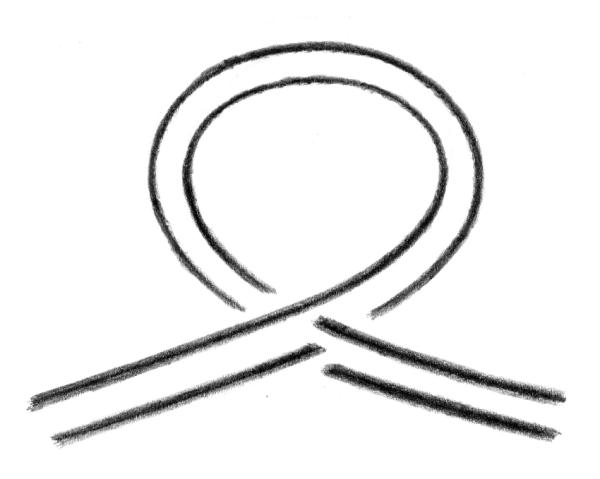
An alternative DNA structure named Form X, which was observed previously by polyacrylamide gel electrophoresis of DNA fragments containing a tract of the CA microsatellite poly (CA) · poly (TG) but had not been characterized, has now been identified as a DNA loop maintained at its base by a semicatenated DNA junction (Fig. 6). Structures containing DNA hemicatenanes had been previously suggested to exist in the cell but had not been isolated before. The possibility to prepare such structures, combined with their remarkable stability, should allow one to study their evolution and their possible function when introduced into living cells.
Figure 6.
Form X: a DNA loop with a semicatenated DNA junction.
Materials and Methods
DNA fragments
The DNA fragments used were from plasmid pE10, which contains a 60 bp tract of poly(CA) · poly(TG) (accession n° X96980) and were prepared by standard techniques. To close the ends of linear DNA fragments, 26 nucleotide-long synthetic hairpin oligonucleotides with appropriate ends were used.
Form X
To prepare Form X, DNA fragments were heat-denatured and allowed to renature in the presence of protein HMG1 or HMG2, as follows: ~ 0.1 to 10 ng of 32P end labelled DNA fragment, in 5 μL of 10 mM Tris-HCl, 1 mM EDTA, pH 7.5, was denatured at 100°C for 2 min, added as quickly as possible to 20 μL of a solution containing the reassociation buffer and ~ 10 ng of HMG protein, and allowed to renature for 45 min. at 37°C. The conditions of reassociation were: 50 mM NaCl, 25 mM Tris-HCl pH 7.5, 1 mM DTT, 1 mM EDTA, 100 μg/mL bovine serum albumin. This reassociation process yields complexes between Form X and HMG1/2, which were purified by electrophoresis in 4% polyacrylamide gels (acrylamide:bis 30:1) in 6.7 mM Tris-acetate, 3.3 mM Na acetate, 1 mM EDTA, at 4°C with buffer recirculation. The complexes were electroeluted, proteins removed by chloroform treatment in 1% SDS and 1 M NaCl, and Form X ethanol precipitated and redissolved in 10 mM Tris-HCl, 1 mM EDTA, 0.1 M NaCl, pH 7.5. The presence of 0.1 M NaCl was found to stabilize Form X, presumably by stabilization of base pairing in the terminal double-stranded regions.
Acknowledgments
Acknowledgements
We would like to thank Luigi Jonk, Nathalie Delgehyr, and Sandrine Jaouen for their help at various stages of this work. We are grateful to Susan Elsevier for critical reading of the manuscript. C.G. would also like to thank Prof. Alexander Rich (M.I.T.), in whose laboratory bands X were first observed. This work was made possible in part by grants from the Association Française contre les Myopathies, the Ligue Nationale Française Contre le Cancer, and the Association pour la Recherche contre le Cancer.
Contributor Information
Claire Gaillard, Email: gaillard@ijm.jussieu.fr.
François Strauss, Email: strauss@ijm.jussieu.fr.
References
- Hamada H, Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982;298:396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Timsit Y, Moras D. Cruciform structures and functions. Q Rev Biophys. 1996;29:279–307. doi: 10.1017/s0033583500005862. [DOI] [PubMed] [Google Scholar]
- Haniford DB, Pulleyblank DE. Facile transition of poly[d(TG) · d(CA)] into a left-handed helix in physiological conditions. Nature. 1983;302:632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Nordheim A, Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci USA. 1983;80:1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C, Strauss F. Association of poly(CA).poly(TG) DNA fragments into four-stranded complexes bound by HMG1 and 2. Science. 1994;264:433–436. doi: 10.1126/science.8153633. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii BP, Johnston BH. Denaturation and association of DNA sequences by certain polypropylene surfaces. Anal Biochem. 1997;251:251–262. doi: 10.1006/abio.1997.2249. [DOI] [PubMed] [Google Scholar]
- Gaillard C, Flavin M, Woisard A, Strauss F. Association of double-stranded DNA fragments into multistranded DNA structures. Biopolymers. 1999;50:679–689. doi: 10.1002/(SICI)1097-0282(199912)50:7<679::AID-BIP1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii BP, Johnston BH. Polypropylene tube surfaces may induce denaturation and multimerization of DNA. Science. 1996;271:222–223. doi: 10.1126/science.271.5246.222. [DOI] [PubMed] [Google Scholar]
- Gaillard C, Strauss F. Polypropylene tube surfaces may induce denaturation and multimerization of DNA - Response. Science. 1996;271:223. doi: 10.1126/science.271.5246.223a. [DOI] [PubMed] [Google Scholar]
- Shure M, Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. . Cell. 1976;8:215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Shure M, Pulleyblank DE, Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4:1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Pil PM, Chow CS, Lippard SJ. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec EB, Holloman WK. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986;44:545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Laurie B, Katritch V, Sogo J, Koller T, Dubochet J, Stasiak A. Geometry and physics of catenanes applied to the study of DNA replication. Biophys J. 1998;74:2815–2822. doi: 10.1016/S0006-3495(98)77988-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Hyrien O. Hemicatenanes form upon inhibition of DNA replication. Nucleic Acids Res. 2000;28:2187–2193. doi: 10.1093/nar/28.10.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K, von Hippel PH, Langowski J. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem Sci. 1995;20:500–506. doi: 10.1016/S0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- Engel JD, Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]