Abstract
We improved the target RNA-primed RCA technique for direct detection and analysis of RNA in vitro and in situ. Previously we showed that the 3′ → 5′ single-stranded RNA exonucleolytic activity of Phi29 DNA polymerase converts the target RNA into a primer and uses it for RCA initiation. However, in some cases, the single-stranded RNA exoribonucleolytic activity of the polymerase is hindered by strong double-stranded structures at the 3′-end of target RNAs. We demonstrate that in such hampered cases, the double-stranded RNA-specific Escherichia coli RNase III efficiently assists Phi29 DNA polymerase in converting the target RNA into a primer. These observations extend the target RNA-primed RCA possibilities to test RNA sequences distanced far from the 3′-end and customize this technique for the inner RNA sequence analysis.
Keywords: ACTB, GAPDH, padlock probe, Phi29 DNA polymerase, PPIA, RNA-primed, target-primed, RCA, RNase III
INTRODUCTION
Padlock probes and rolling circle amplification are widely used for single or multiparallel analysis of different macromolecules and their complexes. Usually the padlock probes for RCA are created by circularization of DNA oligonucleotides on complementary DNA targets (Nilsson et al. 1997; Baner et al. 1998; Lizardi et al. 1998). The proximity ligation and template formation for following RCA were also successfully applied for protein detection (Soderberg et al. 2006; Jarvius et al. 2007; Ericsson et al. 2008). The RCA has been shown as a useful method not only for the characterization of isolated or preformed targets in vitro, but also for the detection and analysis of single DNA, RNA, and protein molecules in situ (Christian et al. 2001; Larsson et al. 2004; Soderberg et al. 2006; Jarvius et al. 2007).
Literature data indicate that T4 DNA and RNA ligases can catalyze the ligation of nicked DNA involved in complementary double-stranded DNA–RNA structures (Nilsson et al. 2000, 2001; Christian et al. 2001). Such DNA padlock probes ligated on RNA targets were successfully used with additional DNA primers to initiate RCA in detection of mRNA expression levels in individual cells (Christian et al. 2001).
Direct detection of RNA molecules by RCA, using target RNA as a primer for RCA, were previously described for microRNA (Jonstrup et al. 2006). The method was upgraded using Turtle Probe (TP) formation on ribosomal RNA or nonpolyadenylated messenger RNA, and it was successfully applied in situ (Stougaard et al. 2007). Other variants of target RNA-primed RCA techniques, coupled with fluorescence quantification or combined with capillary electrophoresis, were used in qualitative and quantitative examination of microRNAs (Cheng et al. 2009; Li et al. 2009). However, since the target RNA–DNA probe duplexes described in these publications are perfectly matched at the 3′-end of RNA targets, this method is limited to detection of only target sequences located at the end of the RNA template. In certain cases, this limitation is absolute, for example, in the detection of eukaryotic messenger RNA molecules, which have poly(A) sequences at the 3′-end (Mijatovic et al. 2000), so individual transcripts cannot be tested. The method is also not suitable for the analysis of inner RNA sequence peculiarities, for example, RNA spliced variants.
The observation that Phi29 DNA polymerase exhibits a 3′ → 5′ single-stranded RNA-specific exonucleolytic activity allowed us to use the enzyme for the conversion of target RNA into a primer for RCA (Lagunavicius et al. 2008). Later, the exoribonucleolytic activity of Phi29 DNA polymerase was successfully applied in vitro and in situ for direct detection and analysis of RNA molecules with the padlock probes complementary for target-RNA inner sequences located close to their 3′-ends and without additional DNA primers required (Lagunavicius et al. 2009). However, the conversion of target RNA into a primer was hindered when padlock probe hybridized to RNA sequence distanced far from the 3′-end of target molecule. In this case, the double-stranded RNA structures present in the 3′-end of the RNA target interfered with the single-stranded 3′ → 5′ RNA exonucleolytic activity of Phi29 DNA polymerase. In this report, we demonstrate that in such unproductive cases, the double-stranded RNA-specific Escherichia coli RNase III can efficiently assist Phi29 DNA polymerase in target-RNA conversion into a primer for RCA. These observations extend the target-RNA-primed RCA possibilities to test RNA sequences distanced far from the 3′-end and customize this technique for the inner RNA sequence analysis.
RESULTS AND DISCUSSION
Previously we demonstrated that the 3′ → 5′ exoribonucleolytic activity of Phi29 DNA polymerase can be used to convert target RNA into a primer prior to switching to polymerization mode and proceeding into RCA (Lagunavicius et al. 2008).
The impact of double-stranded RNA-specific E. coli RNase III on the target-RNA conversion into a primer
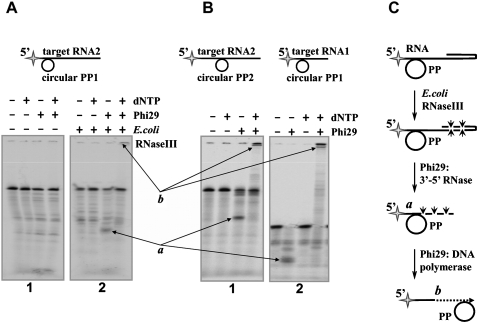
However, in some RNA–padlock probe hybrids, such as in experiments testing the RNA2–PP1 hybrid formed close to the 5′-end of RNA (Fig. 1A, panel 1), neither RNA hydrolysis products nor a specific RCA product were obtained at standard reaction conditions using Phi29 DNA polymerase. Since the duplex formation between the 5′-labeled target RNA2 and preformed circular padlock probe PP1 was verified by electrophoretic mobility shift assay (data not shown), we presumed that the absence of RCA products, most likely, is related to the presence of RNA secondary structures hindering the activity of Phi29 DNA polymerase. The bioinformatics analysis of RNA2 sequence on the DINAMelt Server (Markham and Zuker 2005, 2008; http://frontend.bioinfo.rpi.edu/applications/hybrid/quikfold.php) predicted that RNA2 molecule complexed by probe PP1 can form a strong RNA secondary structure at the 3′-end of the region located downstream from the hybridization area (ΔG = −20 kcal/mol), which could interfere with a single-stranded exoribonuclease activity of Phi29 DNA polymerase.
FIGURE 1.
Target RNA conversion into a primer for RCA, the impact of E. coli RNase III. The reactions were carried out under the conditions described in Materials and Methods. The 5′-end-labeled (A) RNA2-PP1 (panel 1), (B) RNA2-PP2 (panel 1), and RNA1-PP1 (panel 2) hybrids were incubated with Phi29 DNA polymerase in the absence or presence of dNTP. (A, panel 2) The experiments with E. coli RNase III were performed using RNA2-PP1 duplex. The RNA degradation and RCA products were analyzed by electrophoresis through denaturing 8% polyacrylamide gels. Different RNA-PP duplexes are shown above the gels. The radioactive label is indicated by the star. The contents of samples are shown above the gel lines. Reaction products are labeled as “a” for the 5′-end-labeled RNA1 hydrolysis product serving as an RNA primer for RCA and “b” for the 5′-end-labeled RCA product. (C) The reaction scheme represents the conversions of target RNA.
Strong RNA secondary structures could be attenuated using DNA probes, which hybridize closer to the 3′-end of target RNA. Indeed, when the RNA2 target was complexed with another DNA molecule, probe PP2, the hybridization region of which was shifted toward the 3′-end by 10 nucleotides (nt), bioinformatics tools predicted a secondary structure in the 3′-end of RNA2 (ΔG = −12 kcal/mol) to be 1.7-fold weaker than the RNA structure predicted in the RNA2–PP1 complex (see above). Experimental data supported this prediction, because, contrary to the RNA2 target in the RNA2–PP1 hybrid (Fig. 1A, panel 1), RNA2 hybridized to preformed circular padlock probe PP2 was partly degraded by Phi29 DNA polymerase and converted into a primer for RCA (Fig. 1B, panel 1, products a,b). However, shifting of the hybridization region is not applicable in real-life practice, where specific RNA sequences, for example, SNPs or spliced sequences, need to be analyzed.
Alternatively, the RNA 3′-end secondary structures could be eliminated by shortening the molecules of target RNA. Bioinformatics analysis of a 48-nt-long RNA oligonucleotide RNA1, a truncated version of oligonucleotide RNA2 (111 nt), predicted that the RNA1 molecule complexed with probe PP1 does not form any stable RNA structures at the 3′-end (ΔG = ∼0 kcal/mol). The prediction was confirmed using RNA1–PP1 complex and Phi29 DNA polymerase in an RCA experiment, and the target RNA1 was completely converted into a primer for RCA (Fig. 1B, panel 2, products a,b). In practical applications the shortening of RNA targets, which have stable double-stranded structures at the 3′-end of the molecules, could be performed by structure-specific RNases. We found experimental evidence that E. coli RNase III, which endonucleolytically digests double-stranded RNA (March and Gonzalez 1990), rendered the earlier unproductive RNA2–PP1 hybrid RCA competent. After digestion of double-stranded RNA structures at the 3′-end of the target by E. coli RNase III, Phi29 DNA polymerase partly converted the RNA2 target into a primer, which later was extended into an RCA product (Fig. 1A, panel 2, products a,b; Fig. 1C).
The impact of double-stranded RNA-specific E. coli RNase III on the detection of target transcripts in vitro
To assess the effect of E. coli RNase III on mRNA detection in vitro, we used GAPDH transcripts in total RNAs isolated from human HeLa cells and tongue and liver tissues from mouse.
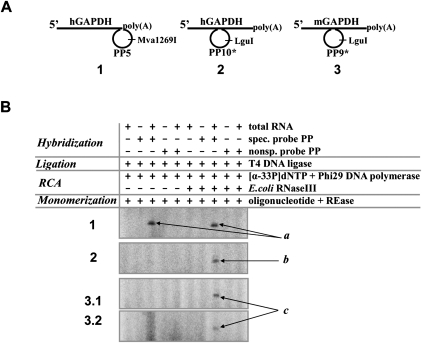
Initially, human GAPDH transcripts were tested using inner RNA sequence at the border of the poly(A) tail and a specific padlock probe PP5, the 5′- and 3′-ends of which were designed to be complementary to the hGAPDH transcript hybridization region (Fig. 2A, duplex 1). A nonspecific probe, PP6, which had a few mismatched nucleotides at the 3′- and 5′-ends, was used as a control probe for ligation specificity. As expected, no influence of E. coli RNase III was noticed using the specific probe PP5. Similar amounts of 70-nt-long RCA monomers, Mva1269I restriction fragments of long RCA product, were obtained either in the absence or in the presence of E. coli RNase III (Fig. 2B, panel 1, products a). Besides, the nonspecific probe PP6 did not generate any labeled specific products (Fig. 2B, panel 1).
FIGURE 2.
Detection of GAPDH transcripts in vitro, the impact of E. coli RNase III. The reactions were carried out with GAPDH-padlock probe duplexes (A, duplexes 1–3) under the conditions described in Materials and Methods. (B) The oligonucleotides PP5 (panel 1) and PP10* (panel 2) were used as specific tools for the hGAPDH transcript detection in the human total RNA isolated from HeLa cells. The oligonucleotides PP6 (panel 1) and PP9* (panel 2) were used as controls for reaction specificity. The oligonucleotides PP9* and PP10* (panels 3.1, 3.2) were used as probes for mGAPDH transcript detection and controls for reaction specificity, respectively, when testing mouse total RNAs isolated from the tongue (panel 3.1) and liver (panel 3.2) tissues. The RCA reactions were carried out in the absence or presence of E. coli RNase III (panels 1–3). The reaction products were monomerized by cleaving them with Mva1269I (panel 1) or LguI (panels 2, 3.1, 3.2) REases and analyzed by electrophoresis through a denaturing 8% polyacrylamide gel. The contents of samples are shown above the gel lines. The reaction products are labeled as “a,” “b,” and “c” for the 70-nt-long labeled monomers of RCA products obtained using transcript-padlock probe duplexes 1, 2, and 3, respectively (A).
However, quite different results were obtained when padlock probe hybridized to RNA sequence distanced far from the 3′-end. As an example, we used a specific padlock probe, PP10*, which hybridizes to the hGAPDH transcript sequence 720 nt away from the poly(A) tail of the transcript (Fig. 2A, duplex 2). Under usual conditions, without E. coli RNase III, Phi29 DNA polymerase did not generate any products (Fig. 2B, panel 2). The addition of E. coli RNase III changed the situation, and RCA monomers, LguI restriction fragments of RCA product, were observed (Fig. 2B, panel 2, product b). In contrast, specific labeled products were absent when nonspecific padlock probe PP9* differing by a single nucleotide in the ligation junction region was used as a control (Fig. 2B, panel 2).
Another series of experiments was carried out with GAPDH transcripts in total RNA isolated from mouse tissues. Mouse GAPDH transcript differs from human GAPDH transcript by a single nucleotide at the hybridization region of the 3′-ends of padlock probes. So, as a specific padlock probe for the mGAPDH transcript detection via ligation-based RCA, we used probe PP9* (Fig. 2A, duplex 3), and probe PP10* was used as a control probe for ligation specificity. As expected, specific RCA products were obtained only when total RNA isolated from the tongue or liver tissues was hybridized to the specific probe, PP9*, and RCA was performed in the presence of E. coli RNase III (Fig. 2B, panels 3.1,3.2, products c). The observed discrepancy between the yields of RCA products generated on total RNAs of different origin could be due to GAPDH mRNA level variations in RNA samples isolated from different mouse tissues, as previously was shown for GAPDH mRNA by Northern blot analysis (Gory et al. 1999). Significant differences in the expression level of GAPDH gene in various human tissues were also previously determined by RT-qPCR, for example, in skeletal muscle total RNA GAPDH cDNA was detected to be in 2.3-fold higher numbers than in liver total RNA (Barber et al. 2005).
In summary, in vitro results led us to conclude that for the detection and analysis of RNA sequences distanced far from the 3′-end, the addition of E. coli RNase III, endoribonuclease specific for double-stranded RNA, can efficiently facilitate target-RNA conversion into primer for RCA by Phi29 DNA polymerase.
The impact of double-stranded RNA-specific E. coli RNase III on the detection of target transcripts in situ
Our previous RNA-primed RCA experiments were carried out using hGAPDH transcript as a target and a specific padlock probe, PP5, which was complementary to the hybridization region overlapping with the poly(A) tail (Fig. 2A, duplex 1; Lagunavicius et al. 2009). The experimental data obtained showed that the samples generated specific RCA products that were subsequently labeled with FITC hybridization probes. The arithmetic mean of the hGAPDH-specific RCA products was ∼24 copies per HeLa cell. Similar absolute values of arithmetic means of hGAPDH transcript quantity were obtained in other cells using another single-cell technique—“digital RT-PCR” (Warren et al. 2006). However, the overall detection efficiency of target GAPDH transcript-primed RCA in situ was only ∼1% relative to the results collected by the RT-qPCR, ∼2.6 × 103 cDNA molecules per HeLa cell (Lagunavicius et al. 2009).
Here we report a more potent procedure for RNA transcript detection in situ. The improvement was achieved (1) by optimizing the circularization of padlock probes and (2) by adding the E. coli RNase III to work side by side with Phi29 DNA polymerase in target transcript-primed RCA.
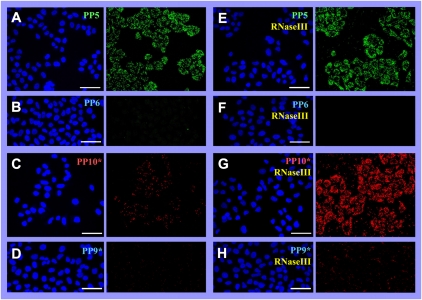
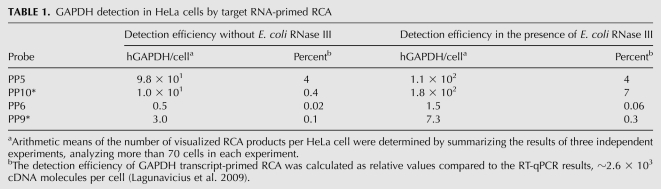
Optimizing the circularization of the specific probe PP5 by adjusting T4 DNA ligase concentration increased the detection efficiency by fourfold compared to the earlier described results. The arithmetic mean of the number of visualized specific RCA products increased from 24 (Lagunavicius et al. 2009) to 98 copies per HeLa cell in this study (the detection efficiency was ∼4%) (Fig. 3A; Table 1).
FIGURE 3.
Detection of individual human GAPDH transcripts in situ, the impact of E. coli RNase III. The reactions in situ were carried out with target transcript–padlock probe duplexes (Fig. 2A, duplexes 1,2) under the conditions described in Materials and Methods. The oligonucleotides (A,E) PP5 [complementary to the hybridization region overlapping with poly(A) tail] and (C,G) PP10* (complementary to the hybridization region in the middle of RNA molecule) were used as specific tools for hGAPDH transcript detection in situ. (B,D,F,H) The oligonucleotides PP6 and PP9* (in the ligation junction regions slightly differing from PP5 and PP10*, respectively) were used as controls for reaction specificity. The RCA reactions were carried in the absence (A–D) or presence (E–H) of E. coli RNase III. The reaction products were hybridized with oligonucleotides containing FITC or Alexa555 labels, and the slides were analyzed using TSC SP5 Confocal Microscope with 63× objectives. The images are presented as DAPI (A–H, left panels) and FITC (A,B,E,F, right panels) or Alexa555 (C,D,G,H, right panels) projections. DAPI stains nuclei blue, FITC or Alexa555 labels specific RCA products green or red, respectively. Scale bars, 50 μm.
TABLE 1.
GAPDH detection in HeLa cells by target RNA-primed RCA
However, using specific probe PP10*, which is complementary to the hybridization region distanced far from the end of the RNA molecule (Fig. 2A, duplex 2) and Alexa555-labeled hybridization probes, we have detected only few fluorescently labeled spots in cell cytoplasm (Fig. 3C). The arithmetic mean of the number of visualized specific RCA products per cell was 10-fold lower in comparison to RCA products obtained with probe PP5 and was only ∼10 copies per cell (the detection efficiency was ∼0.4%) (Table 1). The control samples with nonspecific probes PP6 and PP9* generated backgrounds of ∼0.02% and ∼0.1%, respectively, relative to the RT-qPCR results (Fig. 3B,D; Table 1).
As expected, E. coli RNase III supplementation did not influence the detection efficiency in samples with specific probe PP5 complementary to the hybridization region overlapping with the poly(A) tail (Fig. 3A,E). The arithmetic mean of the number of visualized specific RCA products was ∼1.1 × 102 copies per HeLa cell (Table 1). However, the addition of E. coli RNase III to target RNA-primed RCA with specific probe PP10* complementary to the hybridization region in the middle of the RNA molecule increased the detection efficiency by 18-fold (Fig. 3C,G). The arithmetic mean increased from 10 to 1.8 × 102 copies per HeLa cell (Table 1). Control samples with nonspecific probes PP6 and PP9* generated backgrounds of ∼0.06% and ∼0.3%, respectively, relative to the RT-qPCR results (Fig. 3F,H; Table 1).
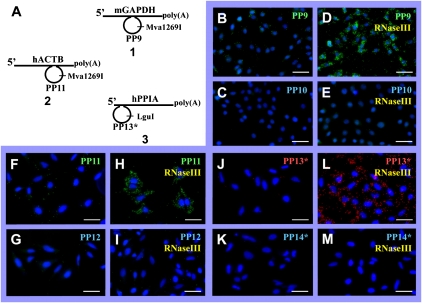
To confirm that the target RNA-primed RCA is applicable to other target RNA molecules, in situ detection of mouse GAPDH and human ACTB and PPIA transcripts was tested. As a specific padlock probe for the mGAPDH transcript detection, we used probe PP9 (Fig. 4A, duplex 1), which is complementary to the same RNA hybridization region as previously used probe PP9* (Fig. 2A, duplex 3). Probe PP10 was used as a control probe for ligation specificity. Testing hACTB and hPPIA transcripts, the specific padlock probes PP11 and PP13* were hybridized to RNA sequences in the middle of the hACTB transcript and at the 5′-end of the hPPIA transcript, 596 nt and 2078 nt away from the poly(A) tail, respectively (Fig. 4A, duplexes 2,3). The control samples with nonspecific probes PP12 or PP14*, which in the ligation junction region differ from PP11 and PP13* in a few nucleotides or a single nucleotide, respectively, were used as control probes for the ligation specificity.
FIGURE 4.
Detection of individual mouse GAPDH and human ACTB and PPIA transcripts in situ, the impact of E. coli RNase III. The reactions in situ were carried out with target transcript–padlock probe duplexes (A, duplexes 1–3) under the conditions described in Materials and Methods. The oligonucleotides (B,D) PP9 (complementary to the hybridization region in the middle of mGAPDH transcript), (F,H) PP11 (complementary to the hybridization region in the middle of hACTB transcript), and (J,L) PP13* (complementary to the hybridization region at the 5′-end PPIA transcript) were used as specific tools for target transcript detection in situ. (C,E,G,I,K,M) The oligonucleotides PP10*, PP12, and PP14 (in the ligation junction regions slightly differing from PP9*, PP11, and PP13, respectively) were used as controls for reaction specificity. The RCA reactions were carried in the absence (B,C,F,G,J,K) or presence (D,E,H,I,L,M) of E. coli RNase III. The reaction products were hybridized with the oligonucleotides containing FITC or Alexa555 labels, and the slides were analyzed using the fluorescence microscope Olympus IX70 with 20× objectives. The images are presented as the overlay of DAPI and FITC (B–I) or Alexa555 (J–M) projections. DAPI stains nuclei blue, FITC or Alexa555 labels specific RCA products green or red, respectively. Scale bars, 25 μm.
Similar to the previous observations with the hGAPDH transcript, the presence of E. coli RNase III significantly influenced detection efficiency of the mGAPDH (by sixfold) (Fig. 4B,D), hACTB (by 11-fold) (Fig. 4F,H), and hPPIA (by 16-fold) (Fig. 4J,L) transcripts when padlock probes hybridized to the RNA sequences distanced far from the 3′-end of the molecules. However, in control samples, its effect on background level was imperceptible (Fig. 4C,E,G,I,K,M).
In summary, the results in situ correlated well with the data obtained in vitro. The E. coli RNase III supplementation significantly improved the target RNA-primed RCA technique when the padlock probe hybridized to the region distanced far from the 3′-end of RNA molecule. On the other hand, the presence of the endoribonuclease specific for double-stranded RNA had no effect on the detection efficiency with the padlock probe oligonucleotide hybridizing near the 3′-end of the RNA target.
Conclusions
We have improved the target RNA-primed RCA technique for in vitro and in situ use. Previously we described the application of 3′–5′ single-stranded RNA exonucleolytic activity of Phi29 DNA polymerase in the target-RNA conversion into a primer and the following amplification of the padlock probe by the polymerization activity of Phi29 DNA polymerase, which generated specific RCA products in vitro and in situ (Lagunavicius et al. 2008, 2009). However, when the padlock probe hybridized to a region distanced far from the 3′-end, strong double-stranded structures present at the 3′-end of target RNA molecules were interfering with the single-stranded exoribonucleolytic activity of Phi29 DNA polymerase and hindering RCA product accumulation. We show that the addition of endoribonucleases specific for the double-stranded RNA can change situation significantly. We demonstrate that in such hampered cases of RCA, the E. coli RNase III efficiently assists Phi29 DNA polymerase in target RNA conversion into a primer. The positive impact of the double-stranded RNA-specific endonuclease was successfully demonstrated on the human and mouse transcripts (hGAPDH, mGAPDH) in the total RNA isolated from human cells or mouse tissues. The results in vitro correlated well with the data obtained in situ. The E. coli RNase III supplementation significantly improved the target RNA-primed RCA technique when tested with the individual human and mouse transcripts (hGAPDH, by 18-fold; mGAPDH, by sixfold, hACTB, by 11-fold; hPPIA, by 16-fold) with their specific padlock probes, which hybridized to the regions distanced far from the 3′-ends of RNA molecules. As a result, with the assistance of E. coli RNase III, the in situ detection of the inner RNA sequences located either near the end or in the middle of the RNA molecule (hGAPDH) became comparably efficient (4% and 7%, respectively). Similar detection efficiencies in situ (1%–10%) were presented previously for the DNA-primed RCA technique on the mitochondrial or chromosomal DNA targets (Larsson et al. 2004; Lohmann et al. 2007).
MATERIALS AND METHODS
Proteins, buffers, and reagents
BSA, DEPC-treated water, DNase I, DNA polymerases (Taq, Phi29), glycogen, RevertAid H Minus First Strand cDNA Synthesis Kit, REases (Mva1269I, LguI), Ribolock RNase inhibitor, T4 DNA ligase; PBS buffer (10×), SSC buffer (20×), Tango buffer (10×), TBE buffer (10×), and RNA Loading Dye Solution (2×) were products of Fermentas UAB. Fetal bovine serum was obtained from Biochrom AG; creatinphosphate, Dulbecco's Modified Eagle's medium without L-glutamine, L-glutamine, gentamycine sulphate, and polyvinyl alcohol (from Sigma); E. coli RNase III (from Ambion and Epicentre); and the RNeasy Mini Kit (from QIAGEN).
Nucleotides, oligonucleotides
[α-33P]dATP (∼3000 Ci/mmol) was purchased from Hartmann Analytic. Unlabeled dNTPs were products of Fermentas UAB. The origin and sequences of oligonucleotides DP-FITC, Mva1269I, PP1, PP2, PP5, PP6, RNA1, and RNA2 have been published previously (Lagunavicius et al. 2008, 2009). The following DNA oligonucleotides were obtained from Metabion:
DP2-Alexa555: 5′-Alexa555-ACTGTGGTGCTCTTCAGGCTG-3′;
LguI: 5′-CTGTGGTGCTCTTCAGGCGT-3′;
PP9: 5′-GCTAAGCAGTTGGTGATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAATGCGCAACCTTGGCCAGGGGG-3′;
PP9*: 5′-GCTAAGCAGTTGGTGCTGTGGTGCTCTTCAGGCGTGTTAGTTTGTACTGGTGACGACCTTGGCCAGGGGG-3′;
PP10: 5′-GCTAAGCAGTTGGTGCTGTGGTGCTCTTCAGGCGTGTTAGTTTGTACTGGTGACGACCTTGGCCAGGGGT-3′;
PP10*: 5′-GCTAAGCAGTTGGTGCTGTGGTGCTCTTCAGGCGTGTTAGTTTGTACTGGTGACGACCTTGGCCAGGGGT-3′;
PP11: 5′-TGCGGTGGACGATGGATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAATGCGCACCGCCTAGAAGCATT-3′;
PP12: 5′-AGCGGTGGACGATGGATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAATGCGCACCGCCTAGAAGCATA-3′;
PP13*: 5′-CTGCTGTCTTTGGGACTGTGGTGCTCTTCAGGCGTGTTAGTTTGTACTGGTGACGGAGCACGAAAATTTT-3′; and
PP14*: 5′-CTGCTGTCTTTGGGACTGTGGTGCTCTTCAGGCGTGTTAGTTTGTACTGGTGACGGAGCACGAAAATTTA-3′.
Target-RNA conversion into a primer for RCA
All manipulations with RNA–PP hybrids were performed exactly as described previously (Lagunavicius et al. 2008), except that some RCA reactions with the RNA2–PP1 hybrid were carried out in the presence of 0.01 U of E. coli RNase III (Epicentre).
Ligation-based target transcript detection in total RNA
Total RNA was isolated from human HeLa cells and tongue and liver tissues from mouse using the RNeasy Mini Kit and treated with DNase I. The presence of target transcripts and the absence of encoding genes were confirmed by RT-PCR experiments (data not shown).
The hybridization reaction with human total RNA was performed in 9.3 μL of reaction mixture [1× Tango, 3 nM phosphorylated oligonucleotide PP5 or PP10* (specific for hGAPDH), 0.25 μg of human total RNA] by heating for 5 min at 65°C and cooling down to room temperature. A free total RNA and its mixture with nonspecific phosphorylated oligonucleotide PP6 or PP9* (in the ligation junction region differing from PP5 and PP10* in a few or a single nucleotides, respectively) and free oligonucleotides were used in other samples as controls for reaction specificity. After hybridization, 2.5 U of T4 DNA ligase, 20 μM ATP, and 10 U of Ribolock were added to each sample, and the ligation reactions were performed for 2 h at 37°C. The RCA reactions were initiated by adding 2.5 U of Phi29 DNA polymerase and 1 mM dNTP (10 μCi [α33-P]) to each hybridization sample, in a final volume of 12.8 μL. The samples were incubated for 3 h at 37°C, and reactions were terminated for 10 min at 70°C. Some RCA reactions were performed in the presence of 0.5 U of E. coli RNase III (Ambion). Testing mGAPDH in the total mouse RNA, all procedures were performed as described above, except that the total RNA was isolated from mouse tongue or liver tissues, and the phosphorylated oligonucleotide PP9* was used as a specific probe, while oligonucleotide PP10* was used as a control, nonspecific probe.
Furthermore, the specificity of RCA products was tested. Complementary oligonucleotides containing Mva1269I or LguI REase recognition site, at 1 μM concentration, were annealed to the RCA products in 1× Tango (PP5, PP6 samples) or 2× Tango (PP9*, PP10* samples) buffer, respectively, in a final volume of 13 μL. The annealing step was performed for 5 min at 95°C, 2 min at 80°C, 2 min at 70°C, 2 min at 60°C, 2 min at 50°C, 2 min at 40°C, 2 min at 30°C, 2 min at 20°C, and 2 min at 10°C. Then each sample was supplemented with 5 U of appropriate REase, and the monomerization reaction was performed for 1 h at 37°C. Before electrophoretic analysis, all samples were treated with 200 μg/mL proteinase K in the presence of 0.4% SDS for 30 min at 37°C. Following incubation, an equal volume of 2× RNA Loading dye solution was added to stop the reaction. The samples were heated for 10 min at 95°C and placed on ice, then loaded onto the 8% (w/v) denaturing polyacrylamide gel (29:1 [w/w] acrylamide/bisacrylamide, 7 M urea, 1× TBE buffer). The gels were analyzed using the Cyclone Storage Phosphor System and OptiQuant Image Analysis Software (Packard Instruments).
Individual transcript detection in situ
The human HeLa or mouse 3T3 cells were cultured in 10-well glass slides (Electron Microscopy Sciences) at a density of 3 × 103 cells/well in Dulbecco's Modified Eagle's medium (without L-glutamine) supplemented with 10% fetal bovine serum, 0.3 mg/mL L-glutamine, and 50 μg/mL gentamycine sulfate for 24 h at 37°C in a CO2 incubator. The cells on the slide were washed with 1× PBS buffer for 3 min and fixed with 4% formaldehyde (prepared freshly from paraformaldehyde) in 1× PBS for 20 min at room temperature. After fixation the cells were washed with 1× PBS buffer for 3 min; dehydrated with 70% (5 min), 85% (3 min), and 99.9% (3 min) ethanol; and air-dried at room temperature. The human transcript hybridization reaction in the HeLa cells was performed in 30 μL/well of hybridization mixture (250 nM phosphorylated oligonucleotide PP5, PP10* [specific for hGAPDH], PP11 [specific for hACTB] or PP13* [specific for hPPIA], 475 mM Tris-HCl at pH 8; 0.95 mM EDTA, 760 mM NaCl) for 2–3 h at 37°C in a humidity chamber. The nonspecific phosphorylated oligonucleotides PP6, PP12 or PP9*, PP14* (in the ligation junction region differing from PP5, PP11 and PP10*, PP13* in a few nucleotides or a single nucleotide, respectively) were used in other samples as controls for reaction specificity. Testing mGAPDH in the 3T3 cells, all procedures were performed as described above, except that the phosphorylated oligonucleotide PP9 was used as a specific probe, while oligonucleotide PP10 was used as a control, nonspecific probe.
Furthermore, the glass slide was washed in pre-heated washing buffer (100 mM Tris at pH 7.5, 150 mM NaCl, 0.05% Tween 20) for 5 min at 37°C, dehydrated by serial washes with increasing concentrations of ethanol (70%, 85%, 99.9%, for 3 min of each) and air-dried at room temperature. The ligation of padlock probes on human transcripts was performed with 30 U of T4 DNA ligase in 30 μL/well of 1× T4 DNA ligase buffer supplemented with 0.1 mg/mL BSA and 60 U of Ribolock for 1 h at 37°C in a humidity chamber. The glass slide was washed in the pre-heated washing buffer, dehydrated, and air-dried. The RCA was performed with 30 U of Phi29 DNA polymerase in 30 μL/well of 1× Phi29 DNA polymerase buffer supplemented with 0.2 mg/mL BSA, 0.25 mM dNTP, and 30 U of Ribolock for 16 h at 37°C in a humidity chamber. Some of the RCA reactions were performed in the presence of 0.1 U of E. coli RNase III (Ambion). The glass slide was washed in the pre-heated buffer, dehydrated, and air-dried. The RCA products were detected by their hybridization with fluorescently labeled oligonucleotides DP-FITC (PP5, PP6, PP9, PP10, PP11, and PP12 samples) or DP2-Alexa555 (PP9*, PP10*, PP13*, and PP14* samples) in 30 μL/well of hybridization mixture (250 nM fluorescent oligonucleotide, 2× SSC buffer, 20% formamide, 5% glycerol) for 2 h at 37°C in a humidity chamber. The glass slide was washed in the pre-heated washing buffer twice for 5 min at 37°C, dehydrated, and air-dried. The slides were mounted in Vectashield mounting medium for fluorescence with 1.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and analyzed with fluorescence microscope Olympus IX70 and Image-Pro Plus software (Media Cybernetics) or TSC SP5 Confocal Microscope (Leica Corp.). Fluorescent-labeled RCA products were calculated using ImageJ 1.41o software (http://rsb.info.nih.gov/ij). The total number of RCA products was obtained by dividing the total fluorescent area by the average value of particle size. The number of particles in the cell was calculated by dividing the total number of RCA products by nuclei count.
ACKNOWLEDGMENTS
This work was supported in part by an EU FP6 Integrated Project “MolTools” award and EU grant for project “Molecular Biology Tools of New Generation” (Nr.VP2-1.3-ŪM-02-K-01-009). We are grateful to Dr. Lolita Zaliauskiene (Fermentas UAB) for cell lines and tissues, Laima Kazilioniene (Fermentas UAB) for the assistance in RNA isolation, and Professor Vida Kirveliene (University of Vilnius) for lending the confocal microscopy. Also we express our appreciation to Dr. Ramune Leipuviene (Fermentas UAB) for critical reading of the manuscript and linguistic help.
Footnotes
Abbreviations: ACTB, β-actin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; hGAPDH, human GAPDH; mGAPDH, mouse GAPDH; PPIA, peptidylprolyl isomerase A; RCA, rolling circle amplification; REase, restriction endonuclease.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2068510.
NOTE ADDED IN PROOF
Parallel to this study, a report was published describing an efficient detection of individual RNA molecules method in situ by RCA-based analysis of their reverse transcription products, cDNAs (Larsson et al. 2010). We describe similar results obtained by a competitive method, the direct detection of RNA transcripts by target RNA-primed RCA.
REFERENCES
- Baner J, Nilsson M, Mendel-Hartvik M, Landegren U 1998. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res 26: 5073–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber RD, Harmer DW, Coleman RA, Clark BJ 2005. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 21: 389–395 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y 2009. Highly sensitive determination of micoRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Ed 48: 3268–3272 [DOI] [PubMed] [Google Scholar]
- Christian AT, Pattee MS, Attix CM, Reed BE, Sorensen KJ, Tucker JD 2001. Detection of DNA point mutations and mRNA expression levels by rolling circle amplification in individual cells. Proc Natl Acad Sci 98: 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson O, Jarvius J, Schallmeiner E, Howell M, Nong RY, Reuter H, Hahn M, Stenberg J, Nilsson M, Landegren U 2008. A dual-tag microarray platform for high-performance nucleic acid and protein analyses. Nucleic Acids Res 36: e45 doi: 10.1093/nar/gkn106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gory S, Vernet M, Laurent M, Dejana E, Dalmon J, Huber P 1999. The vascular endothelial-cadherin promoter directs endothelial-specific expression in transgenic mice. Blood 93: 184–192 [PubMed] [Google Scholar]
- Jarvius M, Paulsson J, Weibrecht I, Leuchowius KJ, Andersson AC, Wahlby C, Gullberg M, Botling J, Sjoblom T, Markova B, et al. 2007. In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol Cell Proteomics 6: 1500–1509 [DOI] [PubMed] [Google Scholar]
- Jonstrup SP, Koch J, Kjems J 2006. A microRNA detection system based on padlock probes and rolling circle amplification. RNA 12: 1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunavicius A, Kiveryte Z, Zimbaite-Ruskuliene V, Radzvilavicius T, Janulaitis A 2008. Duality of polynucleotide substrates for Phi29 DNA polymerase: 3′ → 5′ RNase activity of the enzyme. RNA 14: 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunavicius A, Merkiene E, Kiveryte Z, Savaneviciute A, Zimbaite-Ruskuliene V, Radzvilavicius T, Janulaitis A 2009. Novel application of Phi29 DNA polymerase: RNA detection and analysis in vitro and in situ by target RNA-primed RCA. RNA 15: 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U, Nilsson M 2004. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat Methods 1: 227–232 [DOI] [PubMed] [Google Scholar]
- Larsson C, Grundberg I, Soderberg O, Nilsson M 2010. In situ detection and genotyping of individual mRNA molecules. Nat Methods 7: 395–397 [DOI] [PubMed] [Google Scholar]
- Li N, Jablonowski C, Jin H, Zhong W 2009. Stand-alone rolling circle amplification combined with capillary electrophoresis for specific detection of small RNA. Anal Chem 81: 4906–4913 [DOI] [PubMed] [Google Scholar]
- Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet 19: 225–232 [DOI] [PubMed] [Google Scholar]
- Lohmann JS, Stougaard M, Koch J 2007. Detection of short repeated genomic sequences on metaphase chromosomes using padlock probes and target primed rolling circle DNA synthesis. BMC Mol Biol 8: 103 doi: 10.1186/1471-2199-8-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March PE, Gonzalez MA 1990. Characterization of the biochemical properties of recombinant ribonuclease III. Nucleic Acids Res 18: 3293–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2005. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33: W577–W581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2008. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Mijatovic T, Houzet L, Defrance P, Droogmans L, Huez G, Kruys V 2000. Tumor necrosis factor-α mRNA remains unstable and hypoadenylated upon stimulation of macrophages by lipopolysaccharides. Eur J Biochem 267: 6004–6012 [DOI] [PubMed] [Google Scholar]
- Nilsson M, Krejci K, Koch J, Kwiatkowski M, Gustavsson P, Landegren U 1997. Padlock probes reveal single-nucleotide differences, parent of origin and in situ distribution of centromeric sequences in human chromosomes 13 and 21. Nat Genet 16: 252–255 [DOI] [PubMed] [Google Scholar]
- Nilsson M, Barbany G, Antson DO, Gertow K, Landegren U 2000. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat Biotechnol 18: 791–793 [DOI] [PubMed] [Google Scholar]
- Nilsson M, Antson DO, Barbany G, Landegren U 2001. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res 29: 578–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 12: 995–1000 [DOI] [PubMed] [Google Scholar]
- Stougaard M, Lohmann JS, Zajac M, Hamilton-Dutoit S, Koch J 2007. In situ detection of nonpolyadenylated RNA molecules using Turtle Probes and target primed rolling circle PRINS. BMC Biotechnol 7: 69 doi: 10.1186/1472-6750-7-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Bryder D, Weissman IL, Quake SR 2006. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci 103: 17807–17812 [DOI] [PMC free article] [PubMed] [Google Scholar]