Abstract
Experimental evidence suggests that half or more of the mammalian transcriptome consists of noncoding RNA. Noncoding RNAs are divided into short noncoding RNAs (including microRNAs) and long noncoding RNAs (lncRNAs). We defined complementary DNAs (cDNAs) lacking any positive-strand open reading frames (ORFs) longer than 30 amino acids, as well as cDNAs lacking any evidence of interspecies conservation of their longer-than-30-amino acid ORFs, as noncoding. We have identified 5446 lncRNA genes in the human genome from ∼24,000 full-length cDNAs, using our new ORF-prediction pipeline. We combined them nonredundantly with lncRNAs from four published sources to derive 6736 lncRNA genes. In an effort to distinguish standalone and antisense lncRNA genes from database artifacts, we stratified our catalog of lncRNAs according to the distance between each lncRNA gene candidate and its nearest known protein-coding gene. We concurrently examined the protein-coding capacity of known genes overlapping with lncRNAs. Remarkably, 62% of known genes with “hypothetical protein” names actually lacked protein-coding capacity. This study has greatly expanded the known human lncRNA catalog, increased its accuracy through manual annotation of cDNA-to-genome alignments, and revealed that a large set of hypothetical-protein genes in GenBank lacks protein-coding capacity. In addition, we have developed, independently of existing NCBI tools, command-line programs with high-throughput ORF-finding and BLASTP-parsing functionality, suitable for future automated assessments of protein-coding capacity of novel transcripts.
Keywords: lncRNA, noncoding RNA, transcriptome, hypothetical protein, CPC, ORF-Predictor
INTRODUCTION
Noncoding-RNA (ncRNA) genes are genes that do not encode proteins. They were initially thought to be limited to ribosomal, transfer, spliceosomal, and other essential RNAs, but have been shown to be far more diverse. ncRNAs can be divided into short ncRNAs (which include, but are not limited to, microRNAs) and long ncRNAs (lncRNAs). To date, several hundred human microRNAs have been identified (Griffiths-Jones et al. 2008). The importance of microRNAs as key post-transcriptional repressors is universally acknowledged. Considerably less is known about lncRNA genes, which are an order of magnitude more prevalent than microRNAs (Carninci and Hayashizaki 2007). Since they comprise over half of the transcriptional units (TUs) in mammalian genomes (Carninci et al. 2005), lncRNAs represent a major unexplored component of genomes of great potential biological importance. lncRNAs, similar to mRNAs, are RNA polymerase II-promoted, polyadenylated, and often alternatively spliced (Ginger et al. 2006; Mehler and Mattick 2007). Numerous lncRNA-encoding conserved loci, transcribed in mammals, possess epigenetic signatures similar to those of protein-coding genes, while also presenting strong evidence of regulation—including cis-regulation—of transcription factors and an involvement in transcriptional control that requires further validation (Johnson et al. 2009). lncRNAs are unlikely to simply represent transcriptional noise because they are expressed in a tissue- and developmental-specific manner, and their sequence displays phylogenetic conservation consistent with negative evolutionary selection, which indicates functional constraint (Ponjavic et al. 2007). Known lncRNA roles encompass endogenous antisense mechanisms, transcription factor (TF) nucleocytoplasmic translocation control, coactivation and corepression of specific TFs, and epigenetic regulation (Martianov et al. 2007; Rinn et al. 2007). The spatiotemporally restricted expression patterns of hundreds of other lncRNAs suggest that many other functional roles remain to be discovered (Pollard et al. 2006; Mercer et al. 2008).
In light of the rapidly growing interest in lncRNAs, it is important to develop high-confidence catalogs of noncoding RNAs for experimental and bioinformatic study. In particular, while recent studies have focused on cataloging mouse lncRNAs (Dinger et al. 2008a; Ponjavic et al. 2009), there is no available comprehensive catalog of human lncRNAs. To address this need, we have curated and classified a data set of 6736 human long noncoding RNAs from a variety of sources.
RESULTS
Our ORF-Predictor and BLASTP pipeline identifies 5446 human lncRNA genes largely unique relative to public lncRNA collections
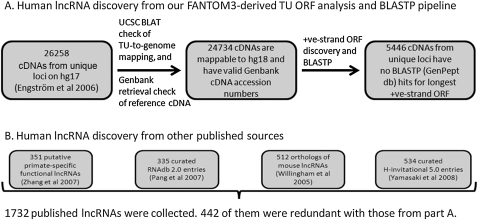
A consistent and accurate definition of protein-coding capacity of transcripts, along with the elimination of transcripts that are redundant relative to other transcripts or ambiguous with respect to the genomic location of their source genes, is an essential foundation for any genome-wide effort to catalog lncRNA genes. Therefore, we developed an open reading frame (ORF)-Predictor/BLASTP pipeline that first automatically delineated all ORFs in all three positive-strand frames for each complementary DNA (cDNA), analyzed all ORFs longer than 30 amino acids using BLASTP, and then pinpointed all cDNAs for which none of the ORFs revealed mammalian conservation in protein BLAST searches. To prepare input for the pipeline, we first ensured that all cDNAs processed by the pipeline originated from specific unambiguous genomic loci. Starting from 26,258 cDNA-supported TUs on the hg17 human genome assembly (Engström et al. 2006), we eliminated TUs not able to be mapped to the hg18 assembly, retaining 24,734 mapped TUs with valid GenBank cDNA accession numbers. Of those 24,734 cDNAs, 5446 (Supplemental Data Set 1) were identified as putative ncRNAs by our ORF-Predictor/BLASTP pipeline.
We compared our 5446 predicted ncRNAs (Fig. 1A) with the 1732 lncRNAs from the four public sources (Fig. 1B). From the four public sources, 442 (Supplemental Data Set 3) ncRNAs were eliminated because of redundancy. Of the 1732 literature-derived lncRNAs (Fig. 1B), 242 were redundant because their exons genomically overlapped with at least one exon on the same strand of—and in the same transcriptional orientation as—one of the 5446 lncRNAs from our pipeline. One hundred sixty-one literature-derived lncRNAs had GenBank accession numbers that exactly matched those of lncRNAs on our list of 5446. Among the four data sets of Figure 1B, 39 were internally redundant but had no counterparts among the lncRNAs from our pipeline. Therefore, the majority of our 5446 lncRNAs were not in the public sources listed in Figure 1B.
FIGURE 1.
lncRNA discovery with our analytical pipeline and lncRNA import from public databases. (A) A set of lncRNAs was predicted by applying our own ORF-Predictor/BLASTP parsing pipeline to a human genome-wide transcriptional unit (TU) catalog (Engström et al. 2006). (B) We retrieved 534 putative lncRNAs from the H-Invitational Database (H-InvDB), 335 from RNAdb, 512 lncRNAs from an early functional study of conserved lncRNAs (Willingham et al. 2005), and 351 primate-specific transcriptionally active region ncRNAs (Zhang et al. 2007). There were 6736 (= 1732 − 442 + 5446) nonredundant lncRNA genes.
For an additional assessment of redundancy between our pipeline's output and existing public lncRNA collections, we consulted the NcRNA Expression Database (NRED) (Dinger et al. 2009). We chose the platform “GNF Atlas 2,” which was the sole human platform available, and confined the selection to ncRNAs classified as “noncoding only” (n = 917). Among the 917, 130 had GenBank accession numbers that exactly matched those of lncRNAs on our list of 5446, and 82 did not match by accession number but were on the same strand as, and partially or wholly genomically overlapped with, at least one of our lncRNAs (Supplemental Data Set 5).
We also compared our data set with a recently published collection of 1177 nonconserved “orphan” genes that are suspected to lack protein-coding capacity (Clamp et al. 2007). The aforementioned collection (18) was based on Ensembl v35 (hg17), while our ncRNA analysis was based on the newer hg18 human genome assembly. For the 382 “orphans,” we used the Ensembl track of the UCSC Genome Database to directly retrieve the hg18 map location. For the other 795 orphans, we retrieved cDNA sequences from Ensembl (v35). In cases where an Ensembl gene identification number referred to multiple transcripts, we randomly selected one of those transcripts as the reference transcript. Then, we performed BLAT to determine the highest-confidence genomic map location of each orphan gene. Surprisingly, none of the 1177 orphans overlapped with any of our 5446 ncRNAs.
Recently, Khalil et al. (2009) identified 3289 human large intergenic noncoding RNA (lincRNA) genes from human and mouse H3K4me3 (K4, promoter)–H3K36me3 (K36, transcribed region) chromatin domains outside of protein-coding genes. To assess the potential redundancy between our lncRNA data set and the lincRNA catalog, we identified all genomic overlaps between the 2514 Khalil et al. (2009) lincRNAs and our lncRNAs. Only 402 lincRNAs overlapped our lncRNAs (Supplemental Data Set 11), and since lincRNA transcription orientation was not known, we estimate that half (201) of the lincRNAs matched our lncRNAs. Summarily, on the basis of our multiple assessments of redundancy between our pipeline's output and a wide repertoire of public lncRNA data sets, it appears that our lncRNA discovery pipeline has a substantial false-negative rate, and that expansion of the nonredundant human lncRNA catalog can be accomplished by the synthesis of several independent computational approaches to lncRNA identification.
Independent evidence that members of the predicted lncRNA catalog have little protein-coding capacity
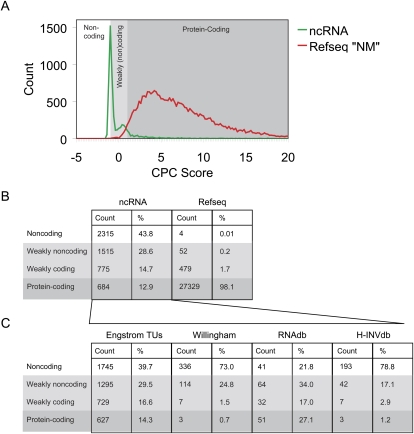
As stated above, the lncRNA catalog was populated by predicted noncoding transcripts from a variety of sources. The majority was obtained by filtering TUs from the Engström et al. (2006) data set through a stringent negative BLASTP filter, while the noncoding status of transcripts from other data sets was ascertained by a variety of methods of varying stringency (Okazaki et al. 2002; Imanishi et al. 2004; Pang et al. 2007). Given this variety of noncoding filtering methods, it is important to use an independent test to validate the coding status of all transcripts. A number of such methods exist (Dinger et al. 2008b). We chose to employ the recently developed Coding Potential Calculator (CPC) tool (Kong et al. 2007), which combines a variety of parameters in conjunction with a support vector machine to predict the coding potential of a given transcript. CPC was used to assess the coding potential of lncRNAs in our catalog (Fig. 2A,B). As a reference, we carried out the same analysis on the RefSeq catalog of human protein-coding genes. The results attested to the quality of the lncRNA discovery pipeline: 87.1% of lncRNAs were classified as noncoding, weakly noncoding, or weakly coding, in contrast to 1.9% of the RefSeq genes.
FIGURE 2.
Independent assessment of the protein-coding capacity of our lncRNA catalog. cDNA sequences of ncRNAs were submitted to the Coding Potential Calculator (CPC) (Kong et al. 2007). All sequences are assigned a score based on their estimated protein-coding capacity: <−1, noncoding; from −1 to 0, weakly noncoding; from 0 to 1, weakly coding; and >1, coding. A similar analysis was also carried out for 27,864 coding RefSeq genes, having accession numbers commencing with NM. CPC scores for each transcript are plotted as a continuous distribution in A, and the fractions of transcripts are broken down by classification (our ncRNA genes versus NCBI RefSeq genes) in B, as well as by data source (for ncRNA genes only) in C.
More than half of the human lncRNA genes overlap or reside near known protein-coding genes
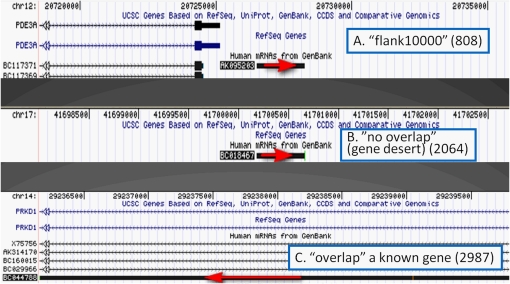
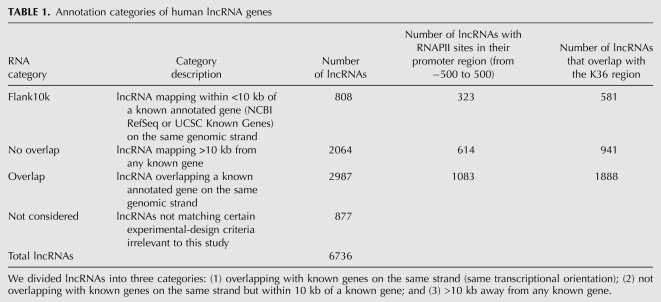
Computational identification of lncRNA candidates supported by public transcriptome data overlooks the complex genomic landscape of how those genes are related to their genomic neighbors and whether they or their neighbors in fact encode proteins. To characterize these relationships, we defined three categories of lncRNAs (Fig. 3): (1) “Flank10k,” lncRNA genes mapping within 10 kb of a known annotated gene (NCBI RefSeq or UCSC Known Genes) on the same genomic strand; (2) “no overlap,” lncRNA genes mapping >10 kb from any known gene; and (3) “overlap,” lncRNAs overlapping a known annotated gene on the same genomic strand. We analyzed 5859 putative ncRNAs (4687 of the 5446 lncRNAs from our pipeline, and 1172 of the 1732 lncRNAs from published sources; exclusions comprised lncRNAs not matching certain experimental-design criteria irrelevant to this study). There were 808, 2064, and 2987 lncRNAs belonging to the Flank10k, no overlap, and overlap classes, respectively (Table 1). As this analysis concerned only the positional associations between lncRNA genes and known annotated genes on the same strand, we also performed a Flank10k analysis of the “lncRNA gene–annotated gene” pairs such that the lncRNA gene and the annotated gene were on opposite strands (Supplemental Data Set 9). In addition to our 808 same-strand Flank10k loci, we isolated 397 head-to-head (divergent) lncRNA gene-annotated gene pairs as well as 144 tail-to-tail (convergent) lncRNA gene-annotated gene pairs, with no overlap between the lncRNA gene and the annotated gene, and with less than 10 kb of genomic distance separating the lncRNA gene from the oppositely oriented annotated gene in all cases. The excess of divergent over convergent gene pairs may merely reflect the known genomic excess of adjacent gene pairs driven by putative bidirectional promoters (Trinklein et al. 2004), and may in part reflect specifically the incidence of lncRNA gene-coding gene bidirectional pairs in mammalian genomes (Engström et al. 2006). Of lncRNA genes, 2064 (35%) reside in genomic regions >10 kb away from any protein-coding genes, and therefore likely act by mechanisms other than cis-regulation of their protein-coding neighbors. These no-overlap genes, due to the lack of any positional association with known protein-coding genes, likely represent bona fide, standalone lncRNAs that may function by mechanisms distinct from any cis-regulation of nearby genes.
FIGURE 3.
Examples of lncRNA stratification based on genomic context. (A) Flank10k (lncRNA gene maps within 10 kb of a known gene); (B) no overlap (lncRNA gene is >10 kb away from any known gene); and (C) overlap (lncRNA gene is encoded on the same strand of the genome as a known gene, and overlaps at least a part of that known gene). One representative UCSC Genome Browser snapshot for each category is shown. The lncRNA from our list is highlighted by the red arrow, which also indicates the direction of transcription of the lncRNA.
TABLE 1.
Annotation categories of human lncRNA genes
lncRNA genes within 10 kb of protein-coding genes are generally standalone transcriptional units
We closely inspected all Flank10k lncRNA genes in order to determine whether they were standalone genes, rather than misannotated extensions of nearby protein-coding genes. It is common for protein-coding gene UTRs to be long and to wholly encompass entire full-length cDNAs, which might be mistakenly interpreted as lncRNAs, even though they are truncated clones from UTRs of coding genes. To exclude lncRNA-like artifacts arising from such long UTRs, we checked each lncRNA mapping within <10 kb of a known protein-coding gene for membership in a transcript (cDNA and/or expressed sequence tag [EST]) contig connecting the lncRNA with the known gene. After checking whether the ncRNAs were connected by exonic sequences of ESTs/cDNAs to their nearby genes, we found that the majority of the Flank10k candidates were actual, distinct lncRNA genes unconnected to their protein-coding neighbors (717/808) (Supplemental Data Set 2). Only 84 out of 808 were extensions of coding flanking genes, and seven were extensions of noncoding flanking genes. These seven rare exceptions underscore the occasional misannotation of coding-gene UTRs as candidate lncRNAs, as well as the utility of cDNA and EST contig-building in revealing such misannotation.
Epigenetic landmarks are consistent with widespread lncRNA transcription
To gauge empirical support for transcriptional activity of lncRNA-encoding loci, we computed all genomic overlaps between our lncRNA genes and two accepted measures of transcriptional activity: all human H3K36me3 domains available at the time of our analysis (Gm12878, HUVEC, K562, and NHEK), and RNA Polymerase II genomic binding sites [antibody “Pol2(b),” cell types HUVEC, K562, and NHEK]. All data originated from the UCSC Genome Browser track “ENCODE Histone Modifications by Broad Institute ChIP-Seq.” Promoter (defined as the −500 to +500 nucleotide interval relative to the transcription start site of the representative GenBank or database cDNA accession) coverage of our lncRNA genes by Pol II binding sites was comparable regardless of the genomic context of the lncRNAs: 30% for no-overlap lncRNA genes; 36% for lncRNA genes overlapping known genes; and the highest, 40%, for the Flank10k lncRNA genes. This is consistent with our annotation-supported contention that, despite their proximity to known protein-coding genes, these Flank10k lncRNA transcription units are standalone (Supplemental Data Set 7). K36 domain coverage support of lncRNA genomic spans ranged from 46% (no-overlap lncRNAs) to 72% (Flank10k lncRNAs), signifying that even lncRNAs far away from known genes are frequently supported by K36 data (Supplemental Data Set 8). A limitation of these analyses is that the majority of the cDNA and EST data supporting expression of these lncRNAs originates from cell types different from those used in the Broad Institute histone modification and Pol II occupancy study.
Protein-coding capacity evaluation of known genes in the vicinity of lncRNA genes reveals that hundreds of known hypothetical-protein genes are noncoding
We noted that certain RefSeq and UCSC Known Genes database entries lacked descriptive gene names, and were labeled solely as encoding “hypothetical proteins.” We collectively defined hypothetical-protein genes, as well as genes possessing solely numerical identifiers, as uninformatively named genes. Hypothetical-protein genes may not actually encode functional proteins, and therefore may themselves be lncRNA genes (Clamp et al. 2007). This concerned us because of the impact on the accuracy of the annotations of the lncRNAs in our data set. Specifically, overlap lncRNA candidates should remain annotated as bona fide lncRNA genes if their overlapping known genes do not encode proteins. Similarly, Flank10k lncRNA candidates that could be joined to their neighboring known genes via cDNA or EST contigs should remain annotated as bona fide lncRNA genes, if their neighboring genes—despite being included in known gene catalogs—in fact lack protein-coding capacity. We hence set out to check all uninformatively named genes throughout our data set for protein-coding capacity, as described below (see Materials and Methods). Nearly two-thirds (866/1397; 62%) of uninformatively named, known, presumed-coding genes were shown to lack protein-coding capacity by our method (Fig. 4; Supplemental Data Set 2). Only 531 (38%) were consistent with the public annotations designating them as protein coding. Of the uninformatively named known genes within 10 kb of our lncRNAs, 93 out of 164 (57%) were determined by our method (Fig. 4) to encode noncoding RNAs and 71 were protein-coding genes. This result further emphasizes the utility of our ORF-Predictor/BLASTP pipeline for effective identification of lncRNAs.
FIGURE 4.
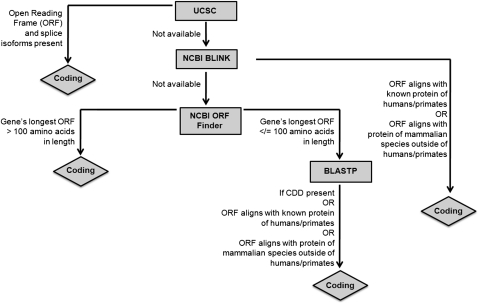
Protein-coding capacity testing of uninformatively named known genes that overlapped, or were located <10 kb away from, lncRNA genes. This is a flowchart of our approach toward the definition of protein-coding capacity of uninformatively named known genes overlapping, or in proximity to, lncRNA genes. This is a manual-curation approach.
In order to comprehensively profile genome-wide expression of lncRNAs, it is important to define the extent to which lncRNAs are represented on prefabricated commercially available microarray platforms. We intersected our data set of 5446 lncRNA genes discovered by the BLASTP and ORF-Predictor pipeline with the UCSC Genome Database genomic mappings of Affymetrix U133 (A and B) and Illumina hWG-6 (v3) probe sets. Of these lncRNAs, 3116 (57%) lacked Affymetrix U133 representation, and 4100 (75%) lacked Illumina hWG6 representation (Supplemental Data Set 4). Because we defined Affymetrix representation as any same-strand overlap between the entire genomic span of a U133 consensus exemplar sequence and our lncRNA, the actual percentage of our lncRNAs lacking U133 representation might be even higher if only true exon-to-exon overlaps are included. We conclude that the majority of human lncRNAs may not be represented by commercial microarray probe sets, and accordingly that the commercial microarray probe sets retain a substantial bias in favor of protein-coding genes.
DISCUSSION
To facilitate the process of identifying lncRNAs, we developed the ORF-Predictor, a high-throughput standalone equivalent of the NCBI ORF-Finder. We have produced a stringently compiled data set of cDNA-supported TUs, 5446 of which represent lncRNAs according to the output of our ORF-Predictor and BLASTP parsing pipeline. The extent of redundancy between this collection of lncRNAs and multiple public data sets is low.
We also evaluated lncRNAs based on their proximity to nearby known genes. Existing ncRNA databases classify ncRNAs in different ways (Liu et al. 2005; Dinger et al. 2009) not including the proximity to known genes. Some databases, such as RNAdb and fRNAdb, are themselves collections of different ncRNA data sets. Published ncRNA databases can be heterogeneous, and are not always anchored via GenBank accession numbers directly to primary transcriptome data. Our contribution to the emerging lncRNA field has been to construct an inclusive and nonredundant lncRNA catalog incorporating our findings and those of the published databases, as well as to stratify the cataloged lncRNAs based on their genomic position relative to known genes. The proximity of lncRNAs to known protein-coding genes may serve as a predictor of cis-regulation by those lncRNAs of the known genes (Guttman et al. 2009).
Our analysis reveals that over 65% of lncRNA genes are located within 10 kb of known, mostly protein-coding, genes. This suggests that cis-regulatory relationships may exist between the lncRNAs and the known genes. Various experimental studies suggest that gene-proximal (i.e., Flank10k class) lncRNAs are likely to regulate their coding genomic neighbor in cis (or vice versa) (Rinn et al. 2007; Faghihi et al. 2008; Guttman et al. 2009). Additionally, the lncRNA gene and its proximal genomic neighbor might be regulated by a common control event upstream in the regulatory network, for example, by nearby binding of a specific transcription factor at a single site that controls both the lncRNA gene and its neighbor within a particular regulatory program. Moreover, we have determined that 959 of 1561 hypothetical-protein genes overlapping or within 10 kb of our lncRNA genes, and annotated as protein-coding in the UCSC Known Genes and/or the NCBI RefSeq databases, actually lack protein-coding capacity by the FANTOM3 criterion (Dinger et al. 2008b). This result is consistent with our discovery of thousands of cDNA/EST-supported lncRNA genes, because both results indicate that existing database known-gene annotations are incomplete and unreliable, respectively.
Putative lncRNA genes may represent host genes that are biologically processed into shorter functional RNAs such as microRNAs, piRNAs, snoRNAs, or snRNAs. To investigate this possibility, we examined all lncRNA genes in our catalog for the internal presence of known small-RNA genes. We compared the genome mappings of the complete genomic spans (from the 5′ gene boundary to the 3′ gene boundary) for 4687 lncRNAs from our pipeline with all mapped genomic locations of known RNA genes from the RNA Genes and sno/miRNA tracks of the UCSC Genome Database. Thirty of the lncRNA genes overlapped with known RNA genes of those two classes; 27 were lncRNAs overlapping known genes and three were lncRNAs within <10 kb of known genes. This analysis revealed that less than 1% of our lncRNA genes serve as known small-RNA host genes, although it certainly does not exclude the possibility that additional lncRNAs in our list are precursors of heretofore unknown snRNA, snoRNA, microRNA, or other small-RNA functional molecules.
The abundance, and diversity, of lncRNA genes necessitate access to a suitable expression analysis platform. Due to misannotation of lncRNA genes as hypothetical-protein genes in public databases, we expected that a subset of lncRNA genes would be represented on conventional catalog microarrays designed for expression profiling of protein-coding genes. We investigated this possibility by examining the human probe sets underlying the Affymetrix U133 A and B human gene expression analysis arrays, as well as the Illumina hWG6-v3 array. Of 5446 lncRNA genes analyzed, 3116 (57%) were not represented on the Affymetrix arrays, and 4100 (75%) were not represented on the Illumina array. Therefore, lncRNA genes are undersampled by commercial platforms. Our data can be used to mine existing microarray result repositories such as NCBI GEO for lncRNA expression. Nevertheless, custom arrays and/or high-throughput transcriptome sequencing will be necessary to quantitate the expression of the lncRNome in a less biased fashion.
Our study does not support the protein-centric assumption that the ORF of any novel cDNA encodes a hypothetical protein. We show that numerous cDNA-encoded hypothetical proteins in the NCBI GenPept database fail a test for protein-coding capacity and therefore might not encode proteins, as large numbers of unique or highly evolutionarily divergent proteins without protein database matches are unlikely to exist. The protein-centric automatic assignment of hypothetical-protein identifications to novel ORFs inferred from cDNAs may merit reconsideration. However, accurate in silico determination of protein-coding capacity remains an unsolved problem (Dinger et al. 2008b).
The extensive experimental evidence for thousands of lncRNA genes in the human genome establishes a framework for future large-scale functional analysis of the long noncoding transcriptome. A combination of in silico and laboratory-based approaches will be needed to test lncRNA genes for function. In particular, functional analysis of lncRNA genes residing in disease candidate regions (Schaefer et al. 2009) or in linkage disequilibrium with disease-associated SNPs (Ishii et al. 2006) should reveal entry points for investigating the phenotypic impact of the long noncoding transcriptome.
MATERIALS AND METHODS
Retrieval of human lncRNA sequences
Human lncRNAs were curated from two sources (Fig. 1). One subset was predicted by our ORF-Predictor/BLASTP-parser pipeline (for details, see the next section) from a human genome-wide TU catalog (Engström et al. 2006). Another subset was comprised of four public ncRNA data sets: H-Invitational database (H-InvDB) (Yamasaki et al. 2008), RNAdb (Pang et al. 2007), a human–mouse conserved lncRNA set used in a high-throughput functional screen (Willingham et al. 2005), and primate-specific ncRNAs stratified by sequence conservation and the probability of forming stable secondary structures (Zhang et al. 2007). It is important to note that each lncRNA selected by us for this study represented a distinct transcriptional unit (gene), and that, therefore, all subsequent analyses were at the TU (gene) level, not at an individual-transcript level.
Identification of lncRNA sequences by our ORF-Predictor/BLASTP pipeline
We developed the program ORF-Predictor in Perl to find all the ORFs of cDNAs, which start with ATG and end with TAA, TGA, or TAG on the positive strand. ORF-Predictor considers all three positive-strand translations of each cDNA input. It is different from the NCBI ORF-Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) in several ways. (1) ORF-Predictor only finds ORFs on the positive strand of the input, while the NCBI ORF-Finder finds ORFs on both strands. (2) Since all our input sequences are full-length cDNAs, ORF-Predictor solely considers ORFs that start with an ATG and end with a stop codon (TAA/TGA/TAG), while the NCBI ORF-Finder also finds incomplete ORFs containing a start or stop codon, but not necessarily both. (3) ORF-Predictor processes batch sequence inputs. In contrast, the NCBI ORF-Finder is web based, does not have a batch-input capacity, and is not available as a standalone executable program, which limits its throughput to one user-submitted sequence at a time. This tool was developed because the NCBI ORF-Finder is web based and has no exact command line alternative.
Our BLASTP result parsing tool is a Perl script that parses BLASTP outputs and automatically determines which outputs reveal the lack of mammalian protein sequence conservation for each query. BLASTP result parsing was used to search query hits to mammalian targets. Mammalian hits whose length is longer than 30 amino acids and whose e-value is less than 0.001 were accepted as bona fide protein-coding hits; otherwise, a “no protein homology” result was returned. NCBI BLASTP was run with these parameters: -p = BLASTP, -e=−10.0, -G = −1, -E = −1, -F = T, -I = F, -X = 15, -f = 11, -g = T, - Q = 1, -M = BLOSUM62, -W = 3, -z = 0, -K = 0, -P = 0, -Y = 0, -S = 3, -y = 0.0, -Z = 0, -n = F, -w = 0, -t = 0, -B = 0, -V = F, and database nr.
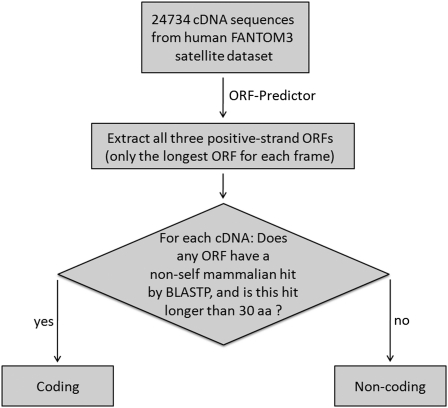
To apply our pipeline, we first used ORF-Predictor to find all the ORFs in all three possible reading frames on the positive strand of each cDNA (from Engström et al. 2006). Then we ran BLASTP on all ORFs longer than 30 amino acids, for each cDNA. For each ORF we used our BLASTP result parsing module to search the hits. If we found any hits matching our protein-coding criteria for any ORFs, then we regarded the cDNA from which the ORF came as the coding RNA; otherwise, it was regarded as a long noncoding RNA (Fig. 5). Even if the ORF with the protein-coding BLASTP hits was not the longest possible positive-strand ORF, the cDNA was still labeled protein coding.
FIGURE 5.
De novo lncRNA identification with our ORF-finding and BLASTP-parsing pipeline. This flowchart illustrates our use of the ORF-Predictor, which we developed, along with NCBI BLASTP to gauge the protein-coding capacity of any cDNA. This is an automated approach.
Elimination of redundant sequences
Because our data set consisted of our own lncRNA pipeline results along with an integration of four public lncRNA data sources, we needed to eliminate redundant lncRNAs. First, we searched each GenBank accession number from the combined data set, detecting all GenBank accession numbers occurring more than once. We eliminated all lncRNAs from four data sets whose NCBI GenBank accession numbers were redundant with respect to any FANTOM lncRNAs. Second, we mapped all remaining GenBank accession numbers of lncRNAs, from our pipeline and from the four public data sets, to the HG18 human genome assembly using the all_mrna file of the UCSC Genome Database.
Of the 5646 lncRNAs with GenBank accession numbers, 5570 mapped to one genomic location per lncRNA according to the UCSC Genome Database all_mrna HG18 data set. An additional 76 lncRNAs with GenBank accession numbers were absent from the UCSC all_mrna data set, lacking UCSC-precomputed genomic mappings. One lncRNA with a GenBank accession number had an ambiguous mapping to multiple genomic locations in the all_mrna data set; we arbitrarily chose one of the two coordinate sets for that lncRNA. For the 76 unmappable lncRNAs, as well as for all lncRNAs from public data sets that lacked GenBank identifiers, we ran UCSC BLAT on the FASTA sequences to determine the genomic position.
We checked each lncRNA, at its unique genomic mapping location as established in the preceding paragraph, for same-strand genomic span overlap with all other lncRNAs. If there was same-strand overlap, regardless of intron or exon localization of the overlap, then the two lncRNAs were regarded as redundant. We first checked genomic position redundancy among all members of the four public lncRNA data sources. Then we compared the Engström et al. (2006) lncRNAs with the nonredundant lncRNAs from the four public lncRNA data sets, and in the cases of redundancies we always retained the Engström et al. (2006) GenBank accession number.
Classification of lncRNA genes in terms of their proximity to known genes
We determined the genomic position of each nonredundant lncRNA gene relative to the genomic positions of its nearest known genes from the RefSeq and UCSC Known Genes databases. We divided our lncRNA genes into three categories: (1) lncRNAs overlapping with known genes on the same strand; (2) lncRNAs not overlapping with known genes on the same strand but within 10 kb of known genes (we considered the shortest possible distance between a boundary of the known gene and the closer of the two boundaries of its neighboring lncRNA gene); and (3) lncRNAs more than 10 kb away from known genes on the same strand. Strand orientation was taken into account because it has been well documented that sense and antisense transcripts are distinct at cis-antisense loci. Distance stratification was performed because of recent data (Guttman et al. 2009) suggesting that lncRNAs near known genes are more likely to be functional, as well as because of our concern that artifactual lncRNAs representing extended untranslated regions (UTRs) of protein-coding genes would be contaminants of our lncRNA data set.
Independent quality assessment of lncRNA candidates reveals that most lncRNAs are encoded by distinct transcriptional units separated by large genomic distances
We sorted all lncRNA mappings by orientation and genomic position, and we extracted all same-strand pairs of the two nearest (adjacent) lncRNAs, regardless of the intervening distance. We defined the nearest (adjacent) lncRNAs as the two lncRNAs on the same chromosome and on the same strand, with no other lncRNAs between them. For each lncRNA, we calculated two distances: one to the nearest upstream same-strand lncRNA, and one to the nearest downstream same-strand lncRNA. We found that most lncRNAs are located far away from other same-strand lncRNAs. Of particular note is the finding that the majority of lncRNA–lncRNA same-strand genomic distances (5303 out of 6681; 79%) were greater than 100 kb; 94% of the distances were greater than 10 kb (Supplemental Data Set 10). These considerable genomic distances between same-strand lncRNA genes suggest that annotation artifacts, which split up single lncRNA genes with insufficient or truncated cDNA and EST support into multiple apparent lncRNA genes, are unlikely, because such artifactual split genes should not be separated by very large genomic distances far exceeding typical genomic sizes of human genes.
Manual curation of lncRNA candidates near known genes and definition of high-confidence standalone lncRNA genes
For each lncRNA candidate within 10 kb of a known protein-coding gene on the same strand, we visually checked, in the UCSC Genome Browser, for the existence of a same-direction EST and/or cDNA contig connecting the lncRNA and the known gene, with exon overlaps of the tiled cDNAs or ESTs. If the exonic sequences of cDNAs and/or ESTs bridged the lncRNA to its flanking gene, then we annotated the lncRNA as putatively connected to the flanking gene. Otherwise, we defined the lncRNA gene as a high-confidence standalone lncRNA gene.
Protein-coding capacity of known genes that had uninformative names and resided within 10 kb of lncRNA candidates
We defined a gene with an uninformative name as any known gene that had a purely alphanumeric name (e.g., “FLJ number,” “KIAA number”), a name that included the phrase hypothetical protein, or a name that consisted solely of a GenBank accession number. We examined the ORF, as visually indicated by the vertical dimension of exonic-sequence rectangles in the UCSC Known Genes and RefSeq tracks of the UCSC Genome Browser, for each such known gene. If there were at least two protein-coding reference transcripts visible in either or both of those UCSC tracks, and if the start and/or stop codon locations on the genome matched between any two or more protein-coding reference transcripts, then we labeled the gene as protein coding. Otherwise, we checked whether NCBI BLINK (precomputed BLASTP) was available for the putative protein sequence encoded. If NCBI BLINK was available, then we considered hits spanning more than a contiguous one-half of the protein sequence query. BLINK self-hits (human protein database entries identical to the query sequence) were excluded. If all BLINK hits were confined to primates and corresponded to uninformative gene names, we considered the query gene to be noncoding. If any BLINK hits were to more distant lineages, and/or if any BLINK hits had informative descriptive gene names, we considered the query gene to be protein coding. If NCBI BLINK was not available, we used the NCBI ORF-Finder to determine the longest positive-strand ORF of the uninformatively named known gene, based on a full-length cDNA corresponding to the UCSC Known Genes or RefSeq reference structure of the uninformatively named known gene. If the ORF encoded a protein over 100 amino acids in length, it was automatically labeled protein coding. There are several lines of computational evidence favoring the assertion that cDNAs whose ORFs are shorter than 100 amino acids lack protein-coding capacity (Dinger et al. 2008b); our analysis of the Engström et al. (2006) data set (data not shown) supports this assertion further, as we determined that 92% of the 18,358 Engström et al. (2006) cDNAs with ORFs >100 amino acids, 15,780 of which corresponded to RefSeq genes, also have conserved ORFs. Therefore, to examine the protein-coding capacity of shorter-than-100-amino acid ORFs, we manually performed NCBI BLASTP searches with conserved domain database (CDD) queries. Genes with CDD domain hits and/or coding BLASTP protein database hits were labeled protein coding. (The definition of protein-coding versus noncoding BLASTP hits was identical to that used for BLINK hits above.) Figure 4 summarizes our protocol for verifying the protein-coding capacity of genes that are allegedly protein coding in public databases but have uninformative names.
Validating protein-coding capacity of transcripts using the CPC tool
cDNA sequences were obtained for our lncRNA catalog, and for the protein-coding (NM_# accessions only) component of the NCBI RefSeq known-gene catalog. For ncRNAs, all sequences were successfully extracted and submitted to the CPC, except for 297 predicted noncoding RNAs from the study of Zhang et al. (2007), whose genomic span and strand are uncertain. A similar analysis was also carried out for 27,864 coding RefSeq genes, having accession identification numbers commencing with “NM.” All ncRNA and RefSeq cDNA sequences were submitted in FASTA format to the CPC web server (http://cpc.cbi.pku.edu.cn/) (Kong et al. 2007). Based on the recommendations of Kong et al. 2007, the protein-coding capacity of ncRNA and RefSeq transcripts were classified by their resultant CPC score: <−1, noncoding; from −1 to 0, weakly noncoding; from 0 to 1, weakly coding; and >1, coding.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
POTENTIAL CONFLICT OF INTEREST DISCLOSURE
Intellectual property protection is being applied for with respect to the technology described in the paper and the supplemental data sets. We have the potential to financially benefit from successful commercialization of any such intellectual property.
ACKNOWLEDGMENTS
This work was funded by NIH NIDA R03 1R03DA026021-01 (to L.L.), Wayne State University New PI Start-Up Budget Grant 338171 (L.L.), and by Genome Institute of Singapore competitive intramural research funding (to L.W.S.). We thank Jason Blythe for technical assistance.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1951310.
REFERENCES
- Carninci P, Hayashizaki Y 2007. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev 17: 139–144 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci 104: 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. 2008a. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS 2008b. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol 4: e1000176 doi: 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS 2009. NRED: A database of long noncoding RNA expression. Nucleic Acids Res 37: D122–D126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, et al. 2006. Complex loci in human and mouse genomes. PLoS Genet 2: e47.doi: 10.1371/journal.pgen.0020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C 2008. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med 14: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM 2006. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci 103: 5781–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol 2: e162 doi: 10.1371/journal.pbio.0020162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. 2006. Identification of a novel noncoding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51: 1087–1099 [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Jia H, Vanisri RR, Pandey T, Lu ZH, Buckley NJ, Stanton LW, Lipovich L 2009. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G 2007. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y, Bu D, Zhao Y, Chen R 2005. NONCODE: An integrated knowledge database of noncoding RNAs. Nucleic Acids Res 33: D112–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A 2007. Repression of the human dihydrofolate reductase gene by a noncoding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS 2007. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 87: 799–823 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573 [DOI] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Dinger ME, Engström PG, Lenhard B, Mattick JS 2007. RNAdb 2.0—an expanded database of mammalian noncoding RNAs. Nucleic Acids Res 35: D178–D182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443: 167–172 [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP 2009. Genomic and transcriptional co-localization of protein-coding and long noncoding RNA pairs in the developing brain. PLoS Genet 5: e1000617 doi: 10.1371/journal.pgen.1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE, Loos BG, Jepsen S, Schreiber S 2009. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet 5: e1000378 doi: 10.1371/journal.pgen.1000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM 2004. An abundance of bidirectional promoters in the human genome. Genome Res 14: 62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Yamasaki C, Murakami K, Fujii Y, Sato Y, Harada E, Takeda J, Taniya T, Sakate R, Kikugawa S, Shimada M, et al. 2008. The H-Invitational Database (H-InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res 36: D793–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pang AW, Gerstein M 2007. Comparative analysis of genome tiling array data reveals many novel primate-specific functional RNAs in human. BMC Evol Biol 7 (Suppl 1): S14 doi: 10.1186/1471-2148-7-S1-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]