Abstract
Hepatocellular carcinoma (HCC) is a deadly cancer, whose incidence is increasing worldwide. Albeit the main risk factors for HCC development have been clearly identified, such as hepatitis B and C virus infection and alcohol abuse, there is still preliminary understanding of the key drivers of this malignancy. Recent data suggest that genomic analysis of cirrhotic tissue - the pre-neoplastic carcinogenic field - may provide a read-out to identify at risk populations for cancer development. Given this contextual complexity, it is of utmost importance to characterize the molecular pathogenesis of this disease, and pinpoint the dominant pathways/drivers by integrative oncogenomic approaches and/or sophisticated experimental models. Identification of the dominant proliferative signals and key aberrations will allow for a more personalized therapy.
Pathway-based approaches and functional experimental studies have aided in identifying the activation of different signaling cascades in HCC (e.g. epidermal growth factor, insulin-like growth factor, RAS, MTOR, WNT-ßcatenin, etc.). However, the introduction of new high-throughput genomic technologies (e.g. microarrays, deep sequencing, etc.), and increased sophistication of computational biology (e.g. bioinfomatics, biomodeling, etc.), opens the field to new strategies in oncogene and tumor suppressor discovery. These oncogenomic approaches are framed within emerging new disciplines such as systems biology, which integrates multiple inputs to explain cancer onset and progression. In addition, the consolidation of sophisticated animal models, such as mosaic cancer mouse models or the use of transposons for mutagenesis screens, have been instrumental for the identification of novel tumor drivers. We herein review some classical as well as some recent fast track approaches for oncogene discovery in HCC, and provide a comprehensive landscape of the currently known spectrum of molecular aberrations involved in hepatocarcinogenesis.
Overview
Hepatocellular carcinoma (HCC) represents a major health problem worldwide [1]. Despite its global importance, liver cancer is understudied compared to other major lethal cancers, and hence, our knowledge of the genomic alterations implicated in HCC initiation and progression is still fragmentary. This is partly due to the high complexity and heterogeneity of HCC cancer genomes, which hamper a straightforward identification of new cancer genes by genomic approaches alone. There is also only a limited arsenal of model systems available to study the development of the disease, and to test potential intervention strategies. In this scenario, HCC is probably one of the tumor types where a more complete understanding of the underlying genetic alterations could have a major impact on the development of new treatment strategies. Because of their therapeutic relevance as potential new targets, this review will first summarize the current knowledge of signaling pathways in HCC. Next, we will briefly comment the role of P53 due to its paradigmatic role as a tumor suppressor in HCC, identified using a gene-based approach. Finally, we will discuss new integrative oncogenomic approaches for an accelerated cancer gene discovery in HCC.
Signaling pathways in hepatocellular carcinoma
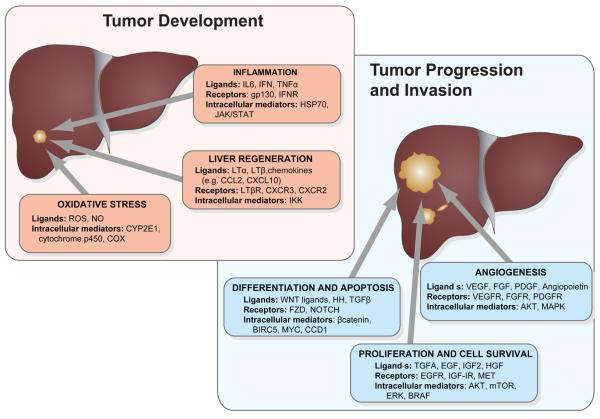
Activation of several signaling pathways, both in cirrhotic tissue and in overt HCC, has been implicated in human hepatocarcinogenesis [2] (Fig. 1). Their relevance resides in their capacity to act as targets for new therapies. In fact, activation of RAS/MAPK and VEGF signaling (i.e. angiogenesis) in HCC stimulated initial evaluations of the multi-kinase inhibitor sorafenib for the treatment of human HCC. As a result of recent positive investigations, sorafenib is currently the sole systemic therapy approved for the treatment of advanced HCC [3,4].
Figure 1. Signaling pathways frequently dysregulated in hepatocellular carcinoma.
Pathways are clustered according to the biological process they have been mostly implicated in HCC development and progression, including cell proliferation/survival, differentiation, inflammation, oxidative stress and angiogenesis.
Proliferation signaling pathways
Epidermal growth factor (EGF) signaling is one of the most thoroughly evaluated signaling cascades in human HCC. Robust data demonstrate its activation in a subset of HCC, up-regulation of EGF being one of its mechanisms of activation [5]. Genetic evidence was also provided by a recent study showing gains of chromosome 7 in human HCC, where EGF is located [6]. Some complementary evidence points to EGF as a key driver in HCC. A case-control study identified a significant increase in HCC risk associated with a specific single nucleotide polymorphism (SNP) of the EGF gene [7]. Interestingly, the same investigation showed that this SNP increased EGF mRNA stability in cell lines; thus, markedly enhancing signaling through the EGF receptor (EGFR). Additionally, EGF was ranked among the top up-regulated genes included in a gene signature that was able to identify HCC patients treated with surgical resection at high risk of developing a late recurrence (de novo HCC) [8]. Unsupervised mutagenic screening in mice also detected EGFR as a relevant gene during HCC development [9]. There are some consistent data regarding a selective EGF-blockade in experimental models. Gefitinib, an EGFR tyrosine kinase inhibitor, was able to significantly decrease HCC incidence in a genotoxic model of HCC [10]. Another study showed anti-proliferative and pro-apoptotic effects following EGFR blockade in human HCC xenografts [2]. In addition, robust evidence suggests the prominent role of TGFA as a major paracrine activator of EGF signaling in HCC [11]. TGFA contributes to proliferation and the invasion phenotype of tumor cells, and it has been shown to generate liver tumors upon co-activation with other oncogenes in animal models [e.g, double-transgenic mice: MYC/TGFA [12]. Finally, preliminary data obtained in phase II clinical trials using an EGFR specific inhibitor (i.e. erlotinib) in HCC [13,14], are currently being expanded into phase III investigations.
The insulin-like growth factor (IGF) signaling system is an essential regulator of growth and development [15]. The biological actions of the axis comprise a complex network of molecules whose main components are two high affinity mitogenic ligands: IGF1 and IGF2. The type 1 IGF receptor (IGF1R) has tyrosine kinase activity, the type 2 IGF receptor (IGF2R) is involved in the internalization and degradation of IGF2, and at least six high-affinity IGF-binding proteins (IGFBPs) which modulate the amount and bioactivity of locally available IGFs. Despite its role in normal physiology, the IGF axis is involved in the pathogenesis of several human malignancies, including breast, colon, prostate, lung, and liver [16]. In HCC, the most frequently described aberrant feature concerning this pathway is over-expression of IGF2, even found in preneoplastic lesions [11]. This mitogen, highly expressed during embryonic development, is strongly down-regulated after birth due to tight epigenetic regulation of the P2-P4 fetal promoters. Reactivation of IGF2 expression involves loss of specific imprinting and hypomethylation [17]. Allelic losses of IGF2R have been detected in 60-70% of HCC, being inactivating mutations in the remaining allele also reported[18]. In addition, reduced expression of IGFBP-3 associated to promoter hypermethylation has been reported in human HCC samples [19]. A recent study found aberrant activation of IGF1R in 21% of early stage hepatitis C related HCC, and provided preclinical evidence of antineoplastic activity following IGF1R selective blockade using a monoclonal antibody [20]. The potential role of the HBx viral protein as an inducer of IGF-IR expression has been also suggested [21].
Hepatocyte growth factor (HGF) is a potent mitogen for hepatocytes, and the unique ligand for the MET receptor. MET activation by HGF induces disruption of intercellular links and allows cells to migrate from their primary location to adjacent surroundings. The HGF/MET system is a major player of cell migration during embryogenesis and recent data has shown that the HGF/MET system is involved in reactivation of the “invasive program” during tumor progression and metastasis [22]. In addition, inhibition of MET expression using RNA-interference in cancer cell lines harboring MET amplifications decreased cell proliferation, survival, and invasion as well as delayed tumor growth in xenograft models [23]. MET activation in cancer may occur by several mechanisms including ligand-receptor autocrine stimulation, somatic and germ-line mutations, or MET amplification, allowing MET activation even in the absence of HGF [24].
In HCC, several investigators identified over-expression of MET (20-48%) when compared to surrounding non-tumoral tissue. For example, a study found a correlation between MET over-expression and a higher incidence of intrahepatic metastases, as well as a reduced survival following tumor resection [25]. By contrast, decreased HGF expression has been reported in HCC [25], suggesting a marginal role for HGF in this malignancy. A recent study took advantage of a comparative functional genomics approach on 242 HCC samples for identifying a gene signature correlated with HCC metastatic potential. This signature included genes associated with oxidative stress, cell motility, cytoskeletal organization, and angiogenesis [26]. Overall, MET has emerged as a very attractive target for anticancer therapies, albeit there is no approved drug so far. Moreover, there is compelling evidence of antineoplasic activity (i.e. decreased proliferation, migration and invasion) following MET down-regulation in experimental models of HCC [27].
Upon EGFR, IGFR or MET activation, extracellular signals can be transduced through either AKT or MAPK signaling pathways. Molecules belonging to both signaling cascades (e.g., AKT or KRAS) have been identified as bona fide oncogenes in human cancer. The RAS cascade transduces extracellular signals initiated by growth factor receptor activation that in turn activate RAF, MEK and ERK and result in proliferative and anti-apoptotic signals [28]. RAS mutations in HCC are found at low frequency [29]. Interestingly, recent studies show frequent over-expression of RAS in human HCC [30,31]. In addition, down-regulation of RAS negative regulators has been reported, such as hypermethylation of the RAS association family 1 gene A (RASSF1A) and its homologue NORE1A [30]. There is no pure anti-RAS therapy approved, and only sorafenib has shown some effective signaling blockage, either alone or in combination with rapamycin (RAS and MTOR blockade in experimental models) [31].
AKT has been described as a predictor of tumor recurrence after surgical resection in a large cohort of Japanese patients [32], suggesting a possible implication in the invasive phenotype. MTOR is one of the most important molecules downstream AKT. Two distinct MTOR complexes have been described, one closely implicated in protein translation, mTOR complex 1 (MTORC1), whereas the other, mTOR complex 2 (MTORC2), is the primary one responsible for the phosphorylation of AKT at serine 473, conferring its full activation [33]. There are intriguing feedback-loops in the mTOR pathway, which, at least in part, explains why the exact implications of each MTOR complex in the molecular pathogenesis of cancer are not fully understood [34]. However, it seems that MTORC2 integrity could be necessary to sustain the oncogenic phenotype related to loss of PTEN, at least in prostate cancer [35]. As recently reported, MTORC1 plays a pivotal role in human HCC [5]. Evidence from different studies has demonstrated MTORC1 activation in a subset of HCC patients, showing prognostic implications for some of them in terms of patient tumor recurrence after surgery [5]. Moreover, the anti-neoplasic properties of MTOR inhibitors (e.g., rapamycin and its analogs) in experimental models of HCC further increase the relevance of this pathway [36]. All this compiled evidence established the proof-of-principle to conduct clinical trials evaluating mTOR inhibitors in human HCC.
Pathways involved in liver development and cell differentiation
The ultimate identity of the specific target cell for transformation in HCC is still elusive [37]. Despite recent progresses, the involvement of altered embryonic cellular features such as self-renewal, plasticity, asymmetric division, pluripotency and cellular fate in human cancer remains enigmatic [38]. In accordance with the cancer stem cell hypothesis, a number of studies found dysregulation of pathways involved in cellular differentiation in HCC, such as the WNT canonical pathway. Besides its role in liver development and differentiation [39,40], WNT signaling has also been implicated in cell proliferation and metabolism [41]. Unlike colon cancer, APC mutations are infrequent in HCC, whereas CTNNB1 and AXIN1 mutations are frequently found in HCC [2]. Mutations prevent ß-catenin ubiquitination and subsequent degradation. Nuclear accumulation of ß-catenin induces transcription of several genes related to cell differentiation and proliferation. In addition, unsupervised clustering of gene expression data from two different studies enabled the identification of a subclass of patients characterized by activation of genes related to WNT signaling [6,42]. Hence, there is enough evidence to assume the implication of this signaling cascade in HCC, although it is still unknown whether non-canonical activation of this pathway may also be relevant during human hepatocarcinogenesis. There are currently several efforts evaluating different blockade strategies for the WNT pathway, being stabilization of AXIN one of the more promising [43]. Nevertheless, there are no clinical trials in late phases evaluating selective blockade of any of its components.
Another pathway involved in proliferation control that is found abnormally activated in HCC is the Hedgehog signaling pathway (HH). This cascade is structurally very similar to WNT, although different mechanisms account for the dysregulation of HH. Over-expression of Sonic or Indian ligands, and/or inactivation of a tumor suppressor, the HH inhibitory protein, have been identified as responsible for HH activation in experimental models of HCC as well as in human samples [44,45]. Several HH inhibitors are currently under preclinical evaluation in HCC.
Pathways involved in inflammation
The role of cascades involved in inflammation has been described in early stages of the disease. Inflammation is a hallmark of liver cirrhosis, and hence, several inflammation-related pathways have been evaluated as potential targets for HCC chemoprevention. Antiviral therapies with interferon failed in a recent large clinical study (n = 1005) to show efficacy in terms of HCC-prevention in patients with hepatitis C-related liver disease who failed to respond to conventional antiviral therapies [46]. Nonetheless, other information pinpoints interleukin 6 (IL-6) signaling as an appealing target for HCC prevention. IL-6, a multifunctional cytokine with different functions in the immune system, justifies gender disparities in HCC incidence (i.e. much higher incidence rate in males than females [47]). Estrogens suppress IL-6 production and therefore limit chemically induced liver carcinogenesis.
Enrichment in IL-6 downstream signals has also been found in a gene signature associated with poor survival and de novo tumor formation in patients with HCC treated with surgical resection [8]. Additionally, a recent report identified IL-6 signaling through STAT3 as a major pathway in maintaining stem cell like features in HCC [48], highlighting the potential role of this cascade as a novel target in cancer stem cell renewal. Currently, different anti-IL-6 humanized monoclonal antibodies are under evaluation in inflammatory conditions, but their role in HCC is yet to be determined. There is some degree of crosstalk between IL-6 and TGFB signaling, a pleiotropic pathway with potentially antagonistic actions. Comparative functional genomics have allowed the identification of a subgroup of HCC patients with a significant enrichment in late responsive TGFB genes. Thus patients expressing these genes also had more invasive tumors and poor survival [49]. All these data have encouraged efforts aiming to modulate TFGB signaling, in part due to its clear involvement in metastasis. Downstream of IL-6, signals can be transduced through the JAK/STAT signaling cascade. Nuclear localization of STAT3 results in trans-activation of its target genes (e.g. protein inhibitors of activated STATS [PIAS], the SH2-containing phosphatases [SHP], and the suppressors of cytokine signaling [SOCS]), implicated in cell growth, proliferation, differentiation and survival. Constitutive activation of JAK/STAT has been detected in a subset of HCC, mostly due to promoter methylation of STAT inhibitors such as CIS, SOCS-1, SOCS-2, SOCS-3, and SHP1 [30]. Additionally, treatment with de-methylating agents resulted in growth suppression and strongly induced apoptosis in HCC cell lines [30].
There is strong evidence suggesting a linking role of the NF-Kappaß cascade in inflammation and liver oncogenesis [50]. Recent progress regarding drug-mediated inhibition of this pathway provides new perspectives for primary prevention strategies in cirrhotic patients. AEG1 has been identified as a downstream molecule in this pathway, and could also act as a novel molecular target in this setting [51]. Additionally, aberrant activation of the lymphotoxin pathway in mice livers recapitulates the stages of fibrosis and inflammation that precedes human liver cancer, providing a novel family of potential therapeutic targets [52,53].
Pathways involved in neoangiogenesis
High vascularization is a hallmark of human HCC. Tumor cells release pro-angiogenic factors in response to hypoxic conditions and nutrient deprivation and thus activate endothelial cells. Multiple preclinical studies have revealed vascular endothelial growth factor (VEGF), and fibroblast growth factors (FGFs) as major drivers underlying pro-angiogenic signals in human HCC. VEGF is considered the most potent angiogenic factor in HCC, and it is often found over-expressed in tumor specimens, as well as its receptors VEGFR-1 and VEGFR-2. Moreover, high VEGF expression levels have been consistently associated with a more aggressive disease [54], and VEGF serum levels have also been identified as a predictor of poor prognosis after resection [55]. The FGF receptor family includes 4 transmembrane receptors (FGF1-4) and 1 soluble receptor (FGF5) and at least 23 known ligands. There is evidence that links FGF signaling dysregulation and malignant transformation of several cancers. In HCC, clinical studies have shown over-expression of FGFs in human samples and correlation with tumor angiogenesis [56]. Several studies have reported crosstalk between FGF and VEGF signaling, indicating a possible synergistic effect of both cascades promoting angiogenesis [57]. Besides these pathways, angiopoetins (Ang) have also been involved in normal and aberrant vascular formation through its interaction with the receptor Tie-2. Despite recent controversial data, Ang-2 has been described as a promoter of tumor angiogenesis in HCC, particularly in the presence of VEGF [58]. Finally, experimental models have suggested a role for another pro-angiogenic cascade, platelet derived growth factor (PDGF) signaling, in HCC development [59]. Indeed, PDGF receptor C transgenic mice developed fibrosis and HCC with long latency [60]. Results of the recently published phase III clinical trial with sorafenib (i.e. B-Raf, VEGFR and PDGFR inhibitor) further suggest an important role of anti-angiogenic agents in the therapeutic management of HCC [4].
The P53 tumor suppressor
The P53 tumor suppressor has been found mutated in more than 50% of all human tumors. Owing to its role in preserving genetic stability, P53 was designated as the guardian of the genome. Upon DNA damage inducing cellular stresses, P53 becomes activated and prevents further damage and oncogenic transformation by either triggering apoptosis or by inducing a transient cell cycle arrest to allow DNA repair [61]. The p53 protein is a tetrameric transcription factor, and, in addition to its roles in apoptosis- and cell cycle regulation, P53 was shown to impact cellular differentiation, developmental processes and senescence. Over the past couple of years it was shown that P53 plays a major role in hepatocarcinogenesis. Strikingly, there is a high variability regarding the frequency of P53 mutations and its mutational spectrum in different geographical areas. In areas with high Aflatoxin B1 food contamination (sub-Saharan Africa and China) there is a high percentage of HCCs revealing a recurrent point mutation at the third position of codon 249 resulting in a G:C to T:A transversion [62]. Interestingly, it was shown that the 249Ser mutation is an early event in AFB1-induced hepatocarcinogenesis, with a high frequency of 249Ser mutations already being found in non tumoral regions of livers with chronic AFB1 exposure [62]. In strong contrast, P53 mutations are believed to represent a late, progression-associated event in hepatocarcinogenesis in non-AFB1 related HCCs. For example, different P53 mutations have been found in nodules in HCCs leading to disease progression, and overall P53 mutations are preferentially found in less differentiated HCCs. One major risk factor for the development of hepatocellular carcinoma is chronic infection with hepatotropic viruses like hepatitis B and C. Different viral proteins have been shown to impact hepatocarcinogenesis in experimental systems. In the past couple of years, a lot of attention was paid to the hepatitis B virus X protein (HBx), one of the most frequently integrated viral genes in the host genome [63,64]. Besides many other functions, HBx was shown to bind P53 and to block P53 sequence specific DNA-binding and P53 dependent transcription, being ultimately capable of blocking P53-mediated apoptosis.
Cancer genome screening and validation strategies
Current efforts in cancer gene discovery mostly rely on genomic approaches alone. Recent advances in genome scanning technologies have enabled to examine cancer genomes at a high resolution, thus making it feasible to catalogue every gene whose mutation occurs in human cancer [65]. Regions of copy number alteration can be identified by high-resolution array-based comparative genomic hybridization (CGH) or SNP arrays. Regions of chromosomal amplification mostly harbor oncogenes whereas deleted regions contain tumor suppressor genes [66]. In addition, high-throughput sequencing technologies are available for detection of somatic point mutations selected during tumor evolution [67,68]. However, such approaches are expensive and yield candidates based on statistical criteria only. The situation is even more complicated because due to genomic instability, gene linkage and spontaneous mutagenesis, cancers also contain many passenger mutations that are not contributing to the tumor phenotype. Moreover, it is difficult to pinpoint relevant tumor suppressors in large deletions, as some genes are haploid sufficient tumor suppressors, so that loss of even one allele can promote tumorigenesis, even without a corresponding mutation in the remaining wild-type allele. In conclusion, candidate genes identified through genomic approaches require stringent functional validation before expensive drug development efforts can be justified. In this regard, functional characterization of cancer genes is often a tedious process, and it is not always obvious which assays will reveal the putative oncogenic activity of a previously uncharacterized gene. Furthermore, although cell culture systems are fast and tractable, they do not recapitulate important factors like the tumor microenvironment, and so, they do not survey all relevant gene activities.
For proving new candidate cancer genes it has become gold standard to generate transgenic and knockout mice, and to study the respective genes in a relevant context in vivo [69]. Importantly, as such genetically engineered mouse models (GEMM) allow to generate a detailed roadmap of the route taken by the tumor cell to reach its destination, cancer mouse models are suitable for modeling intermediate stages of tumor formation, which is essential for a complete understanding of the disease and for the development of new treatment strategies. This is in strong contrast to the traditional studies of human cancers, which only provide a snapshot of the final result. GEMM also play an important role in preclinical drug testing, as it is becoming more evident that subcutaneously grown tumors in immunodeficient mice, which were derived from cultured tumor cell lines, may not predict the treatment responses of human tumors, and may hence contribute to the high numbers of ineffective compounds entering clinical trials [70].
Over the last years, GEMM have provided elegant and powerful approaches to validate potential oncogene and tumor suppressor genes in HCC (Table 1A) [12, 60, 71-90]. Currently, mouse models of liver cancer are based mainly on classic transgenic approaches, tetracycline-regulated transgene expression or chemically induced carcinogenesis (thoroughly reviewed elsewhere [91]). These models have provided important insights into the molecular mechanisms of hepatocarcinogenesis, although, they have several limitations. First, the use of tissue-specific promoters generates a bias towards certain epithelial and may thus alter the cell of origin. Furthermore, the homogeneous expression of an oncogene (or deletion of a tumor suppressor gene) in all cells within a tissue creates a field effect such that all cells have altered gene expression, a situation that clearly does not mimic the situation of spontaneous tumorigenesis in humans. Second, it is very cost and time-intensive to generate germ-line transgenic and knockout animals, and production of compound mutant animals can involve complicated intercrossing strategies that are extremely slow. Third, germ-line transmission of transgenes or targeted knock-out alleles in some cases produce prenatal abnormalities resulting in embryonic lethality or developmental compensation in the resulting tissue.
Table 1.
Relevant oncogenes and tumor suppressor genes identified through (A) selective dysregulation in animal models, or through (B) functional oncogenomic approaches.
| Gene name | Oncogene / Tumor suppressor gene | Biological activity/Pathway | Reference |
|---|---|---|---|
| A) Long established HCC cancer genes which have been functionally validated in vivo | |||
|
| |||
| MYC | Oncogene | Proliferation control | [12, 71-73] |
| EGF | Oncogene | EGFR signaling (mitogenic signaling) | [78] |
| TGFA | Oncogene | EGFR signaling (mitogenic signaling) | [12, 77] |
| CTNNB1 | Oncogene | Wnt-signaling | [75, 79] |
| APC | Tumor suppressor gene | Wnt-signaling | [74, 80] |
| PTEN | Tumor suppressor gene | PI3K/Akt/mTOR signaling | [76, 80] |
| AKT | Oncogene | PI3K/Akt/mTOR signaling | [73] |
| HGF* | Oncogene | Growth factor | [81, 82] |
| MET | Oncogene | Growth factor receptor | [83, 84] |
| PDGFR | Oncogene | Growth factor receptor | [60] |
| RAS (NRAS; HRAS) | Oncogene | Ras/MAPK signaling | [71, 73, 75, 85] |
| P53 | Tumor suppressor gene | Stress response, cell cycle inhibition | [73, 86] |
| E2F1 | Oncogene | Cell Cycle | [87] |
| CCND1 | Oncogene | Cell Cycle | [88, 89] |
| Telomerase | Oncogene | Cell senescence | [90] |
|
| |||
| B) HCC cancer genes recently identified and validated through integrative functional genomics | |||
|
| |||
| YAP | Oncogene | Hippo Signaling pathway | [73, 97] |
| cIAP1 (BIRC2) | Oncogene | Regulation of Cell death | [73] |
| XPO4 | Tumor suppressor gene | Nuclear transport | [80] |
| SET* | Tumor suppressor gene/oncogene | Histone chaperone | [80] |
| NRSN2 | Tumor suppressor gene | Largely unknown | [80] |
| GJD4 (CX40.1) | Tumor suppressor gene | Gap junction protein | [80] |
| DDX20 | Tumor suppressor gene | RNA helicase | [80] |
| FGF6 | Tumor suppressor gene | Fibroblast growth factor family | [80] |
| FSTL5 | Tumor suppressor gene | Follistatin like protein | [80] |
| ZBBX [FLJ23049] | Tumor suppressor gene | Largely unknown | [80] |
| GLO1 | Tumor suppressor gene | Metabolism | [80] |
| BTBD9 | Tumor suppressor gene | Largely unknown | [80] |
| EIF5A2 | Oncogene | Translation control, mRNA-stability | [80] |
| UBE2H** | Oncogene | Ubiquitylation | [9] |
| EGFR | Oncogene | EGFR signaling [mitogenic signaling] | [9] |
| AEG1 | Oncogene | WNT and NF-Kß signaling | [51] |
| VEGF | Oncogene | Angiogenesis | [6] |
Dual role, context-dependent activity has been suggested.
Only in vitro validation.
A strategy that addresses some of the problems with germline transgenic mice is the use of so called chimeric mouse models, which are based on the genetic manipulation and retransplantation of stem or progenitor cells. Such models were initially developed for studying hematopoietic malignancies and have proven very successful to get new insights into mechanisms of tumor initiation, progression and treatment response in blood cancer [92-95]. More recently such models were also developed for the study of HCC [73,96]; these mosaic HCC mouse models are based on the ex-vivo manipulation of liver progenitor cells followed by the seeding of these cells into normal recipients. They allow studying compound genetic lesions in HCC at a fraction of the time that would be needed when using standard transgenic or knockout approaches.
Integrative oncogenomic approaches for accelerated cancer gene discovery in HCC
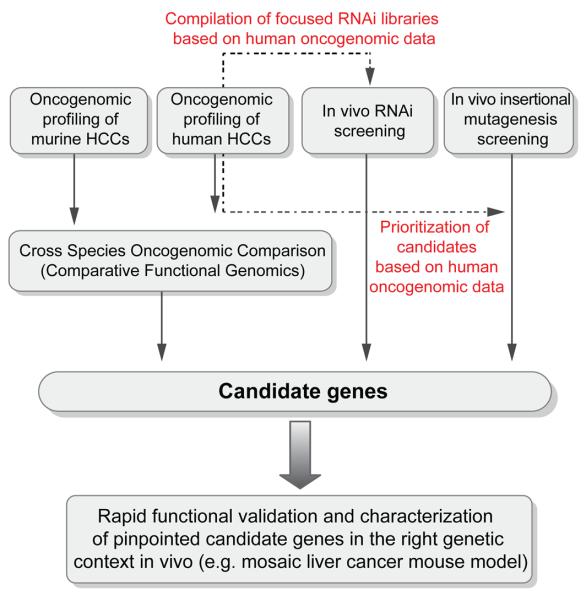
To speed up the process of cancer gene discovery in HCC, recent studies took advantage of integrative approaches, where oncogenomic analyses of human HCC were coupled with screening approaches in innovative mosaic cancer mouse models. The most relevant validated examples are summarized in Table 1B [9, 51, 72, 73, 97]. For example, high resolution array-CGH profiling of progenitor cell derived murine HCCs of the genotype c-MYC; P53−/−, revealed that these tumors spontaneously acquire genomic amplification of the 9qA1 chromosomal region, which is syntenic to the human 11q22 region. Interestingly, this region has been found amplified in different human tumors. Delineation of the minimal region of overlap between human and mouse HCCs and subsequent analyses of expression levels for all genes embedded in the overlapping region, pinpointed cIAP1 and YAP as potential driver genes of the 11q22 amplicon in human HCC. Interestingly, when both genes were functionally tested in the mouse model, they did not only accelerate hepatocarcinogenesis individually, but also showed a strong collaboration to accelerate the development of liver tumors. Therefore, these genes were validated as new oncogenes in HCC and were found to be co-driver genes of the 11q22 amplicon [98]. Importantly, a subsequent study reported the expression of the YAP oncogene as an independent prognostic factor in human HCC [98].
The discovery of RNA-interference has revolutionized “loss of function genetics”. RNAi-based delivery strategies have shown antitumoral activity in animal models of HCC [99], and dysregulation of miRNA have also proved high prognostic predictive value in this disease [100]. It was recently shown that stable RNAi can be efficiently used in mosaic cancer mouse models [73, 80, 101]. For example, the use of stable RNAi in murine hepatocarcinomas that bore the cIAP1 and YAP embedding 9qA1 amplicon quickly allowed the validation of these genes as promising therapeutic targets in HCC [73]. As microRNA-based shRNAs can be driven by polymerase II promoters, available systems for tetracycline regulated gene expression can be combined with this technology. This allows to reversibly regulate gene expression in vivo. The use of reversible RNAi against the P53 tumor suppressor has recently enabled the identification of P53 loss as a major requirement for the maintenance of murine liver carcinomas [89]. Similar RNAi-based approaches were recently used to functionally validate DLC-1 as a bona fide tumor suppressor included in 8p, a chromosomal region that is found deleted in up to 50 % of human HCC [102].
Importantly, RNAi technology cannot only be used on a single gene level, but genome wide collections of siRNAs, shRNAs and shRNAmir are available and have been successfully used for the identification of unbiased and comprehensive collections of new gene targets in different biological systems [80]. Importantly, the feasibility of performing multiplex RNAi screens in vivo was recently demonstrated [80]. In this study, an oncogenomics-based shRNAmir library was generated by compiling shRNAs against all genes that were found embedded in focal deletions [<5 MB] of 98 human HCCs. A total of 362 candidate tumor suppressor genes were found and corresponding mouse orthologs for 301 of them were identified. More than 630 microRNAs against these genes were available in the Codex shRNAmir library [http://codex.cshl.edu/scripts/newmain.pl] and these hairpins were screened in the above described mosaic liver cancer mouse model for their ability to accelerate tumorigenicity of immortalized liver progenitor cells of the background MYC; P53 −/−. This integrated approach proved highly effective, resulting in the identification and functional validation of 13 new tumor suppressor genes.
The top-scoring candidate tumor suppressor from the screen, exportin 4, mediates the nuclear export of SMAD3, EIF5A1, and EIF5A2. XPO4 loss seems to be protumorigenic by promoting the nuclear accumulation of its key substrates. Interestingly, XPO4 deletions are relatively common, thus suggesting that this may be an important mechanism of oncogenesis. It was observed that suppression of XPO4 stimulates TGF-β signaling [80], which can promote invasion and metastasis in late stage liver cancer. Similarly, EIF5A2 over-expression occurs in many tumor types [103] and it could be demonstrated that XPO4 loss enhances proliferation through EIF5A2, which is per se oncogenic in mice [80]. Importantly, the XPO4-EIF5A2 signaling circuit appears relevant in HCC and other tumor types.
It is noteworthy that most of the identified genes have never been linked to cancer before. Examples for identified genes are an FGF (FGF6), an RNA helicase (DDX20/GEMIN3), a metabolic enzyme (GLO1), and GJD4 (CX40.1), a gap junction protein. Functional validation assays show that apparently all these genes act as tumor suppressors in vivo. Some of the genes, for example FSTL5, NRSN2 (C20ORF98), and ZBBX (FLJ23049), are largely uncharacterized, however, based on the functional data obtained from the mouse model and on the notion that all of them were found somatically altered in human cancer, these genes very likely represent important tumor suppressor genes in HCC. It is obvious that more work will be required to understand how each of these genes suppresses tumorigenesis, and, given the unexpected nature of each gene, such studies may uncover new pathways or principles relevant to human HCC. Some of the identified genes point towards new strategies for cancer therapy. For example, FGF6 and FSTL5 encode secreted proteins whose systemic administration might restore tumor suppressor function and serve as new biological anticancer therapies.
The described results establish the feasibility of performing functional in vivo RNAi screens and illustrate how combining cancer genomics, RNAi and mosaic mouse models can facilitate the functional annotation of the cancer genome (Fig. 2). Such integrative approaches may complement existing large-scale approaches like the human cancer genome project. Another appealing example for in vivo screening approach to identify new cancer genes in HCC was recently demonstrated by Keng et al. [9]. In their study the authors used a liver directed insertional mutagenesis screen, which was accomplished by hepatocyte-specific expression of a mutagenic transposon to screen for new cancer genes in HCC. This screen was performed either in a wild type- or in a sensitized background with a mutant p53 allele. In both backgrounds, preneoplastic nodules could be detected starting 160 days after liver-specific activation of the mutagenic transposon. The vast majority of animals only contained preneoplastic nodules or hepatic adenomas, but in some animals [> 440 days of age] also full-blown liver carcinomas were found. Using pyrosequencing, the authors identified common insertion sites in the genomic DNA from enucleated preneoplastic foci. A total of 19 common insertion sites were identified and two affected genes were chosen for functional validation, UBE2H and a truncated EGFR. While definite experimental evidence that these genes cause HCC is still missing, the authors were able to show that over-expression of the candidate oncogene UBE2H accelerates proliferation of murine hepatoma cell line in vitro. The truncated EGFR was functionally tested resulting in the development of preneoplastic lesions in repopulated mouse livers. While the long latency until tumor development and the low penetrance to full blown HCC certainly represent limitations of the model, it is yet noteworthy that some of the 19 candidate HCC cancer genes could be found altered in recently published datasets of human HCC. Therefore, it will be of high interest to subject these candidate genes to thorough functional validation in different HCC mouse models to ultimately probe their contribution to the development of HCC.
Figure 2. Integrative oncogenomic strategies for accelerated cancer gene discovery in hepatocellular carcinoma.
Schematic representation of functional oncogenomics based strategies for the discovery of candidate genes involved in HCC pathogenesis.
With the advent of novel high-throughput technologies, such as deep sequencing and integrative genomic analysis, and the consolidation of sophisticated animal models, such as mosaic cancer mouse models or the use of transposons for mutagenic screen, we are at the dawn of a new era in cancer gene discovery. These technologies are identifying novel oncogenes and tumor suppressor genes, which along with the already known drivers obtained from classical GEMM (Table 1) will provide a more comprehensive picture of the key molecular alterations of liver cancer.
Acknowledgments
Grant Support: Lars Zender is supported by grants from the German Research Foundation [Emmy Noether Programme] and grants from the Helmholtz Association of German Research Centers (“Helmholtz Impuls und Vernetzungsfond”). Augusto Villanueva is supported by a EASL-Sheila Sherlock Fellowship. Josep Llovet is supported by grants from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (1R01DK076986-01), The Samuel Waxman Cancer Research Foundation and the Spanish National Health Institute (SAF-2007-61898).
ABBREVIATIONS
- Ang
angiopoietin
- c-IAP
cellular-inhibitor of apoptosis
- CGH
comparative genomic hybridization
- CTNNB1
catenin-beta 1
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FGF
fibroblast growth factor
- HGF
hepatocyte growth factor
- HH
hedgehog
- IGF
insulin-like growth factor
- IGFBPs
insulin-like growth factor–binding proteins
- IGF-IR
insulin-like growth factor receptor
- IL-6
interleukin-6
- MAPK
mitogen-activated protein kinase
- MTORC
mTOR complex
- PDGFR
platelet derived growth factor receptor
- PTEN
phosphatase and tensin homolog
- SNP
single nucleotide polymorphism
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- TGF-β
transforming growth factor-beta
- VEGF
vascular endothelial growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Llovet J, Burroughs A, Bruix J. Hepatocellular carcinoma. The Lancet. 2003;362(9399):1907–17. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27(1):55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- 3.Llovet J, Di Bisceglie AM, Bruix J, Kramer BS, Lencioni R, Zhu AX, et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(10):698–711. doi: 10.1093/jnci/djn134. [DOI] [PubMed] [Google Scholar]
- 4.Llovet J, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135(6):1972–83. doi: 10.1053/j.gastro.2008.08.008. 83 e1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68(16):6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe KK, Lemoine A, Finkelstein DM, Kawasaki H, Fujii T, Chung RT, et al. Epidermal growth factor gene functional polymorphism and the risk of hepatocellular carcinoma in patients with cirrhosis. JAMA. 2008;299(1):53–60. doi: 10.1001/jama.2007.65. [DOI] [PubMed] [Google Scholar]
- 8.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene Expression in Fixed Tissues and Outcome in Hepatocellular Carcinoma. N Engl J Med. 2008;359(19):1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27(3):264–74. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffer E, Housset C, Cacheux W, Wendum D, Desbois-Mouthon C, Rey C, et al. Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology. 2005;41(2):307–14. doi: 10.1002/hep.20538. [DOI] [PubMed] [Google Scholar]
- 11.Breuhahn K, Longerich T, Schirmacher P. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene. 2006;25(27):3787–800. doi: 10.1038/sj.onc.1209556. [DOI] [PubMed] [Google Scholar]
- 12.Murakami H, Sanderson ND, Nagy P, Marino PA, Merlino G, Thorgeirsson SS. Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis 1. Cancer Res. 1993;53(8):1719–23. [PubMed] [Google Scholar]
- 13.Philip P. Phase II Study of Erlotinib (OSI-774) in Patients With Advanced Hepatocellular Cancer. Journal of Clinical Oncology. 2005;23(27):6657–63. doi: 10.1200/JCO.2005.14.696. [DOI] [PubMed] [Google Scholar]
- 14.Thomas M, Chadha R, Glover K, Wang X, Morris J, Brown T, et al. Phase 2 study of erlotinib in patients with unresectable hepatocellular carcinoma. Cancer. 2007;110(5):1059–67. doi: 10.1002/cncr.22886. [DOI] [PubMed] [Google Scholar]
- 15.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8(12):915–28. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 16.Sachdev D, Yee D. Disrupting insulin-like growth factor signaling as a potential cancer therapy. MolCancer Ther. 2007;6(1):1–12. doi: 10.1158/1535-7163.MCT-06-0080. [DOI] [PubMed] [Google Scholar]
- 17.Tang SH, Yang DH, Huang W, Zhou HK, Lu XH, Ye G. Hypomethylated P4 promoter induces expression of the insulin-like growth factor-II gene in hepatocellular carcinoma in a Chinese population. Clin Cancer Res. 2006;12:4171–7. doi: 10.1158/1078-0432.CCR-05-2261. [DOI] [PubMed] [Google Scholar]
- 18.De Souza AT, Hankins GR, Washington MK, Orton TC, Jirtle RL. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat Genet. 1995;11(4):447–9. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 19.Hanafusa T, Yumoto Y, Nouso K, Nakatsukasa H, Onishi T, Fujikawa T, et al. Reduced expression of insulin-like growth factor binding protein-3 and its promoter hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2002;176(2):149–58. doi: 10.1016/s0304-3835(01)00736-4. [DOI] [PubMed] [Google Scholar]
- 20.Tovar V, Alsinet C, Villanueva A, Hoshida Y, Chiang D, Sole M, et al. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol. 2009 doi: 10.1016/j.jhep.2010.01.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SO, Park JG, Lee YI. Increased expression of the insulin-like growth factor I (IGF-I) receptor gene in hepatocellular carcinoma cell lines: implications of IGF-I receptor gene activation by hepatitis B virus X gene product. Cancer Res. 1996;56(16):3831–6. [PubMed] [Google Scholar]
- 22.Takayama H, Larochelle WJ, Sharp R, Otsuka T, Kriebel P, Anver M, et al. Diverse tumorigenesis associated with aberrant development in mice overexpressing hepatocyte growth factor/scatter factor. PNAS. 1997;94(2):701–6. doi: 10.1073/pnas.94.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67(5):2081–8. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
- 24.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7(6):504–16. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 25.Ueki T, Fujimoto J, Suzuki T, Yamamoto H, Okamoto E. Expression of hepatocyte growth factor and its receptor c-met proto-oncogene in hepatocellular carcinoma. Hepatology. 1997;25(4):862–6. doi: 10.1002/hep.510250413. [DOI] [PubMed] [Google Scholar]
- 26.Kaposi-Novak P. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006 Jun 1;116(6):1582–95. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvi A, Arici B, Portolani N, Giulini SM, De PG, Barlati S. In vitro c-met inhibition by antisense RNA and plasmid-based RNAi down-modulates migration and invasion of hepatocellular carcinoma cells. Int J Oncol. 2007;31(2):451–60. [PubMed] [Google Scholar]
- 28.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–2. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 29.Karreth FA, Tuveson DA. Modelling oncogenic Ras/Raf signalling in the mouse. Curr Opin Genet Dev. 2009;19(1):4–11. doi: 10.1016/j.gde.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvisi D, Ladu S, Gorden A, Farina M, Conner E, Lee J, et al. Ubiquitous Activation of Ras and Jak/Stat Pathways in Human HCC. Gastroenterology. 2006;130(4):1117–28. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Newell P, Toffanin S, Villanueva A, Chiang DY, Minguez B, Cabellos L, et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J Hepatol. 2009;51(4):725–33. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103(2):307–12. doi: 10.1002/cncr.20774. [DOI] [PubMed] [Google Scholar]
- 33.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17(6):596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6(9):729–34. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 35.Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15(2):148–59. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treiber G. mTOR inhibitors for hepatocellular cancer: a forward-moving target. Expert Rev Anticancer Ther. 2009;9(2):247–61. doi: 10.1586/14737140.9.2.247. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z. Significance of CD90+ Cancer Stem Cells in Human Liver Cancer. Cancer Cell. 2008;13(2):153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8(6):462–72. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 39.Zaret K. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9(5):329–40. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- 40.Zaret K, Grompe M. Generation and Regeneration of Cells of the Liver and Pancreas. Science. 2008;322(5907):1490–4. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson MD, Monga SP. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45(5):1298–305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 42.Boyault S, Rickman DS, de RA, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 43.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–20. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 44.Sicklick JK, Li YX, Jayaraman A, Kannangai R, Qi Y, Vivekanandan P, et al. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis. 2006;27(4):748–57. doi: 10.1093/carcin/bgi292. [DOI] [PubMed] [Google Scholar]
- 45.Tada M, Kanai F, Tanaka Y, Tateishi K, Ohta M, Asaoka Y, et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin Cancer Res. 2008;14(12):3768–76. doi: 10.1158/1078-0432.CCR-07-1181. [DOI] [PubMed] [Google Scholar]
- 46.Lok AS, Seeff LB, Morgan TR, Di Bisceglie AM, Sterling RK, Curto TM, et al. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136(1):138–48. doi: 10.1053/j.gastro.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naugler W, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy A, et al. Gender Disparity in Liver Cancer Due to Sex Differences in MyD88-Dependent IL-6 Production. Science. 2007;317(5834):121–4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 48.Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28(7):961–72. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47(6):2059–67. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elsharkawy AM, Mann DA. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46(2):590–7. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- 51.Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119(3):465–77. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haybaeck J, Zeller N, Wolf MK, Weber A, Wagner U, Kurrer MO, et al. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16(4):272–3. doi: 10.1016/j.ccr.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villanueva A, Savic R, Llovet JM. Lymphotoxins: Novel targets for HCC. Cancer Cell. 2009;16:272–273. doi: 10.1016/j.ccr.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li XM, Tang ZY, Zhou G, Lui YK, Ye SL. Significance of vascular endothelial growth factor mRNA expression in invasion and metastasis of hepatocellular carcinoma. J Exp Clin Cancer Res. 1998;17(1):13–7. [PubMed] [Google Scholar]
- 55.Poon RT, Ho JW, Tong CS, Lau C, Ng IO, Fan ST. Prognostic significance of serum vascular endothelial growth factor and endostatin in patients with hepatocellular carcinoma. Br J Surg. 2004;91(10):1354–60. doi: 10.1002/bjs.4594. [DOI] [PubMed] [Google Scholar]
- 56.Imura S, Miyake H, Izumi K, Tashiro S, Uehara H. Correlation of vascular endothelial cell proliferation with microvessel density and expression of vascular endothelial growth factor and basic fibroblast growth factor in hepatocellular carcinoma. J Med Invest. 2004;51(3-4):202–9. doi: 10.2152/jmi.51.202. [DOI] [PubMed] [Google Scholar]
- 57.Yoshiji H, Kuriyama S, Yoshii J, Ikenaka Y, Noguchi R, Hicklin DJ, et al. Synergistic effect of basic fibroblast growth factor and vascular endothelial growth factor in murine hepatocellular carcinoma. Hepatology. 2002;35(4):834–42. doi: 10.1053/jhep.2002.32541. [DOI] [PubMed] [Google Scholar]
- 58.Mitsuhashi N, Shimizu H, Ohtsuka M, Wakabayashi Y, Ito H, Kimura F, et al. Angiopoietins and Tie-2 expression in angiogenesis and proliferation of human hepatocellular carcinoma. Hepatology. 2003;37(5):1105–13. doi: 10.1053/jhep.2003.50204. [DOI] [PubMed] [Google Scholar]
- 59.Campbell JS, Johnson MM, Bauer RL, Hudkins KL, Gilbertson DG, Riehle KJ, et al. Targeting stromal cells for the treatment of platelet-derived growth factor C-induced hepatocellular carcinogenesis. Differentiation. 2007;75(9):843–52. doi: 10.1111/j.1432-0436.2007.00235.x. [DOI] [PubMed] [Google Scholar]
- 60.Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. PNAS. 2005 Mar;1102(9):3389–94. doi: 10.1073/pnas.0409722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 62.Aguilar F, Harris CC, Sun T, Hollstein M, Cerutti P. Geographic variation of p53 mutational profile in nonmalignant human liver. Science. 1994;264(5163):1317–9. doi: 10.1126/science.8191284. [DOI] [PubMed] [Google Scholar]
- 63.Hofseth LJ, Saito S, Hussain SP, Espey MG, Miranda KM, Araki Y, et al. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. PNAS. 2003;100(1):143–8. doi: 10.1073/pnas.0237083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21(3):201–16. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 65.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29(6):1087–91. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452(7187):553–63. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 69.Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108(2):135–44. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 70.Sharpless N, Depinho R. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–54. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 71.Sandgren EP, Quaife CJ, Pinkert CA, Palmiter RD, Brinster RL. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989;4(6):715–24. [PubMed] [Google Scholar]
- 72.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–7. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 73.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125(7):1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, et al. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. PNAS. 2004;101(49):17216–21. doi: 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harada N, Oshima H, Katoh M, Tamai Y, Oshima M, Taketo MM. Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 2004;64(1):48–54. doi: 10.1158/0008-5472.can-03-2123. [DOI] [PubMed] [Google Scholar]
- 76.Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113(12):1774–83. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH, Merlino GT. TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990;61(6):1137–46. doi: 10.1016/0092-8674(90)90076-q. [DOI] [PubMed] [Google Scholar]
- 78.Tonjes RR, Lohler J, O'Sullivan JF, Kay GF, Schmidt GH, Dalemans W, et al. Autocrine mitogen IgEGF cooperates with c-myc or with the Hcs locus during hepatocarcinogenesis in transgenic mice. Oncogene. 1995;10(4):765–8. [PubMed] [Google Scholar]
- 79.Tward AD, Jones KD, Yant S, Cheung ST, Fan ST, Chen X, et al. Distinct pathways of genomic progression to benign and malignant tumors of the liver. PNAS. 2007;104(37):14771–6. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–64. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horiguchi N, Takayama H, Toyoda M, Otsuka T, Fukusato T, Merlino G, et al. Hepatocyte growth factor promotes hepatocarcinogenesis through c-Met autocrine activation and enhanced angiogenesis in transgenic mice treated with diethylnitrosamine. Oncogene. 2002;21(12):1791–9. doi: 10.1038/sj.onc.1205248. [DOI] [PubMed] [Google Scholar]
- 82.Sakata H, Takayama H, Sharp R, Rubin JS, Merlino G, LaRochelle WJ. Hepatocyte growth factor/scatter factor overexpression induces growth, abnormal development, and tumor formation in transgenic mouse livers. Cell Growth Differ. 1996;7(11):1513–23. [PubMed] [Google Scholar]
- 83.Tward AD, Jones KD, Yant S, Kay MA, Wang R, Bishop JM. Genomic progression in mouse models for liver tumors. Cold Spring Harb Symp Quant Biol. 2005;70:217–24. doi: 10.1101/sqb.2005.70.058. [DOI] [PubMed] [Google Scholar]
- 84.Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153(5):1023–34. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. PNAS. 2005;102(47):17059–64. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klocke R, Bartels T, Jennings G, Brand K, Halter R, Strauss M, et al. Lack of p53 accelerates hepatocarcinogenesis in transgenic mice constitutively overexpressing c-myc in the liver. FASEB J. 2001;15(8):1404–6. doi: 10.1096/fj.00-0487fje. [DOI] [PubMed] [Google Scholar]
- 87.Conner EA, Lemmer ER, Omori M, Wirth PJ, Factor VM, Thorgeirsson SS. Dual functions of E2F-1 in a transgenic mouse model of liver carcinogenesis. Oncogene. 2000;19(44):5054–62. doi: 10.1038/sj.onc.1203885. [DOI] [PubMed] [Google Scholar]
- 88.Deane NG, Lee H, Hamaamen J, Ruley A, Washington MK, LaFleur B, et al. Enhanced tumor formation in cyclin D1 x transforming growth factor beta1 double transgenic mice with characterization by magnetic resonance imaging. Cancer Res. 2004;64(4):1315–22. doi: 10.1158/0008-5472.can-03-1772. [DOI] [PubMed] [Google Scholar]
- 89.Deane NG, Parker MA, Aramandla R, Diehl L, Lee WJ, Washington MK, et al. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 2001;61(14):5389–95. [PubMed] [Google Scholar]
- 90.Farazi P, Depinho R. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006 Sep 1;6(9):674–87. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 91.Newell P, Villanueva A, Friedman SL, Koike K, Llovet JM. Experimental models of hepatocellular carcinoma. J Hepatol. 2008;48(5):858–79. doi: 10.1016/j.jhep.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436(7052):807–11. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92(10):3780–92. [PubMed] [Google Scholar]
- 94.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1(3):289–98. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 95.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109(3):335–46. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 96.Zender L, Xue W, Cordon-Cardo C, Hannon GJ, Lucito R, Powers S, et al. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–61. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Cur rBiol. 2007;17(23):2054–60. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 98.Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009 doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361(15):1437–47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, et al. An epiallelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33(3):396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 102.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22(11):1439–44. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clement PM, Johansson HE, Wolff EC, Park MH. Differential expression of eIF5A-1 and eIF5A-2 in human cancer cells. FEBS J. 2006;273(6):1102–14. doi: 10.1111/j.1742-4658.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]