Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human tumor virus that encodes 12 precursor microRNAs (pre-miRNAs) that give rise to 17 different known ∼22-nucleotide (nt) effector miRNAs. Like all herpesviruses, KSHV has two modes of infection: (1) a latent mode whereby only a subset of viral genes are expressed and (2) a lytic mode during which the full remaining viral genes are expressed. To date, KSHV miRNAs have been mostly identified via analysis of cells that are undergoing latent infection. Here, we developed a method to profile small RNAs (∼18–75 nt) from populations of cells undergoing predominantly lytic infection. Using two different next-generation sequencing platforms, we cloned and sequenced both pre-miRNAs and derivative miRNAs. Our analysis shows that the vast majority of viral and host 5p miRNAs are co-terminal with the 5′ end of the cloned pre-miRNAs, consistent with both being defined by microprocessor cleavage. We report the complete repertoire (25 total) of 5p and 3p derivative miRNAs from all 12 previously described KSHV pre-miRNAs. Two KSHV pre-miRNAs, pre-miR-K12-8 and pre-miR-K12-12, encode abundant derivative miRNAs from the previously unreported strands of the pre-miRNA. We identify several novel small RNAs of low abundance, including viral miRNA-offset-RNAs (moRNAs), and antisense viral miRNAs (miRNA-AS) that are encoded antisense to previously reported KSHV pre-miRNAs. Finally, we observe widespread antisense transcription relative to known coding sequences during lytic replication. Despite the enormous potential to form double-stranded RNA in KSHV-infected cells, we observe no evidence for the existence of abundant viral-derived small interfering RNAs (siRNAs).
Keywords: Kaposi's sarcoma-associated herpesvirus (KSHV), antisense, microRNA (miRNA), microRNA-offset-RNAs (moRNAs)
INTRODUCTION
Two classes of small RNAs that are effectors of RNA silencing are microRNAs (miRNAs) and small interfering RNAs (siRNAs) (for review, see Carthew and Sontheimer 2009). miRNAs have been the subject of enormous interest due to the regulatory roles they play in numerous and disparate biological processes, including viral infection (for review, see Sullivan and Ganem 2005; Nair and Zavolan 2006; Samols and Renne 2006; Sarnow et al. 2006; Gottwein and Cullen 2008; Grey et al. 2008; Pfeffer 2008; Sullivan 2008). miRNAs are small, ∼22-nucleotide (nt) RNAs that bind to mRNAs and regulate gene expression in a post-transcriptional manner (for review, see Bartel 2004; Kim 2005; Vasudevan et al. 2008). miRNAs are derived from longer primary transcripts (pri-miRNAs) that can be thousands of nucleotides long (Cullen 2004). pri-miRNAs contain at least one ∼90-nt sequence that folds into a characteristic hairpin secondary structure element, which is recognized and excised by the multi-protein nuclear Microprocessor complex, giving rise to an ∼60-nt hairpin called a precursor miRNA (pre-miRNA) (Bartel 2004; Kim 2005). The pre-miRNA is then exported to the cytoplasm, whereupon it is recognized and cleaved by a complex of proteins containing the RNase III-like protein Dicer. Dicer cleavage results in a transient, imperfectly complementary double-stranded ∼22-nt RNA, of which one strand becomes stably incorporated into the RNA-induced silencing complex (RISC). Typically, one strand is preferentially retained in RISC (the guide strand), and the other strand (the star [*] strand) is barely detectable due to degradation. Once a miRNA is active within RISC, the complex binds to target mRNA transcripts and affects protein expression.
Recently, two additional classes of small RNA derivatives have been reported from pre-miRNA loci. microRNA-offset-RNAs (moRNAs, or moRs) were originally described as an abundant class of small RNAs present in the simple chordate Ciona intestinalis, where almost 50% of all described miRNA loci also express moRNAs (Shi et al. 2009). moRNAs arise from the base of the pre-miRNA hairpin stem, are ∼22-nt long, and are thought to be processed by the RNAi RNase III components. moRNAs have also been reported to exist in mammalian cells, albeit the frequency of loci that encode them, and their absolute abundance are considerably lower than in C. intestinalis (Langenberger et al. 2009). Additionally, antisense miRNAs (AS-miRNAs) have been described that derive from hairpins formed from complementary transcripts to existing pre-miRNAs. AS-miRNAs are not simply “noise” derivatives of antisense transcription, since several studies report a role for AS-miRNAs in regulating homeotic gene expression in Drosophila and possibly mammals (Stark et al. 2008; Tyler et al. 2008). Recently, AS-miRNAs have been reported in two herpesviruses, MCMV and HSV-1 (Buck et al. 2007; Dolken et al. 2007; Umbach et al. 2008).
siRNAs are ∼21-nt RNAs that are loaded into RISC and can also effect gene silencing. Unlike most miRNAs, siRNAs direct a specific, irreversible cleavage of their target mRNAs. siRNAs are thought to be RNAi effectors that guard against invasive genetic elements such as transposons and viruses (Ding and Voinnet 2007; Carthew and Sontheimer 2009). Synthetic siRNAs have shown great utility as laboratory reagents for knocking down the expression levels of particular genes of interest in mammalian cells, including viral gene products (Elbashir et al. 2001; Gitlin et al. 2002). While all miRNAs are generated from a single-stranded primary transcript (with subportions folding into imperfect double-stranded hairpins), endogenous siRNAs are generated from longer, perfectly complementary double-stranded RNA (dsRNA) transcripts. In mammalian cells, there are hundreds of known endogenous transcripts that code for miRNAs; however, there are only a few clear examples of endogenously derived siRNAs (Babiarz et al. 2008; Watanabe et al. 2008).
So far, there have been no reports of viral-specific siRNAs being produced during infection of mammalian cells (Cullen 2006). However, some viruses encode miRNAs within their genomes. Viral miRNAs have been identified from viruses with DNA genomes, including those of the Herpes, Polyoma, and Asco virus families (Nair and Zavolan 2006; Samols and Renne 2006; Gottwein and Cullen 2008; Grey et al. 2008; Hussain et al. 2008; Sullivan 2008; Sullivan and Cullen 2009). Most viral miRNAs have been identified by sequencing of small RNA libraries (Pfeffer et al. 2004; Cai et al. 2005, 2006; Dunn et al. 2005; Samols et al. 2005; Burnside et al. 2006; Dolken et al. 2007; Yao et al. 2007; Umbach et al. 2008), but computational methods have played a key role in the discovery of numerous viral miRNAs (Grey et al. 2005; Pfeffer et al. 2005; Sullivan et al. 2005, 2009; Cui et al. 2006; Grundhoff et al. 2006; Buck et al. 2007; Sullivan and Grundhoff 2007; Hussain et al. 2008; Seo et al. 2008). Viral miRNAs are capable of targeting host- or viral-encoded transcripts, some of which have been shown to function in various pro-viral processes such as evading the host immune response and autoregulation of viral gene expression (Sullivan 2008). However, the function of most of the approximately 140 known viral miRNAs remains unknown.
Most of the known viral miRNAs derive from members of the Herpes family. All herpesviruses undergo two characteristic modes of replication, latent and lytic. During latent infection, only a small subset of viral gene products is expressed. Alternatively, the full repertoire of viral genes is expressed during lytic infection, culminating in the production of infectious virions and lysis of the cell. Most herpesviral miRNAs were identified from cells that were predominantly undergoing latent infection. However, miRNAs have been detected from viruses undergoing productive lytic infection with HCMV, MCMV, HSV-1, HSV-2, MDV-1, and MDV-2 viruses (Pfeffer et al. 2005; Burnside et al. 2006; Cui et al. 2006; Yao et al. 2007; Tang et al. 2008). Interestingly, accumulating evidence suggests that some viral miRNAs will play a role in regulating the switch from latent to lytic infection (Grey et al. 2007; Murphy et al. 2008; Cullen 2009).
KSHV is a member of the gamma sublineage of the Herpesvirus family. KSHV is the etiologic agent of rare B cell malignancies and Kaposi's sarcoma, a highly vascularized skin lesion found in some people with immunodeficiencies (for review, see Ganem 2007). Like all herpesviruses, KSHV has a large, double-stranded DNA genome (∼160 kb) (Russo et al. 1996). KSHV encodes more than 85 protein-coding genes, and at least 12 pre-miRNAs that give rise to at least 17 different miRNAs (16 different 5p or 3p miRNAs, and a single-nucleotide-edited miRNA) that are highly conserved (Marshall et al. 2007). The pre-miRNAs are clustered in a single ∼4-kb region of the genome and are encoded in a polycistronic fashion from any of several different transcripts made during latent infection (Pearce et al. 2005; Cai and Cullen 2006). Two of the pre-miRNAs found in this cluster, pre-miR-K12-10 and pre-miR-K12-12, are also encoded by an additional transcript that is transactivated during lytic infection (Sullivan and Cullen 2009). Thus, all known KSHV miRNAs are expressed during latent infection, and those miRNAs derived from pre-miR-K12-10 and pre-miR-K12-12 have been shown, or are expected to be further induced during lytic replication. Detailed functional roles of the KSHV miRNAs during bona fide infection are lacking. However, several mRNA targets that are repressed by the KSHV miRNAs have been identified (Gottwein et al. 2007; Samols et al. 2007; Skalsky et al. 2007; Whitby 2009; Ziegelbauer et al. 2009). Some of these targets include host genes that play a role in angiogenesis, transformation, and apoptosis, suggesting that the KSHV miRNA may be involved in the pathogenesis associated with KSHV infection.
Most of the KSHV pre-miRNAs (11 of 12) were originally identified by sequencing small RNA libraries from cells predominantly undergoing latent infection (Cai et al. 2005; Pfeffer et al. 2005; Samols et al. 2005). One exception to this is that the Renne group has performed small-scale sequencing (a few hundred total amplicons) from a library of small RNAs obtained from TPA-treated cells that have an elevated level of lytic infection (Samols et al. 2005). Additionally, one of the KSHV pre-miRNAs, pre-miR-K12-12, was not cloned, but rather was identified by computational methods, and its derivative 5p arm was confirmed by microarray and Northern blot analyses of RNA isolated from cells undergoing latent infection (Grundhoff et al. 2006). Because all of the known KSHV miRNAs cluster within a 4-kb region of the genome that is transcribed during latency, it is reasonable to speculate that additional KSHV miRNAs may exist in the remaining majority of the genome that is actively transcribed during lytic infection. In addition, given that many KSHV transcripts made during lytic replication are expressed at high levels and have head-to-head orientations and overlapping regions, it is possible that lytic replication may trigger an RNAi response that could be probed for by mining for KSHV-specific siRNAs. So far, however, there have been no reports that have examined in depth whether KSHV encodes small RNAs that are specific to the lytic cycle. Therefore, using the 454 and SOLiD platforms (see G Hannon, “Cloning small RNAs for sequencing with 454 technology,” http://genoseq.ucla.edu/images/a/a9/SmallRNA.pdf) (Applied Biosystems 2009), we have deep-sequenced small RNA libraries from cells that are predominantly undergoing latent or lytic infection. Our approach involves making multiple libraries of different size classes allowing us to systematically clone pre-miRNAs as well as derivative miRNAs. This furthermore allows us to identify regions of the genome that are specifically enriched for miRNA/siRNA sizes over other (random degradation) size classes. These studies reveal a surprising wealth of transcription throughout the entire KSHV genome and map the entire repertoire of KSHV miRNAs and star sequences, some of which are abundant but have so far escaped cloning. We also systematically clone and map the viral pre-miRNAs. In addition, we report the first viral-encoded moRNA and KSHV-encoded AS-miRNAs. Surprisingly, KSHV AS-miRNAs appear to be predominantly expressed during lytic infection, representing lytic-specific KSHV miRNAs. Finally, we identify numerous regions of the genome encoding antisense transcripts with the potential to form dsRNAs, yet we observe no convincing evidence of KSHV-specific siRNAs. The implications of these findings to the KSHV replication cycle are discussed.
RESULTS
Small RNA profiling reveals regions of the genome are enriched for 19–23-nt RNAs
We set out to determine if KSHV encodes siRNAs or miRNAs specific to lytic replication. A caveat to studying lytic replication in primary effusion lymphoma (PEL) cells is that the process of viral replication can lead to abundant, nonspecific, general degradation of RNA. We reasoned that one way to differentiate regions of the KSHV genome that were enriched for siRNAs and miRNAs over random degradation fragments would be to compare the number of KSHV 19–23 base-pair (bp) reads versus other size classes present in the libraries for a given region of the genome (Fig. 1B). This logic assumes that regions of the genome that are more prone to random degradation will have increases in other size classes of small RNAs, not just those that correspond to siRNAs and miRNAs. Conversely, regions of the genome enriched for siRNA or miRNA production should show an increase in the ratio of 19–23-mer RNAs over the other size classes.
FIGURE 1.
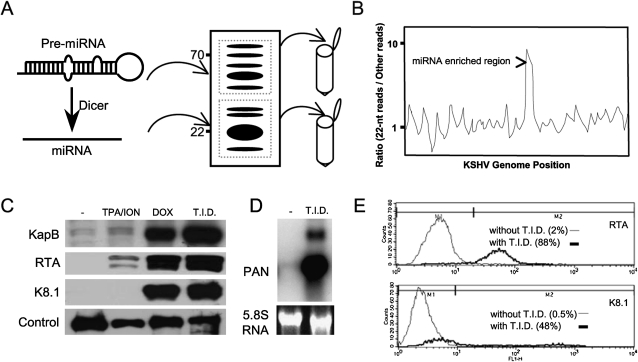
Experimental strategy employed. (A) Two different sizes of small RNAs, 18–25 nt, enriched for miRNAs, and 45–75 nt, enriched for pre-miRNAs, were size-fractionated by denaturing polyacrylamide gel electrophoresis, ligated to linkers, and then converted to cDNA. After amplification, the cDNAs were sequenced by ABI SOLiD. Computational methods were used to map reads corresponding to the KSHV genome (NC_009333.1), and (B) the desired size class (19–23 nt) was plotted over other sizes present in the library to determine regions of the genome enriched for specific classes of small RNAs (hypothetical plot shown). (C) Western immunoblot analysis demonstrating induction of markers for lytic replication with various drug regimens for 48 h. The TREx–RTA BCBL-1 cells are highly inducible for initiation of lytic replication upon treatment with TPA, Ionomycin (ION), and Doxycyclin (DOX) (T.I.D., treatment with all three drugs). KapB, RTA, and K8.1 correspond to KSHV proteins that are known to be induced during lytic replication. (Control) Loading control, a >80-kDa band that cross-reacts with the anti-K8.1 antibody. (D) Northern blot analysis shows that the Polyadenylated Noncoding RNA (PAN), a lytic-specific transcript, is robustly induced when treated with triple drug regimen (T.I.D.) for 48 h. (E) Flow cytometric analysis shows the percent of individual cells expressing RTA or the late lytic protein K8.1. The dark line indicates cells treated with the triple drug regimen, T.I.D.
For most of our experiments, we used an inducible BCBL-1-derived cell line (TREx BCBL-RTA) developed by Jung and colleagues (Nakamura et al. 2003). Recently, this cell line was shown to be highly inducible into lytic replication by the addition of TPA, ionomycin, and doxycylcine (T.I.D.) (Arias et al. 2009). The BCBL-1 cell line (and its derivatives) is infected with KSHV and was originally derived from PEL cells from a patient with advanced HIV disease (Renne et al. 1996). Treatment of the TREx BCBL-RTA cells, as previously described (Arias et al. 2009), results in strong induction of lytic gene expression as judged by multiple assays. Immunoblot analysis for proteins specific to (or enriched for) lytic replication (RTA, Kaposin B, and K8.1) shows robust induction (Fig. 1C). Northern blot analysis shows a strong induction of the noncoding RNA PAN (Fig. 1D), which is only expressed during lytic replication (Sun et al. 1996; Zhong et al. 1996). Finally, flow cytometric analysis of cells for the master lytic switch protein RTA (∼88% cells positive), or a marker of late gene expression, K8.1 (∼48% cells positive), clearly shows a large enrichment in the fraction of cells that are initiating the lytic cascade of gene expression (Fig. 1E).
We next size fractionated total RNA from the latent population of cells or cells enriched for lytic replication. We isolated RNA from two regions of the denaturing gel, including RNA of ∼18–26 nt that should include miRNAs, random degradation fragments, and possibly siRNAs, as well as the RNA that is ∼45–75 nt in length that should include pre-miRNAs, random degradation fragments, and possibly other classes of noncoding RNAs (Fig. 1A). These RNA were ligated to linkers, reverse transcribed, and subjected to deep sequencing using the SOLiD platform (Applied Biosystems 2009). In addition, we examined regular BCBL-1 cells (obtained from ATCC). Untreated BCBL-1 cells predominantly undergo latent infection, whereas treatment with sodium butyrate increases the proportion of cells undergoing lytic infection (Renne et al. 1996). We generated small RNA libraries from both treated and untreated BCBL-1 cells. Immunofluorescent and flow cytometric analyses of sodium butyrate-treated cells for the viral protein K8.1 confirmed an enrichment of cells undergoing lytic infection (data not shown). Total RNA was harvested from each population of cells (treated and untreated), and RNAs in the size range of ∼16–70 nt were isolated. Appropriate linkers were added, and after reverse transcription, the cDNAs were sequenced on a 454 pyrophosphate sequencer (see G Hannon, “Cloning small RNAs for sequencing with 454 technology,” http://genoseq.ucla.edu/images/a/a9/SmallRNA.pdf). We obtained a combined total of approximately 250,000 reads for the 454 analysis of the BCBL-1 cells (∼0.43% mapping to KSHV genome) (Supplemental Table S1) and approximately 98 million reads from SOLiD sequencing of the TREx BCBL-RTA cells (∼5% mapping to the KSHV genome) (Table 1). Because of the significantly greater depth attained by the SOLiD analysis, much of this paper utilizes these data, but several findings are buttressed by the 454 analysis (see below).
TABLE 1.
The mapping results from the ABI small RNA analysis pipeline (v0.5)
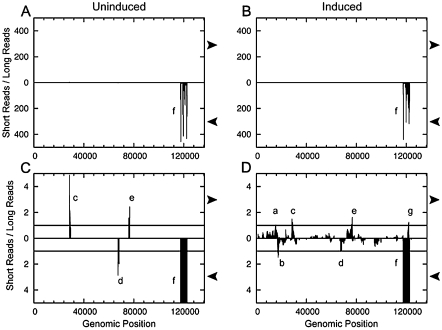
We filtered the results to identify those reads mapping to the KSHV genome. Plotting the fraction of SOLiD reads that were 19–23 nt in length relative to the reads in the larger size class (∼45–75 nt) revealed a single region of the genome greatly enriched for miRNA/siRNA-sized RNAs (Fig. 2A,B). Notably, this part of the genome (nucleotide positions 117,780–122,161), encodes the previously described KSHV polycistronic miRNA cluster. The vast majority of the reads in this region are in the same orientation as the KSHV miRNAs. We can detect distinct start sites of the 5p and 3p miRNAs for all 12 KSHV pre-miRNA loci (Supplemental Fig. S1; discussed below). This result validates the ability of our approach to detect viral miRNAs from cells undergoing latent or lytic infection. Near identical results were obtained (albeit with much less coverage) when the 454 data were analyzed in a similar manner (Supplemental Fig. S2). Combined, these data suggest that no other regions of the KSHV genome encode miRNAs expressed to the same level as the previously described polycistronic miRNA cluster.
FIGURE 2.
Plotting the ratio of 19–23-nt RNAs over other size classes mapping to the KSHV genome identifies regions enriched for small RNAs of interest. (A) Uninduced. (B) Induced. (C) Uninduced, 100× zoom on y-axis. (D) Induced, 100× zoom on y-axis. RNA libraries from the uninduced (A,C) or lytic-induced (B,D) RNA were analyzed. (A,B) Low-resolution view demonstrates the extreme enrichment of reads mapping to the KSHV pre-miRNA loci (labeled “f”). (C,D) Zooming in reveals additional regions enriched for 19–23-nt RNAs (a–e, g). Window analysis of reads that mapped to the KSHV genome was plotted using a window length of 500 nt (W = 500; see Materials and Methods section for more details). Regions with a greater number of short reads than long reads marked “a–f” were selected for further analysis. Arrows at the side of plots indicate the orientation of reads that are plotted (rightward arrows indicate top strand transcripts, leftward-facing arrows indicate the bottom strand transcripts).
Cloning of the KSHV pre-miRNAs and all the derivative 5p and 3p miRNAs
Previous studies have identified 12 KSHV pre-miRNAs (Cai et al. 2005; Pfeffer et al. 2005; Samols et al. 2005; Grundhoff et al. 2006) that give rise to at least 17 different derivative miRNAs (Griffiths-Jones 2006). We mined our SOLiD sequencing data to address three questions: (1) Can we clone the pre-miRNAs that give rise to the known KSHV miRNAs? (2) Can we define the 5p and 3p derivatives of all the KSHV pre-miRNAs? (3) Are any previously missed KSHV miRNA derivatives likely to be biologically active?
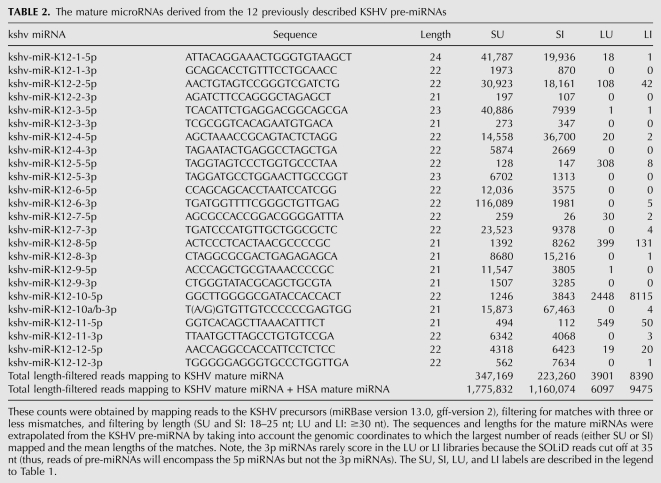
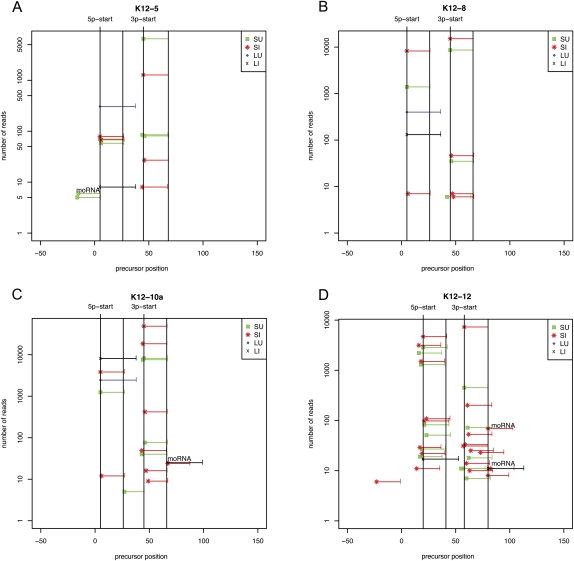
To our knowledge, previous studies have not attempted to systematically clone the pre-miRNAs represented in a population of viral-infected cells. We used computational analysis to identify the amplicons present in our larger size class library (45–75 nt) that map to known human or KSHV pre-miRNA loci (Table 2; data not shown). We next mapped derivative pre-miRNA (from the large uninduced [LU] or large induced [LI] for lytic replication) and miRNA reads (from the short uninduced [SU] or short induced [SI] for lytic replication onto individual, specific miRBase-annotated pre-miRNA sequence positions) (Fig. 3; Supplemental Fig. S4). This analysis identified the KSHV pre-miRNA for 10 of the 12 known pre-miRNAs (Fig. 3; Supplemental Fig. S4). Interestingly, some of the KSHV pre-miRNAs were significantly more abundant than others, but pre-miRNA abundance did not necessarily correlate with miRNA abundance (Figs. 3, 4; Supplemental Fig. S4). Of note, the pre-miR-K12-5 was the fourth most abundant KSHV pre-miRNA that was cloned despite the fact that its derivative miRNAs are among the lowest KSHV miRNAs represented in our library. This observation is consistent with previous reports showing that the pre-miRNA for the BCBL-1-encoded miR-K12-5 is inefficiently processed into miRNAs (Gottwein et al. 2006). For both viral and human pre-miRNAs, the 5′ end of the most abundant pre-miRNA amplicons was almost always co-terminal with the 5′ end of the derivative 5p miRNA. Consistent with the current Microprocessor model (Seitz and Zamore 2006), these data strongly suggest that processing of the pre-miRNA defines the 5′ end of the derivative 5p miRNAs. Thus, pre-miRNAs are “clonable,” and our 45–75-nt libraries (LU and LI) contain pre-miRNAs, while our 19–23-nt libraries (SU and SI) contain derivative 5p and 3p miRNAs. Consistent with this, our 454 library also contained a small number of pre-miRNAs whose sequences were represented in their entirety (Supplemental Fig. S3).
TABLE 2.
The mature microRNAs derived from the 12 previously described KSHV pre-miRNAs
FIGURE 3.
Read coverage of four exemplary KSHV miRNAs and pre-miRNAs. Reads were mapped to a position on the miRBase-annotated hairpins. Note: The miRBase-annotated hairpins are longer than the actual pre-miRNA hairpins that are “liberated” by microprocessor cleavage. Pre-miRs-K12-8 and 12 both express abundant 5p and 3p derivative miRNAs that were missed in previous studies. Pre-miRs-K12-5, 10a, and 12 have flanking moRNAs. The “x” axis indicates the nucleotide position relative to the predicted longer hairpin precursor that is listed in miRBase. The “y” axis indicates the quantity of reads for a particular pre-miRNA derivative and is plotted on a log scale to show lower-abundance derivatives. The horizontal lines (green, red, blue, black) indicate the nucleotide position coverage of a particular read. The four vertical lines denote the start site and end site of the 5p and 3p miRNAs. (SU) The “small” RNA library (19–24 nt) from uninduced cells; (SI) the “small” RNA library from cells induced to initiate lytic replication; (LU) the “large” RNA library (45–75 nt) from uninduced cells; (LI) the “large” RNA library from cells induced to initiate lytic replication. The reads corresponding to the pre-miRNAs (LU, LI) have a maximum length of 35 nt (the upper cut-off of read length available from the current SOLiD platform). Reads corresponding to moRNAs are indicated with text above the relevant horizontal lines.
FIGURE 4.
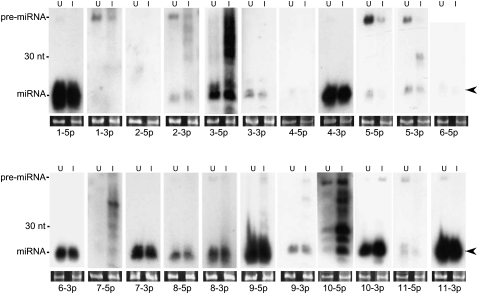
Northern blot analysis of KSHV-encoded miRNAs demonstrates that the 5p and 3p miRNAs are detectable from most KSHV pre-miRNA loci. For a few pre-miRNAs, only one strand was detectable as an miRNA (pre-miR-K12-1, 2, 4, and 6). For the others, the miRNAs were abundant enough that we could detect both the 5p and 3p derivatives. The left lanes (U) correspond to RNA from untreated cells (predominantly latent), the right lanes (I) correspond to RNA from induced cells enriched for lytic replication. Arrows indicate miRNAs migrating at ∼22 nt. The loading control, shown in bottom panels, is ethidium bromide-stained low-molecular-weight RNA.
Some KSHV “star” miRNAs* are abundant and biologically active
Typically, each pre-miRNA gives rise to two predominant products, the 5p and 3p miRNAs, derived from each arm of the pre-miRNA hairpin. Typically one arm (the “miRNA guide strand”) is favored to be stably incorporated within RISC and is more abundant than the other arm derivative. The less abundant derivative, (the “star miRNA [*] passenger strand”) is not typically thought to be the driver of biological activity. However, unlike most host-derived miRNAs, several viral pre-miRNAs give rise to abundant 5p and 3p miRNAs, and at least some viral star miRNAs are biologically active (Sullivan et al. 2005; Seo et al. 2008). As previously mentioned, KSHV is known to encode 17 derivative miRNAs from 12 pre-miRNAs. We searched our libraries to determine if any previously unannotated KSHV miRNA sequences are represented. All 25 5p and 3p derivatives (including the single edited miRNA) were cloned and mapped in our libraries (Fig. 3; Table 2; Supplemental Fig. S4). As expected, many of the miRNA derivatives represented in our libraries that previously escaped detection are of low abundance and are presumably * sequences (miRs-K12-1-3p, 2-3p, 5-5p, 7-5p, 10-5p, and 11-5p). Nonetheless, many of the KSHV miRNA* derivatives are more abundant than some nonstar human miRNAs (data not shown). Furthermore, most KSHV miRNA derivatives represented in our libraries are detectable by Northern blot (Fig. 4). Of note, K12-9-5p was previously annotated as the star derivative (Griffiths-Jones 2006), but is the more abundant amplicon represented in our libraries and is robustly detectable by Northern blot analysis (Fig. 4; Table 2). Interestingly, both K12-8-5p and K12-12-3p had previously escaped detection but are extremely abundant in our libraries and robustly detectable by Northern blot analysis (Figs. 4, 5; Table 2). Combined, these data argue that some KSHV miRNA derivatives that were previously unreported are abundant and might have biological activity.
FIGURE 5.
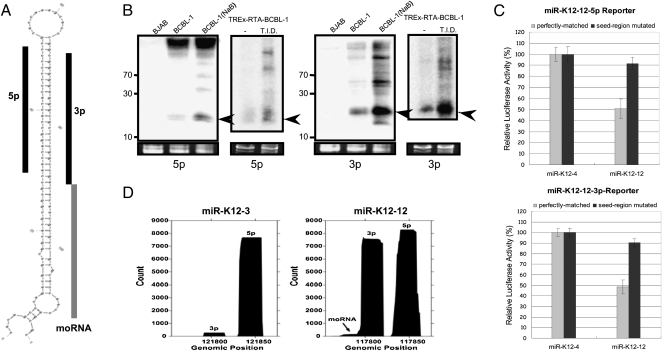
Multiple specific small RNAs derived from pre-miR-K12. (A) Predicted secondary structure of pre-miR-K12-12. The two derivative miRNAs, K12-12-5p (Griffiths-Jones 2006) and K12-12-3p (reported in this study), are indicated with black bars. The K12-12 moRNA is indicated with a gray bar. (B) Northern blot analysis of miR-K12-12 5p and miR-K12-12-3p. Two models of lytic induction were analyzed. RNA from regular BCBL-1 cells that were predominantly undergoing latent infection (untreated) or treated with sodium butyrate (NaB) to induce lytic replication was used. RNA from the nonisogenic, uninfected KSHV-negative BJAB cells serves as a negative control. Additionally, RNA from untreated, predominantly latent, or triple drug-treated T.I.D. (lytic-enriched) TREx-RTA BCBL-1 cells was analyzed. Arrows indicate miRNA band. (C) Luciferase assay demonstrates that both miRNA derivatives of pre-miR-K12-12 are active. Renilla luciferase reporter vector with four copies of miR-K12-12-5p or miR-K12-12-3p target sites were co-transfected with the pre-miR-K12-4 or pre-miR-K12-12 expression vectors. As a negative control, an identical reporter containing mutations in the seed complementary region was also analyzed. A normalization vector (firefly luciferase vector) was also co-transfected as a control for transfection efficiency. (D) Histogram of read coverage from the small induced (18–25, SI) library shows the relative abundance of miR-K12-12-5p, miR-K12-12-3p, and K-12 moRNA. 3p derivative miRNAs correspond to the left peak; 5p derivative miRNAs correspond to right peak. A histogram of reads mapping to pre-miR-K12-3 is shown for comparison.
To test whether K12-12-3p could have biological activity, we constructed luciferase reporters that have binding sites complementary to either the 5p or 3p K12-12 derivatives, or negative control reporters with mutations in the region complementary to the miRNA seed. Transfection of an irrelevant construct encoding miR-K12-4 had little effect on luciferase activity from either the K12-5p and 3p reporters or the negative control reporters (Fig. 5C; data not shown). However, transfection of a vector encoding the K12-12 pre-miRNA reduced luciferase expression from both reporter vectors by ∼50%. An additional negative control experiment, whereby the construct encoding the K12-12 pre-miRNA was transfected with an irrelevant luciferase reporter containing binding sites to the JCV viral miRNA (Seo et al. 2008), showed no effect on luciferase activity (data not shown). Combined, these data argue that the effects of the K12-12 5p and 3p miRNAs are specific. However, we note that until bona fide mRNA targets are identified, the relative activity of K12-12 5p and 3p during infection is uncertain. Thus, the K12-12-3p miRNA is abundant and likely active in KSHV-infected cells.
Discovery of a viral-encoded moRNA
moRNAs are derived from the extreme ends of pre-miRNA loci, and in mammalian cells, are typically of significantly lower abundance than the miRNA derivatives (Langenberger et al. 2009; Shi et al. 2009). So far, moRNAs have not been reported to be encoded by viruses. Most of the KSHV pre-miRNA loci (11 out of 12) did not show abundant read patterns indicative of moRNAs (Fig. 5D; Supplemental Figs. S1, S4). However, a clear exception was pre-miR-K12-12 (Fig. 5D). As is typical for most of the KSHV pre-miRNAs, the histogram of reads covering the pre-miR-K12-12 locus shows distinct peaks for the 5p and 3p derivative miRNAs. Additionally, a small peak, consistent with a moRNA derivative, is evident at the 3′ end of pre-miR-K12-12 (Fig. 5D). The number of reads corresponding to the moRNA was relatively few, around 100 total reads. This number is at the lower end of what we observe for the host miRNAs that are represented in our libraries, and much lower than the K12-5p and 3p miRNAs (consistent with the low processing efficiencies reported for other mammalian-derived moRNAs) (Langenberger et al. 2009). Repeated attempts to express this moRNA using the plasmid construct that expresses pre-miR-K12 (and miRNA derivatives) failed to demonstrate a moRNA-corresponding band via Northern blot analysis (data not shown). This suggests that the K12 moRNA is very inefficiently processed and/or not amenable to exogenous expression and raises the possibility that moRNAs are nonfunctional miRNA processing intermediates. Finally, we note that pre-miR-K12-4, 5, 6, and 10 also contain reads consistent with 3′ or 5′ moRNAs (Fig. 3; Supplemental Fig. S4). We conclude that KSHV encodes multiple, albeit low-abundance, moRNAs.
Discovery of KSHV antisense miRNAs expressed during lytic replication
From our earlier size ratio analysis (Fig. 2A,B), it was clear that no regions of the KSHV genome encoded miRNAs as abundant as the previously described polycistronic miRNA cluster. To explore the possibility that less abundant RNAi-associated RNAs (miRNA/siRNAs) may be expressed during infection, we altered the scale of our previous analysis to include regions of the genome that showed only slight enrichment in 19–23-nt RNAs over other size classes (Fig. 2C,D). As before, 19–23-nt RNAs from the smaller library were compared with other size classes (45–75 nt, from the larger library). At this scale, a general trend across the KSHV genome is evident, whereby most regions are enriched for the non-miRNA-size classes (45–75-nt RNAs). We do not detect regions of the genome that are enriched for 19–23-nt RNAs in both the sense and antisense orientations—that would be indicative of siRNAs. We therefore conclude it is unlikely that abundant siRNAs derived from longer antisense dsRNAs are robustly made during KSHV lytic infection.
During lytic replication, seven regions of the genome show peaks where 19–23-nt RNAs of a single orientation are over-represented during lytic infection (Fig. 2D, “a–g”). We applied an additional filter to weed out potentially artifactual peaks that required each region to also be enriched for 19–23-nt RNAs over >24-nt RNAs present in the small library (another filter for random degradation fragments). This added filter removed region “e” from further consideration. Peak “f” is readily accounted for as the KSHV latent miRNA cluster. Of the remaining peaks, only “d” and “g” had read patterns that were consistent with miRNAs (reads mapping of the appropriate miRNA size and confined to one or a few 5′ and 3′ termini). Peaks “a–c” will be addressed in the Discussion section. Peak “d” maps to the coding region of ORF 45. Northern blot analysis for this region was inconsistent (data not shown), and the flanking regions of this peak are not predicted to fold into a pre-miRNA-like structure (data not shown). Therefore, we abandoned further experiments in this region of the genome.
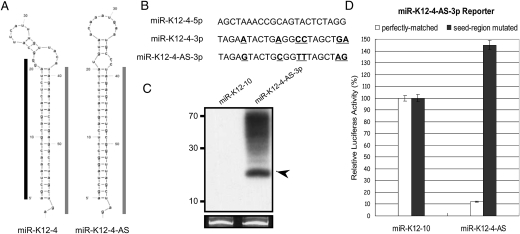
Peak “g” is notable because it encompasses a predominantly 22-nt RNA that maps antisense to miR-K12-4-5p (Fig. 6A). This fact caught our attention since previous reports have identified antisense miRNAs in other herpesviruses (Buck et al. 2007; Dolken et al. 2007; Umbach et al. 2008). As this putative miRNA differs from miR-K12-4-3p by only 6 nt (Fig. 6B), we examined whether there was any preference for sequencing errors in these six residues that would be suggestive of misannotation due to a computational artifact. We observed no preference for mismatches at these particular six nucleotides (data not shown), suggesting that the reads corresponding to the AS-miRNA are not simply sequencing errors of miR-K-12-4-3p. Next, we predicted the secondary structure of transcripts from this region. A hairpin secondary structure is predicted antisense to pre-miR-K12-4 (Fig. 6A). The location of the AS-miRNA maps to a position on the predicted hairpin precursor that is completely consistent with pre-miRNA structure and derivative miRNA processing (Fig. 6A). To determine if transcripts that span this region could be efficiently processed into miRNAs, we subcloned the portion of the genome corresponding to the predicted pre-miRNA hairpin and flanking regions (∼200 bp in total) into a heterologous expression vector. We transfected the vector and harvested total RNA. Northern blot analysis confirmed robust and specific processing of this miRNA (Fig. 6C). We then assayed whether this miRNA has biological activity within the RISC complex. A reporter containing perfectly complementary binding sites, or a negative control reporter containing 5-nt point mutations (complementary to the seed region) was co-transfected with the miRNA-expressing vector. Luciferase assays conducted on lysates harvested from these cells demonstrate robust and specific knockdown of the target reporter (Fig. 6D). These data show that driving transcription of this region of the genome leads to efficient processing of a pre-miRNA and biological activity of the derivative miRNA within RISC. We conclude that this is a bona fide, low-expressed, lytic-replication-specific miRNA. Accordingly, we name this miRNA “miR-K12-4-AS.”
FIGURE 6.
Expression and activity of an antisense miRNA complementary to the pre-miR-K12-4 locus. (A) Predicted secondary structure of the pre-miR-K12-4 and pre-miR-K12-4-AS. Mapped 5p derivative miRNAs are indicated with a black bar; mapped 3p derivative miRNAs are indicated with a gray bar. (B) Sequences of miR-K12-4 and miR-K12-4-AS. Nucleotide differences between K12-4-3p and K12-4-AS-3p are indicated in bold/underline. (C) Northern blot analysis of miR-K12-4-AS. Cells were transfected with a vector expressing either miR-K12-10 or miR-K12-4-AS. RNA was harvested and hybridized with a probe complementary to miR-K12-4-AS. Arrow indicates miRNA band. (D) miR-K12-4-AS is active within the RISC complex. Expression of a plasmid expressing miR-K12-4-AS down-regulates expression of a reporter containing complementary binding sites, but not a control reporter in which the seed complementary region is mutated.
We next asked whether other KSHV pre-miRNA loci might encode AS-miRNAs. Three additional KSHV AS-miRNAs were identified—two from the 5p or 3p arms of the same AS-pre-miRNA, pre-miR-K12-2-AS (Supplemental Fig. S5). We named these for the KSHV miRNA loci they are encoded complementary to: “K12-8-AS-5p,” “K12-2-AS-5p,” and “K12-2-AS-3p.” We also uncovered reads for at least one AS-pre-miRNA, pre-miR-K12-8, complementary to miR-K-12-8 (Supplemental Fig. S5). These results are satisfying because they strongly imply that similar to host miRNAs, the KS AS-miRNAs are derived via normal Microprocessor/Dicer miRNA machinery processing.
Small RNA profiling reveals transcripts throughout the genome originating from both the bottom and top strands
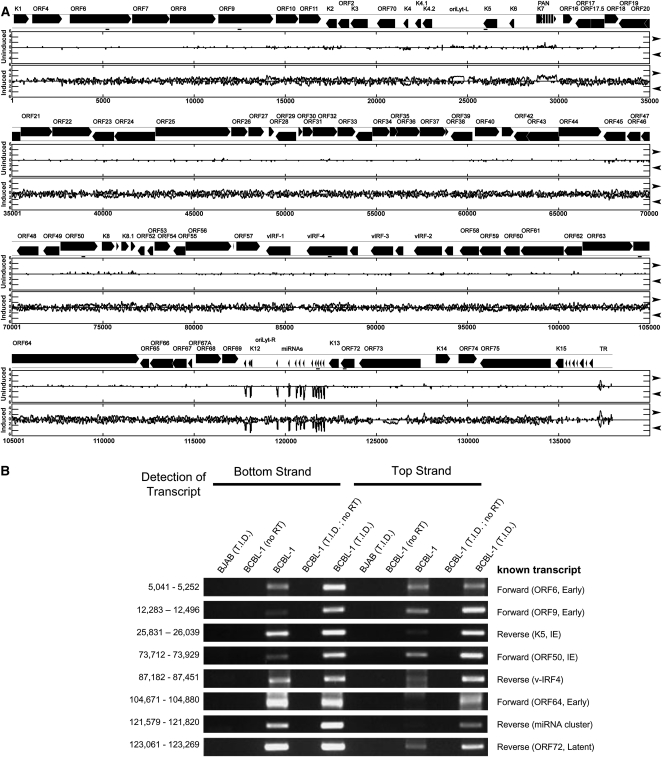
The discovery of KSHV AS-miRNAs is noteworthy since it implies the existence of previously unknown longer precursor transcripts that are encoded antisense and complementary to the KSHV miRNA cluster. To determine if such transcripts exist, we turned to a coverage plot, whereby all the transcripts represented in our libraries were plotted in the bottom or top strand orientations (Fig. 7A). Most of the reads we observe from the latently infected cells correspond to the KSHV miRNA cluster (Fig. 7A). Since much of the genome is only transcriptionally active during lytic replication, it was not surprising to observe a dramatic increase in the number of reads obtained from cells undergoing lytic replication (Fig. 7A). Presumably, many of these are random degradation fragments of larger transcripts. Strikingly, during lytic replication, we observe antisense transcription in regions spanning almost the entire genome. This includes the regions of the genome that encode the latent transcripts and the latent miRNA cluster, which previously were not known to encode antisense transcripts. To confirm that antisense transcription occurs in many regions of the genome, we used directional reverse transcription, followed by PCR. Ten regions of the genome were chosen that were only known to encode transcripts of a single orientation (Supplemental Fig. S6; data not shown). Additionally, for a positive control, we included a region of the genome encompassing a portion of ORF50/RTA known to encode bottom and top strand transcripts (Lukac et al. 1999; Saveliev et al. 2002). Directional RT-PCR confirmed that eight of these regions (including the ORF50/RTA control) encode detectable sense and antisense transcripts during lytic replication (Fig. 7B). Those regions that scored positive for antisense transcription had to meet several criteria: first, there had to be a distinct band of the appropriate size in cells undergoing lytic replication; second, there had to be no band detectable from several different negative controls including: RNA harvested from lytic cells in which RT was omitted from the reaction, RNA harvested from uninfected cells (BJAB) that were treated with the same induction drug regimen, and RNA from cells undergoing lytic replication that was primed with any of 10 different irrelevant RT primers from alternative regions of the KSHV genome (Supplemental Fig. S6). Importantly, directional RT-PCR confirmed that the miRNA cluster region of the genome not only undergoes robust transcription in the same orientation as the miRNAs, but also detectable transcription antisense to the latent KSHV miRNAs (Fig. 7B). Thus, by several criteria, unknown transcripts exist that are antisense to the KSHV miRNA cluster that could give rise to the AS-pre-miRNAs. Furthermore, lytic replication produces numerous previously unknown transcripts that are antisense to the abundant, previously characterized KSHV transcripts.
FIGURE 7.
Antisense transcription is detected throughout the KSHV genome. (A) Coverage plot of all reads mapping to the KSHV genome. Sequenced frequency of reads that mapped to the KSHV genome for the combined short and long fractions of uninduced and induced samples is plotted on a log10 scale on the “y” axis. The “x” axis shows KSHV genomic position. Each graph is divided into top strand transcripts (top, rightward arrow) and bottom strand transcripts (bottom, leftward arrow) as indicated on the right side of the figure. Locations of confirmatory directional RT-PCR products are indicated with a small bar/line below the genome map (positions 5041–5252; 12,283–12,496; 25,831–26,039; 73,712–73,929; 87,182–87,451; 104,671–104,880; 121,579–121,820; 123,061–123,269). Genome annotations are based on NCBI reference sequence NC_009333.1. (B) Directional RT-PCR confirms existence of previously unreported antisense transcription during KSHV lytic infection. A positive control known to have antisense transcription (ORF50; 73,712–73,929) or seven regions previously only known to have transcription in one orientation were tested for antisense transcription. RNA from uninfected BJAB cells was used as a negative control. The orientation of the strand being primed by the RT primer is indicated at the top of the figure. A “no RT” control was included to exclude the possibility of genomic DNA contamination. To control for possible false positives due to RT mis-priming, all regions shown scored negative in PCR assays when any of 10 different control RT primers were used (Supplemental Fig. S6).
DISCUSSION
Identification of previously undescribed abundant miRNA derivatives
In this study, we utilized two different methods to generate our small RNA libraries and sequenced each by either the 454 or SOLiD sequencing techniques. Sequencing at this depth, we were able to identify eight new derivative miRNAs encompassing all 25 derivatives that are known or would be predicted to be encoded by the previously reported 12 KSHV pre-miRNAs (Table 2). Notably, based on Northern blot analysis and relative amplicon numbers, two of the previously unreported KS miRNAs represented in our libraries, K12-8-5p and K12-12-3p, are highly abundant (Figs. 4, 5; Table 2). At present, we do not understand why miR-K12-8-5p has escaped previous detection. miR-K12-12-3p contains a BAN I restriction site that was used in the studies that identified 11 of the 12 abundant KSHV pre-miRNAs and derivatives (Cai et al. 2005; Pfeffer et al. 2005; Samols et al. 2005). Therefore, miRK-12-12-3p was likely dropped out of the libraries during the cloning procedures. Previously, miR-K12-12-5p was uniquely identified by an approach that relied on computational prediction, followed by scoring positive on a custom microarray and then Northern blot analysis (Grundhoff et al. 2006). Because miR-K12-12-3p is a particularly GC-rich miRNA, it was filtered out as part of the pipeline of microarray analysis utilized in this study; therefore, it was not discovered at that time by Northern blot analysis (Grundhoff et al. 2006). In our present study, miR-K12-12-3p is more abundant or at least as abundant as miR-K12-12-5p (Fig. 5B), is fully functional in RISC (Fig. 5C), and may be an important driver of the biological activity associated with this pre-miRNA. What is the function of miR-K12-12-3p? Currently, its function is unknown, but it is notable that the pre-miR-K12-12 and pre-miR-K12-10 derivatives are the only viral miRNAs expressed at noticeably increased levels during lytic activation (Figs. 4, 5B; Cai et al. 2005). These data are consistent with both miRNAs being encoded within the body of the Kaposin transcripts that are up-regulated by RTA transactivation during lytic replication (Sadler et al. 1999; Chang et al. 2002). These observations point to a likely important role during lytic replication or perhaps even a role during the initial times of de novo infection (if these miRNAs are somehow packaged within the virion). Future studies are required to determine the relevant functions of pre-miR-K12-10 and pre-miR-K12-12 derivatives and whether they target viral or host transcripts during KSHV infection.
Other KSHV miRNAs did not display robust changes in the relative amplicons that were consistent between both the 5p and 3p derivatives (Table 2). However, Northern blot analysis clearly demonstrated that some KSHV miRNAs show slightly increased or decreased levels during lytic replication (Fig. 4). Examining host miRNAs that were differentially represented in the SOLiD library by Northern blot analysis revealed that only slightly more than half of the miRNAs changed in abundance as would be expected if relative amplicon number were reflective of actual abundance (data not shown). Thus, relative amplicon abundance shows a general trend of congruence but not an absolute or linear correlation with actual steady-state levels. While some host miRNAs change in abundance during lytic replication, it remains to be determined whether these changes are caused by a specific viral gene product(s), the host innate antiviral response, or rather, represent a bystander effect of the drug regimen used to induce the lytic cycle (or the associated stress of lytic replication). Because of the possible cloning and sequencing biases that may exist between different libraries (Linsen et al. 2009), we take these combined results to mean that changes in relative amplicon number between different libraries represent only a starting point for identifying bona fide changes in miRNA steady-state levels.
Systematic cloning of viral and host pre-miRNAs provides insight into general miRNA processing
Because our experimental strategy profiles a range of RNA size classes, we were able to clone and map the 5′ ends of numerous pre-miRNAs. Using the high-quality-filtered SOLiD data, we were able to identify 10 of the 12 KSHV pre-miRNAs (Table 2). Additionally, using less stringent filters, we were also able to identify the remaining two KSHV pre-miRNAs in our library (data not shown). We also cloned and mapped the 5′ end of numerous human host pre-miRNAs (data not shown). Furthermore, despite the low coverage of our 454 data, we were able to clone and map the entire transcript (including 5′ and 3′ ends) of a few host and viral pre-miRNAs (Supplemental Fig. S3). Several conclusions can be made from these data. First, as would be predicted with the Microprocessor model of miRNA processing (Denli et al. 2004; Gregory et al. 2004; Chendrimada et al. 2005; Seitz and Zamore 2006), the 5′ ends of the pre-miRNAs are almost always co-terminal with the 5′ ends of the 5p miRNAs. Furthermore, our limited in depth searches into 24 viral and host pre-miRNAs (Fig. 3; data not shown) did not identify any examples of nontemplated 5′ modifications. Combined, these data strongly suggest that Drosha cleavage, as part of the Microprocessor complex, determines the final 5′ end of most miRNAs. Our data argue that cloning and profiling of pre-miRNAs are readily achievable and may be of future use for confirmation that candidate miRNAs are, indeed, derived from Microprocessor activity (see discussion below regarding KSHV AS-miRNAs), or for globally profiling post-transcriptional regulation of specific primary miRNA transcripts.
A method for identifying regions of a genome enriched for specific functional classes of RNAs identifies novel KSHV-encoded small RNAs
We developed an experimental strategy for identifying genomic regions enriched for RNAs of a particular size and functional class (Figs. 1A,B, 2). By plotting the ratio of the size class of interest to other sizes in the library (which presumably represents mostly degradation fragments of longer transcripts), we identified a region of the genome enriched for a novel AS-miRNA (e.g., miR-K12-4-AS) (Fig. 6; Supplemental Fig. S5). Such an approach may be particularly useful when dealing with cells undergoing lytic viral replication, as these discrete transcripts were identified amidst a sea of random degradation fragment noise caused by KSHV lytic activation (Fig. 7A). We also identified an additional low-abundance small RNA, the K12-12 moRNA, which we believe to be the first viral moRNA to be reported. Thus, the combination of deep sequencing and size-class analysis was fruitful for identifying novel KSHV-encoded small RNAs.
One question that arises is whether the K12-12 moRNA is the only KSHV-encoded moRNA? In fact, we observe evidence that pre-miRs-K12-4, 5, 6, and 10 also encode moRNAs (Fig. 3C; Supplemental Fig. S4D–F,J,L), albeit at very low abundance. This suggests that additional viral pre-miRNA loci could be processed into moRNAs at low efficiencies (or that cloning biases excluded them from our libraries). At present we have no functional understanding of these viral moRNAs, but the seemingly increased abundance of the K12-12 moRNA suggests this would be a good starting point for future functional studies.
At present we do not understand why miR-K-12-4-AS is represented so much more in our library than the other KSHV AS-miRNAs (>100 reads for miR-K-12-4-AS [on par with several host-encoded miRNAs] versus <10 reads for miR-K-12-2-AS-5p, miR-K-12-2-AS-3p, and miR-K-12-8-AS). There are multiple reasons that could explain this. First, it is possible that only pre-miR-K12-4 can form the proper secondary structure to allow efficient miRNA processing when transcribed from the opposite strand. We note that the predicted secondary structures of the pre-miRNAs for the KSHV AS-miRNAs we cloned appear to be more plausible as Microprocessor substrates than KSHV AS-pre-miRNA derivatives not represented in our libraries (with the caveat that predicting Microprocessor/Dicer substrates in silico is highly error-prone) (data not shown). It is possible that there is a novel promoter upstream of miR-K-12-4-AS that could allow for transcripts that specifically encode this pre-miRNA, but this model is not supported by the close proximity of miR-12-4 to its two closest neighbors, miR-K12-5 (separated by ∼85 nt) and miR-K12-3 (separated by ∼60 nt). Additionally, our studies show that antisense transcripts exist throughout the KSHV latent miRNA cluster (Fig. 7), making this model less likely. Finally, it is possible some of the other KSHV loci might encode more abundant AS-miRNAs, but cloning biases exclude them from our libraries. Our transfection studies demonstrating robust expression and activity of the K12-4-AS-miRNA (Fig. 6C,D) argue that the relatively low number of amplicons we observe during lytic infection is most likely due to a low amount of transcription of the pri-miRNA. Future experiments will address in detail the biogenesis and functions of the KSHV AS-miRNAs during lytic infection.
Currently, the functions (if any) of all of these low-abundance, small KSHV-encoded RNAs are unknown. All are represented by numerous amplicons and have discrete ends suggestive of specific processing. Due to our ability to isolate intermediates in the miRNA biogenesis pathway (pre-miRNAs and 5p and 3p derivatives of the same pre-miRNA) (Supplemental Fig. S4), we have high confidence that the KSHV AS-miRNAs we reported are derived via specific host miRNA machinery processing. For miR-K-12-4-AS we furthermore demonstrate biological activity of this miRNA in an RISC cleavage assay (Fig. 6). The K12-12 moRNA displays the features of other moRNAs (discrete ends mapping to the termini of pre-miRNA hairpins), so it is likely that it shares similar processing with the other known mammalian moRNAs (Langenberger et al. 2009). At present, we do not have sufficient read depth to make the same definitive conclusions about the other putative KSHV moRNAs (K12-4, 5, 6, and 10 moRNAs). Future studies will address the possible functional relevance of these RNAs.
Lytic replication triggers antisense transcription that is detected at discrete loci throughout the KSHV genome
Our analysis demonstrated a striking degree of antisense transcription throughout the entire KSHV genome during lytic replication (Fig. 7). This observation is perhaps not surprising, given the precedence set by lytic infection with other viruses such as adenovirus and human cytomegalovirus (Maran and Mathews 1988; Zhang et al. 2007). However, to our knowledge, this degree of antisense transcription has not been previously reported for KSHV. These transcripts are not reflective of a constant degree of background transcription that is simply up-regulated during lytic infection, since the ratio of antisense transcripts observed (and not just absolute abundance) is dramatically altered during lytic replication (data not shown). Such prevalent amounts of antisense transcripts have a great potential to form dsRNAs. These observations may help to explain why KSHV has evolved numerous countermeasures to deal with the cellular interferon immune response that can be activated by dsRNA (Coscoy 2007; Areste and Blackbourn 2009; Bisson et al. 2009; Lee et al. 2009). In addition, these results suggest that the virus will have to deal with the issue of RNA polymerase exclusion and may, in fact, use antisense transcription to regulate viral gene expression. Despite the depth of our libraries and the success of our methodology for enriching for specific size classes of small RNAs, we did not detect any strong evidence for the existence of abundant viral siRNAs during lytic infection. A few regions of the genome expressed a high ratio of ∼21-nt RNAs relative to other size classes that would be expected for siRNAs (including regions “a”–“c” from Fig. 2D). But these regions mostly encode RNAs in the same orientation as the abundant, annotated, longer transcripts derived from these regions (in fact, peak “c” encompasses reads mapping in the same direction as PAN, the most abundant KSHV transcript made during infection). Thus, our results are not consistent with abundant siRNAs being derived from longer antisense transcripts. The great majority of the genome is not enriched for small RNAs of the siRNA size class, and in the few regions that do contain appropriately sized RNAs, these RNAs are not of opposing orientations. While we cannot rule out the possibility that inverted repeat hairpin formation from single-stranded transcripts leads to some siRNA generation, sequence gazing in these regions failed to demonstrate likely repeat hairpin precursor structures. Therefore, we conclude that viral-specific siRNAs during KSHV infection are either (1) not clonable, (2) of extremely low abundance, or (3) not made. However, in the absence of performing our studies in Dicer knockout cells, we cannot rule out with absolute certainty the possibility that KSHV infection generates low amounts of siRNAs.
Conclusions
In this study, we have identified eight new derivatives from the previously described KSHV-pre-miRNAs. Given that many of these are more abundant than some cellular miRNAs, it is possible that these newly identified miRNAs may have functions relevant to viral infection. We also describe numerous specifically processed small RNAs of low abundance. One of these, miR-K12-4-AS, represents a KSHV miRNA that is specifically expressed during lytic replication and active within the RISC complex. Future studies are required to determine the functional relevance of these RNAs. We demonstrate a striking coverage of antisense transcription throughout the KSHV genome during lytic replication. These data are represented by small RNAs, ranging from 18 to 75 nt in size, that span almost every macroscopic region of the genome. Recently, there has been much excitement with the realization that a majority of the mammalian genome is transcribed—including overlapping sense and antisense transcripts (Maeda et al. 2006; Mattick and Makunin 2006; Birney et al. 2007). As is true with their hosts, it is clear that several DNA viruses are also transcribed in a similar manner. The challenge now is to parse out those noncoding transcripts that are functionally relevant from those that are just noise. Given the emerging similarities between the host and the relatively simpler DNA viral genomes, it seems that viruses will once again serve as valuable models for understanding the hosts they infect.
MATERIALS AND METHODS
Cell culture and RNA isolation
BJAB and BCBL-1 (Renne et al. 1996) cells were maintained in RPMI 1640 medium supplemented with 10% FBS, glutamine, and β-mercaptoethanol. TREx-RTA BCBL-1 cells (Nakamura et al. 2003) were maintained in DMEM medium supplemented with 20% FBS and Hygromycin B (50 μg/mL). To induce KSHV lytic replication in BCBL-1 cells, tetradecanoyl phorbol acetate (TPA; final concentration 20 ng/mL) or sodium butyrate (final concentration 3 mM) was added to the medium 48 h before RNA isolation. To induce KSHV lytic replication in TREx-RTA BCBL-1, Doxycyclin (2 μg/mL), TPA (20 ng/mL), and Ionomycin (500 ng/mL) (referred to as “T.I.D.”) were added to the medium (Arias et al. 2009). Induction of BCBL-1 cells into the lytic cycle was confirmed by flow cytometric analysis and immunofluorescence microscopy staining using antibody that recognizes K8.1, a late gene product, as the marker. Induction of TREx-RTA BCBL-1 cells into the lytic cycle was confirmed by flow cytometric analysis for RTA and K8.1; Northern blot analysis for polyadenylated nuclear RNA; and Western immunoblot analysis for RTA, K8.1, and Kaposin B. Total RNA was isolated by using PIG-B solution as described (Weber et al. 1998).
Small RNA library generation and computational analysis of sequencing reads for 454
Four hundred micrograms of total RNA from uninduced or sodium butyrate-treated BCBL-1 cells was separated by a 8% urea denaturing polyacrylamide gel, and the ∼16–70-nt band was excised, crushed, and soaked in 20 mL of 0.3 M NaOAc (pH 5.2) overnight at 4°C. The supernatants were concentrated via centrifugation with the vivaspin 15R concentrator (Sartorius) at 3000g for ∼30 min at 4°C. The RNA was then ethanol-precipitated by adding 100% ethanol and air-dried. The 3′ ends of the size-fractionated RNA were ligated to the linker (5′-rAppCTGTAGGCACCATCAAT/3ddC/-3′) using T4 RNA ligase (Ambion) overnight at 4°C. Ligated RNAs were separated from unincorporated linkers by a 15% denaturing polyacrylamide gel and extracted as described (Sullivan and Ganem 2005). The 3′ modified RNAs were then treated with tobacco acid pyrophosphatase (Epicentre) to remove the cap (if present) on the 5′ end, and then ethanol-precipitated by adding 100% ethanol. An adapter (ACGGAATTCCTCACTGAG, where RNA nucleosides are underlined) was ligated to the 5′ ends of RNA using T4 RNA ligase (Ambion) overnight at 4°C. After ethanol precipitation by adding 100% ethanol, the RNA was reverse-transcribed using the primer ATTGATGGTGCCTACAG in a 40-μL reaction with 200 units of SuperScript II reverse transcriptase (Invitrogen) for 1 h at 42°C. The RT product was PCR-amplified using the forward primer ACGGAATTCCTCACTGAG and the reverse primer ATTGATGGTGCCTACAG for 15 cycles and then amplified using the forward primer GCCTCCCTCGCGCCATCAGACGGAATTCCTCACTGAG and the reverse primer GCCTTGCCAGCCCGCTCAGATTGATGGTGCCTACAG for 12 cycles. The PCR samples were then directly analyzed via 454 deep sequencing (Roche) (Margulies et al. 2005).
To create a KSHV density maps, the KSHV genome (NC_009333) was divided into 2000-bp segments, each overlapping neighboring segments by 1000 bp, to create a BLAST database. Total sequencing reads were first BLASTed against the KSHV genome and then size-filtered into 19–23-nt or non-19–23-nt classes (∼16–18-nt and ∼25–70-nt) based on the number of nucleotide matches. Each subset was then BLASTed against the split KSHV genome database. The threshold of the E-value for this and following BLAST searches was 10−4. These parameters will detect sequences with up to 2-nt mismatches. The number of sequencing reads that matched to each binned genomic segment was plotted on an XY scatter chart corresponding to the location in the genome. To check the relative abundance of KSHV miRNAs, total sequencing reads were first BLASTed against the KSHV genome, size-filtered into the 19–23-nt class based on the number of nucleotide matches, and then BLASTed against all miRBase-annotated (Griffiths-Jones 2006) KSHV-miRNAs. To check the relative abundance of cellular miRNAs, 18–25-nt sequencing reads were BLASTed against the human pre-miRNA database acquired from miRBase. Thus, the number of reads listed for a particular cellular miRNA (Supplemental Table S1) encompasses both the 5p and 3p miRNAs derived from that pre-miRNA.
Small RNA library generation and computational analysis of sequencing reads for SOLiD
Three hundred fifty micrograms of total RNA from uninduced or T.I.D.-treated TREx-RTA BCBL-1 cells was separated by a 15% urea denaturing polyacrylamide gel, and the ∼18–25-nt and ∼45–75-nt size ranges were excised according to the RNA size markers, crushed, and soaked in 30 mL of 1 M NaCl overnight at 4°C. The supernatants were concentrated via centrifugation with the Vivaspin 15R concentrator (Sartorius) at 3000g for ∼30 min at 4°C. The RNA was then ethanol-precipitated by adding 100% ethanol and air-dried. The four RNA populations were then dissolved and converted into cDNA libraries suitable for SOLiD sequencing using a commercially available kit (Small RNA Expression Kit; Ambion). Briefly, degenerate double-stranded sticky-end adapters were ligated to the single-stranded RNA, with 5′- and 3′-specific adapters, distinguished by sequence, so RNA sequence orientation can be determined upon sequencing. The RNA was reverse-transcribed and treated with RNase H, and fragments of the appropriate length were selected by PAGE. An additional barcode sequence was appended to the 3′ end of these single-stranded cDNAs via PCR of these RNase H–treated samples with primers complementary to the 5′ and 3′ adapters. A final agarose gel purification was performed to isolate full-length library product. During library construction, the location of gel excisions was adjusted to account for the longer 45–70-nt population.
ABI SOLiD sequencing was performed according to the manufacturer's instructions on the SOLiD version 3.0 instrument and associated protocols. Quantitative PCR was used to measure each barcoded cDNA library, which were then pooled in equimolar amounts. Emulsion PCR was performed on this pool according to the manufacturer's instructions to generate beads ready for deposition and sequencing on the ABI SOLiD. The five base barcodes of each sequencing bead were read first, followed by 35 bases of the corresponding cDNA sequence. Software provided by ABI and custom Perl scripts were used to separate the reads based on their barcode sequences and discard any reads with eight or more bases with a quality value of less than 8, resulting in the number of reads shown in Table 1.
The reads for each sample were then processed by the small RNA analysis pipeline (v0.5) available at solidsoftwaretools.org. Briefly, this pipeline (1) removed reads matching human ribosomal or tRNA and reads containing no template; (2) matched each read to the known human and KS microRNA precursor regions in the human and KSHV genomes (NCBI36 and EMBL U75698.1, respectively, as annotated by the Sanger miRBase version 13.0, gff-version 2) wherein the match starts with a short seed and progressively maps until finding the adapter sequence; and (3) matched all remaining reads to the human and KS genomes (NCBI36 and NC_009333.1, respectively). Reads not falling within the expected size ranges (18–24 nt for small RNA, >30 nt for large RNA) were removed from further analysis to minimize artifacts from degradation, low sequence quality, or inaccurate mapping. Custom Perl scripts were then used to extract start site and length information for each read and to extract mature miRNA counts from the mappings to miRBase precursors.
Read size class analysis
Two size classes were defined as small (19–23 nt) from short fractions and large (45–75 nt) from long fractions. The number of mapped reads of each size class was summed in a sliding window of size W nucleotides across the genome in 1-nt increments. Windows with less than 20 short reads or less than 1 long read were discarded. The value for each window was then computed as the number of small reads divided by the number of large reads.
Western immunoblot analysis
Cells were lysed with RIPA buffer (0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA, 150 mM NaCl, and 10 mM Tris at pH 7.2). Fifty micrograms of the total lysate was separated in 12% SDS-polyacrylamide gels and transferred to PVDF membranes (Millipore). Primary antibodies used in this paper are rabbit anti-RTA antibody (a gift from Don Ganem, UCSF), rabbit anti-Kaposin B antibody (a gift from Craig McCormick, UCSF), and mouse anti-ORF K8.1 A/B antibody (Advanced Biotechnologies, Inc.). Blots were probed with a 1:1000 of primary antibody in 5% dehydrated milk in Tris Buffered Saline (TBS) and 1:3000 dilution for the HRP-conjugated secondary antibodies (Invitrogen). Blots were washed in TBS multiple times, incubated with chemiluminescent substrate (SuperSignal West Pico; Thermo Scientific) according to the manufacturer's protocol, and exposed to autoradiography film for visualization of bands.
Northern blot analysis
Small RNA Northern blot analysis was performed as described (Grundhoff et al. 2006). The probe sequences used in this paper are listed in Supplemental Table S2. Northern blot analysis for Polyadenylated Noncoding RNA (PAN) was conducted using total RNA that was separated on a 1% agarose formaldehyde gel and then transferred using Whatman TurboBlotter Rapid Downward Transfer Systems. Probes and hybridization for PAN were conducted via an identical strategy used for the small RNA Northern blot analysis (see Supplemental Table S2).
Flow cytometry
Cells were collected 48 h after mock treatment or treatment with lytic replication-inducing drugs. Cell were washed with cold PBS buffer, fixed, and permeabilized with ice-cold 1:1 acetone/methanol for 5 min, and then blocked with 1% BSA containing 1× phosphate-buffered saline (PBS) solution for 10 min. Primary antibodies against RTA and K8.1 were added to the solution and incubated for 1 h at room temperature. Cells were washed with 1× PBS buffer three times and stained with Alexa Fluor 488-conjugated secondary antibodies (Invitrogen). The working concentration of primary and secondary antibodies was a 1:1000 dilution. Stained samples were analyzed by flow cytometry with a FACSCalibur flow cytometer. The data were analyzed with Cell Pro Software (Becton Dickinson). Dead cells, as determined from aberrant forward/side scatter, were eliminated from the analysis.
pre-miRNA structure prediction
Secondary structures of pre-miRNAs were predicted by the Mfold RNA folding prediction web server (Mathews et al. 1999; Zuker 2003).
Vector construction, transfection, and luciferase assay
The pcDNA3.1-pre-miR-K12-10, pcDNA3.1-pre-miR-K12-12, pcDNA3.1-pre-miR-K12-4, and pcDNA3.1-pre-miR-K12-4-AS expression vectors were made by cloning an ∼200-bp fragment with the entire pre-miRNA hairpin and flanking regions into the pcDNA3.1 vector. The miR-K12-12-5p reporter, miR-K12-12-3p reporter, and miR-K12-4-AS-3p reporter were made by cloning four copies of the complementary sequences of the miRNAs into the pcDNA3.1dsRluc vector (Seo et al. 2008). The seed-region-mutated reporters contain engineered point mutations from the second nucleotide to the sixth nucleotide complementary to the 5′ end of the relevant miRNA. The primers used for cloning are listed in Supplemental Table S2. All sequences were confirmed by Sanger sequencing. 293T cells were plated in 12-well plates and transfected with Lipofectamine 2000 reagent (Invitrogen). Cells were transfected with miRNA expression vectors along with the reporter. The pcDNA3.1Luc2CP vector was also co-transfected as a control for transfection efficiency. Cells were collected 48 h after transfection and analyzed with the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's instructions. The luciferase assays were read on a Lumioskan Ascent luminometer (Thermo Electronic Cooperation). Results for the miRNA reporters are presented with the Renilla luciferase levels normalized by firefly luciferase levels.
Directional RT-PCR
The total RNA from BJAB or induced TREx-RTA BCBL-1 cells was treated with Turbo DNase (Ambion) according to the manufacturer's instructions. Three hundred nanograms of DNase-treated RNA was reverse-transcribed to cDNA with avian myeloblastosis virus (AMV) reverse transcriptase (Finnzymes) using specific RT primers for 50 min at 64°C and for 10 min at 60°C. One-twentieth of the RT product was used for PCR amplification using specific primers and analyzed by agarose gel electrophoresis. No RT control was included to confirm that the bands detected were not due to contaminating DNA. All the RT primers and PCR primers are listed in Supplemental Table S2.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the members and friends of the Sullivan laboratory and Alan Silverman (Roche) for helpful conversations regarding this work; Don Ganem (UCSF), Britt Galunsigner (UC Berkeley), and Craig McCormick (Dalhousie University) for numerous KSHV reagents; Jae Jung (USC) for the kind gift of the TREx-RTA-BCBL-1 cells; and the Whitely and Dudley laboratories (UT Austin) for use of their luminometers. This work was supported by UT Austin start-up funds, a UT Austin Institute for Cellular and Molecular Biology fellowship, and the Runners-up Prize for the 2007 Roche Gigabase Sequencing Grant competition to CSS.
NOTE ADDED IN PROOF
Li et al. (2009) recently reported a new class of small, ∼17-nt RNAs encoded by KSHV. Unfortunately, due to size fractionation, such small RNAs were likely omitted from our libraries. While this manuscript was under review, in agreement with our results, Umbach and Cullen (2010) detected sequencing reads consistent with KSHV-encoded moRNAs and AS-miRNAs.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1967910.
REFERENCES
- Applied Biosystems 2009. Whole-genome discovery and profiling of small RNAs using the SOLiD System. BioTechniques 46: 232–234 [Google Scholar]
- Areste C, Blackbourn DJ 2009. Modulation of the immune system by Kaposi's sarcoma-associated herpesvirus. Trends Microbiol 17: 119–129 [DOI] [PubMed] [Google Scholar]
- Arias C, Walsh D, Harbell J, Wilson AC, Mohr I 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog 5: e1000334 doi: 10.1371/journal.ppat.1000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson SA, Page AL, Ganem D 2009. A Kaposi's sarcoma-associated herpesvirus protein that forms inhibitory complexes with type I interferon receptor subunits, Jak and STAT proteins, and blocks interferon-mediated signal transduction. J Virol 83: 5056–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P 2007. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol 81: 13761–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, Green PJ, Jain N, Isaacs G, Morgan RW 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J Virol 80: 8778–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol 80: 2234–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci 102: 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2: e23 doi: 10.1371/journal.ppat.0020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Shedd D, Gradoville L, Cho MS, Chen LW, Chang J, Miller G 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J Virol 76: 3168–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat Rev Immunol 7: 391–401 [DOI] [PubMed] [Google Scholar]
- Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol 80: 5499–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2004. Transcription and processing of human microRNA precursors. Mol Cell 16: 861–865 [DOI] [PubMed] [Google Scholar]
- Cullen BR 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol 7: 563–567 [DOI] [PubMed] [Google Scholar]
- Cullen BR 2009. Viral and cellular messenger RNA targets of viral microRNAs. Nature 457: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O 2007. Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol 81: 13771–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol 7: 1684–1695 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Ganem D 2007. Kaposi's sarcoma-associated herpesvirus. In Fields virology (ed. Knipe DM and Howley PM), 5th ed, pp. 2875–2888 Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Gitlin L, Karelsky S, Andino R 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418: 430–434 [DOI] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cai X, Cullen BR 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol 80: 5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450: 1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol 79: 12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog 3: e163 doi: 10.1371/journal.ppat.0030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Hook L, Nelson J 2008. The functions of herpesvirus-encoded microRNAs. Med Microbiol Immunol 197: 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S 2006. miRBase: The microRNA sequence database. Methods Mol Biol 342: 129–138 [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Taft RJ, Asgari S 2008. An insect virus-encoded microRNA regulates viral replication. J Virol 82: 9164–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN 2005. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Langenberger D, Bermudez-Santana C, Hertel J, Hoffmann S, Khaitovich P, Stadler PF 2009. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics 25: 2298–2301 [DOI] [PubMed] [Google Scholar]
- Lee HR, Kim MH, Lee JS, Liang C, Jung JU 2009. Viral interferon regulatory factors. J Interferon Cytokine Res 29: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B 2009. Characterization of viral and human RNAs smaller than canonical microRNAs. J Virol 83: 12751–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Lukac DM, Kirshner JR, Ganem D 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol 73: 9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, et al. 2006. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet 2: e62 doi: 10.1371/journal.pgen.0020062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maran A, Mathews MB 1988. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology 164: 106–113 [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V, Parks T, Bagni R, Wang CD, Samols MA, Hu J, Wyvil KM, Aleman K, Little RF, Yarchoan R, et al. 2007. Conservation of virally encoded microRNAs in Kaposi sarcoma–associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis 195: 645–659 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV 2006. Noncoding RNA. Hum Mol Genet 15: R17–R29 [DOI] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc Natl Acad Sci 105: 5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair V, Zavolan M 2006. Virus-encoded microRNAs: Novel regulators of gene expression. Trends Microbiol 14: 169–175 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol 77: 4205–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M, Matsumura S, Wilson AC 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J Virol 79: 14457–14464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S 2008. Viral miRNAs: Tiny but mighty helpers for large and small DNA viruses. Future Virol 3: 291–298 [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734–736 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2: 269–276 [DOI] [PubMed] [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2: 342–346 [DOI] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci 93: 14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol 73: 5722–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Renne R 2006. Virus-encoded microRNAs: A new chapter in virus–host cell interactions. Future Virol 1: 233–242 [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol 79: 9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog 3: e65 doi: 10.1371/journal.ppat.0030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA 2006. MicroRNAs: Expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol 4: 651–659 [DOI] [PubMed] [Google Scholar]
- Saveliev A, Zhu F, Yuan Y 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299: 301–314 [DOI] [PubMed] [Google Scholar]
- Seitz H, Zamore PD 2006. Rethinking the microprocessor. Cell 125: 827–829 [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS 2008. Evolutionarily conserved function of a viral microRNA. J Virol 82: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Chen CJ, Sullivan CS 2009. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383: 183–187 [DOI] [PubMed] [Google Scholar]
- Shi W, Hendrix D, Levine M, Haley B 2009. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol 16: 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 81: 12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev 22: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS 2008. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet 9: 503–507 [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Cullen BR 2009. Noncoding regulatory RNAs of the DNA tumor viruses. In DNA tumor viruses (ed. Damania B and Pipas JM), Chap. 25, pp. 645–681 Springer Science, New York [Google Scholar]
- Sullivan CS, Ganem D 2005. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol 79: 7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff A 2007. Identification of viral microRNAs. Methods Enzymol 427: 3–23 [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435: 682–686 [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D 2009. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology 387: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Miller G 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci 93: 11883–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci 105: 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC 2008. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev 22: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol 84: 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA 2008. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 7: 1545–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Weber K, Bolander ME, Sarkar G 1998. PIG-B: A homemade monophasic cocktail for the extraction of RNA. Mol Biotechnol 9: 73–77 [DOI] [PubMed] [Google Scholar]
- Whitby D 2009. Searching for targets of viral microRNAs. Nat Genet 41: 7–8 [DOI] [PubMed] [Google Scholar]
- Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Sewer A, Zavolan M, Nair V 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation to those encoded by MDV-1. J Virol 81: 7164–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Raghavan B, Kotur M, Cheatham J, Sedmak D, Cook C, Waldman J, Trgovcich J 2007. Antisense transcription in the human cytomegalovirus transcriptome. J Virol 81: 11267–11281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Wang H, Herndier B, Ganem D 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci 93: 6641–6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet 41: 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]