Abstract
The hepatitis C virus (HCV) genomic RNA contains an internal ribosome entry site (IRES) in its 5′ untranslated region, the structure of which is essential for viral protein translation. The IRES includes a predicted pseudoknot interaction near the AUG start codon, but the results of previous studies of its structure have been conflicting. Using mutational analysis coupled with activity and functional assays, we verified the importance of pseudoknot base pairings for IRES-mediated translation and, using 35 mutants, conducted a comprehensive study of the structural tolerance and functional contributions of the pseudoknot. Ribosomal toeprinting experiments show that the entirety of the pseudoknot element positions the initiation codon in the mRNA binding cleft of the 40S ribosomal subunit. Optimal spacing between the pseudoknot and the start site AUG resembles that between the Shine–Dalgarno sequence and the initiation codon in bacterial mRNAs. Finally, we validated the HCV IRES pseudoknot as a potential drug target using antisense 2′-OMe oligonucleotides.
Keywords: hepatitis C virus (HCV), internal ribosome entry site (IRES), pseudoknot, RNA structure, translation initiation, initiation codon
INTRODUCTION
Hepatitis C virus (HCV) is a major public health problem, infecting ∼180 million people worldwide and causing chronic liver problems including cirrhosis and hepatocellular carcinoma (Ghany et al. 2009). New treatments are needed for HCV infection as the current combination of pegylated interferon-α and ribavirin has adverse side effects as well as limited efficacy with some viral genotypes (Deutsch and Hadziyannis 2008). The 5′ untranslated region (UTR) of the viral genomic RNA contains an internal ribosome entry site (IRES), the structure of which is required for viral protein synthesis (Tsukiyama-Kohara et al. 1992; Wang et al. 1993). Due to its significance in the viral life cycle and its high degree of conservation across genotypes, the IRES is an attractive drug target for new HCV therapeutics. A complete structural and mechanistic understanding of how the HCV IRES functions in translation initiation will facilitate the targeted discovery of inhibitors.
The HCV IRES adopts an ion-dependent three-dimensional fold with specific secondary and tertiary structural elements (Brown et al. 1992; Kieft et al. 1999). The RNA is capable of binding directly to 40S ribosomal subunits and eukaryotic initiation factor 3 (eIF3) (Pestova et al. 1998; Sizova et al. 1998; Kieft et al. 2001). Assembly studies suggest that the IRES binds first to 40S subunits and that this complex then recruits eIF3 and the ternary complex (TC) of eIF2-GTP-Met-tRNAimet (Ji et al. 2004; Otto and Puglisi 2004). While recruitment of 40S subunits occurs before eIF3 in HeLa extracts (Otto and Puglisi 2004), the IRES may bind to the 40S subunits that are preassociated with eIF3 and/or the TC in cells. GTP hydrolysis by the TC, mediated by eIF5, enables the 60S ribosomal subunit joining with the help of eIF5B, forming an elongation-competent 80S ribosome (Pestova et al. 1998; Locker et al. 2007). The IRES may also facilitate an alternative, eIF2-independent pathway of translation initiation under conditions of increased eIF2α phosphorylation during infection (Pestova et al. 2008; Terenin et al. 2008).
Significant progress has been made in mapping various functions of the IRES to specific structural domains (Fig. 1A). The basal portion of domain (dom) III is required for 40S recruitment (Kieft et al. 2001), while regions in dom IV and dom II are also contacted by the 40S subunit (Kolupaeva et al. 2000; Kieft et al. 2001; Spahn et al. 2001). The apical IIIabc domain contributes to IRES affinity for eIF3 (Sizova et al. 1998; Kieft et al. 2001). Dom II binds in the tRNA exit site, inducing a 40S conformational change (Spahn et al. 2001) and promoting eIF5-mediated GTP hydrolysis (Locker et al. 2007).
FIGURE 1.
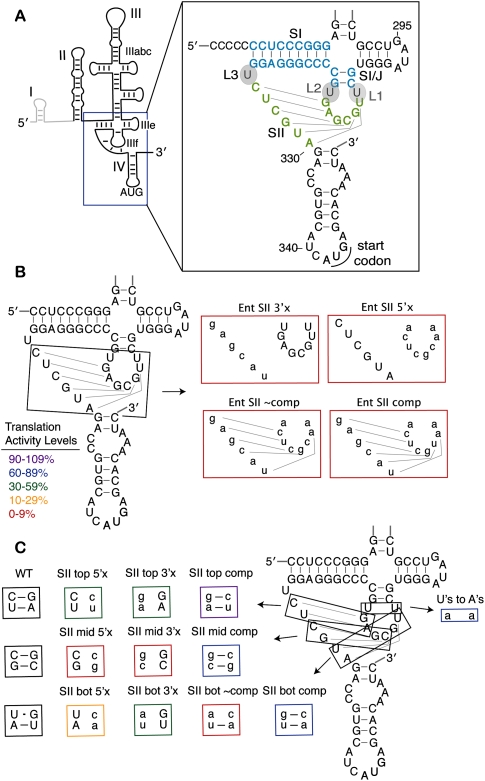
Stem II base pairs form in the HCV IRES pseudoknot and contribute to IRES translation efficiency. (A) A line diagram of the predicted HCV IRES secondary structure with the major domains and the AUG start codon labeled. Dom I is shown in gray and is not included in the IRES-luciferase reporter constructs. The inset shows the sequence and predicted secondary structure of the pseudoknot with the stem and loop nomenclature indicated. SI is shown in light blue and SII is shown in light green. (B) Mutations of the entire (Ent) SII sequence, as previously reported in Kieft et al. (2001) and (C) mutations of SII made two base pairs at a time. Mutated nucleotides are shown in lowercase and the mutants are boxed in colors corresponding to their translation activity levels as defined in B. Disruptive mutations are named according to whether the 5′ or 3′ side of SII (in the primary sequence) was mutated to its complement.
The specific tertiary structure that is formed in the basal portion of dom III has been subject to debate. On the basis of predicted base pairing, a pseudoknot structure was proposed in which the IIIf loop nucleotides base pair with nucleotides in a sequence just upstream of the AUG-containing dom IV (Wang et al. 1995). The proposed pseudoknot consists of two stems, SI and SII, as depicted in Figure 1A. Base pairing within a stem can be experimentally verified if mutation of either side of a stem inhibits activity and mutation of both sides to introduce compensatory base pairs rescues activity. The importance of base pairing in SI has been experimentally verified for both HCV and the related IRES in classical swine fever virus (CSFV) (Wang et al. 1994; Fletcher and Jackson 2002). Similarly, base pairing in SII of the CSFV IRES is critical for activity (Rijnbrand et al. 1997; Fletcher and Jackson 2002). In the HCV IRES, initial studies also showed modest rescue of HCV-mediated translation with compensatory base-pairing mutations in SII (Wang et al. 1995). Subsequently, however, HCV IRES mutants containing compensatory base-pairing mutations throughout the entirety of SII were inactive (Kieft et al. 2001). This result suggested that the specific sequence within the predicted SII, instead of—or in addition to—secondary structure alone, is needed for IRES function.
To resolve this discrepancy and further expand our understanding of the pseudoknot's function, we constructed a large set of mutants (n = 35) to structurally and mechanistically probe how the pseudoknot contributes to the activity of the HCV IRES. The translation activities from two base-pair mutations in SII of the HCV IRES pseudoknot offer strong evidence for functionally important base pairing throughout the 6-nucleotide (nt) stem, suggesting that the full pseudoknot forms as predicted. The SII structure contributes to AUG positioning by the IRES as analyzed by toeprinting analysis using purified human 40S ribosomal subunits. The global structure of the pseudoknot, not just of SI or SII, is required for robust IRES function. Across a wide panel of mutants, translation activity correlates well with the AUG positioning ability. Furthermore, an IRES with a compromised pseudoknot has a more stringent requirement for the proper distance between the AUG and the pseudoknot, supporting a model in which the conformation of the pseudoknot positions the mRNA open reading frame (ORF) in the 40S binding cleft. Based on these findings, we validated the pseudoknot as a possible HCV IRES drug target using antisense 2′-OMe oligonucleotides.
RESULTS
Stem II forms within the HCV IRES pseudoknot
The proposed pseudoknot structure in the HCV IRES is adjacent to dom IV, a stem–loop that contains the AUG start codon (Fig. 1A). The pseudoknot is predicted to form two stems, SI and SII. These are separated by three single uridine loops, L1–L3 (Fig. 1A). Despite conservation of a predicted pseudoknot structure across related hepacivirus/pestivirus (HP) IRESs (Hellen and de Breyne 2007), there have been differing results for the HCV and CSFV IRESs regarding the formation of stem II of the pseudoknot (Wang et al. 1995; Rijnbrand et al. 1997; Kieft et al. 2001; Fletcher and Jackson 2002). Mutation of either side of the predicted stem II severely inhibited activity in both systems, whereas compensatory mutations to restore base pairing showed no restoration of activity in the HCV pseudoknot when the entire SII sequence was mutated at once (Fig. 1B, Ent SII; Kieft et al. 2001). When mutations were made two base pairs at a time in the CSFV IRES, however, substantial rescue of activity was observed for compensatory base-pairing mutations (Fletcher and Jackson 2002).
To reconcile this discrepancy, mutations analogous to those made in CSFV were introduced into the HCV IRES, changing two nucleotides at a time throughout SII to their complements, as shown in Figure 1C. Translation activity of an IRES-firefly luciferase (FF luc) reporter lacking dom I (Fig. 1A) was measured in salt-adjusted rabbit reticulocyte lysate (RRL) previously optimized for authentic IRES translation (KE Berry, B Peng, D Koditek, N Pagratis, J Perry, J Parish, W Zhong, JA Doudna, I-h Shih, in prep.). RNA concentration and the time of the translation reaction were adjusted to fall within the linear range of the translation signal (data not shown). Disruption of two base pairs at a time on either side of SII in the HCV pseudoknot led to a reduction of 2%–40% activity relative to wild type (WT) (Fig. 1C; Table 1). Mutations in the middle base pairs were the most deleterious, and mutations in the top base pairs were the least detrimental.
TABLE 1.
Translation activities of IRES mutants
A significant increase in activity (to 60%–91% of WT levels) was observed for all three sets of base pairs when compensatory mutations were made on both sides of SII (Fig. 1C; Table 1). Thus, SII base pairing is important for activity in the HCV IRES as previously shown for the related CSFV IRES and initially suggested for the HCV IRES (Wang et al. 1995; Rijnbrand et al. 1997; Fletcher and Jackson 2002). After confirming the lack of activity for the Ent SII comp mutant (Fig. 1B; Kieft et al. 2001; data not shown), we wished to determine if this was due to the uridines (U's) in L1 and L2 being changed to their complementary adenosines (A's). To test this possibility, we made only these loop mutations in the WT IRES background. The U's to A's mutant retained 85% activity (Fig. 1C; Table 1), indicating that the sequence of L1 and L2 is not required for high translation activity.
Stem II base pairing contributes to AUG positioning
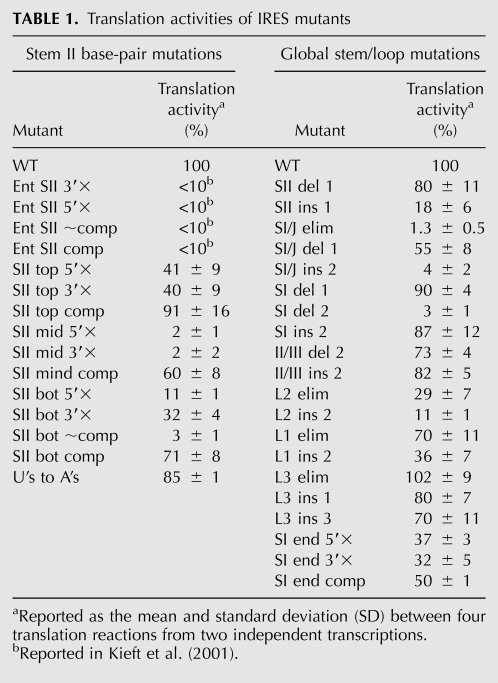
Previous reports have suggested that SII base pairing is important for AUG positioning in the CSFV IRES (Pestova et al. 1998). To test this possibility in the HCV IRES, ribosomal toeprinting was utilized to analyze the effect of the HCV IRES pseudoknot stem II on AUG positioning. Primer extension was conducted on IRES-FF luc mRNA under native conditions using a primer that hybridizes to nucleotides 18–37 of the FF luc ORF. Primer extension in the absence of 40S subunits yielded prominent stops at U239, A321/G318, and U302 (Fig. 2A). These stops correspond to the 3′ end of SII, the middle of SI, and the 3′ end of IIIe, respectively (Fig. 2B), and likely result from the secondary and/or tertiary structure within the IRES. In the presence of 40S subunits, new strong stops appeared at nucleotides 343–346, corresponding to the nucleotides at and immediately downstream from the AUG start codon (Fig. 2A). During translation initiation, this AUG binds to the P-site of the 40S ribosomal subunit. No P-site (+2–5, relative to the AUG) toeprint was observed for a mutant in which dom II was deleted (ΔdomII) (Fig. 2A), consistent with previous reports that this mutant does not stably position the adjacent ORF in the mRNA binding cleft (Pestova et al. 1998; Kolupaeva et al. 2000; Otto and Puglisi 2004).
FIGURE 2.
Stem II base pairing contributes to AUG positioning by the IRES. (A) Establishment of primer extension and toeprinting stops for WT IRES in the absence and presence of 40S subunits and comparison to ΔdomII IRES. Primer extension stops were mapped based on dideoxy sequencing reactions. (B) Location of major primer extension stops mapped onto the pseudoknot predicted secondary structure. Primer extension inhibition and toeprinting of (C) the middle and top SII base pair mutants, (D) the bottom SII base pair mutants, and (E) the entire SII base pair mutants. (F) Primer extension reactions of SII mid-5′× IRES-FF luc mRNA (50 nM) in the presence of increasing concentrations of 40S ribosomal subunits. The positions of the major stops are indicated at the right of each gel and are defined in A and B.
While predominant stops at the leading edge of the 40S subunit have been reported previously using either purified rabbit 40S subunits or rabbit reticulocyte lysate (Pestova et al. 1998; Kolupaeva et al. 2000; Otto and Puglisi 2004; Locker et al. 2007), the P-site (+2–5) stop in this system was previously found to be as strong or stronger than the leading edge stop (Fraser et al. 2009). The +2–5 stop in the P-site can be interpreted as an intermediate in the translation initiation pathway in which the AUG is positioned in the P-site, but the downstream ORF is not locked into the mRNA entry tunnel to give rise to a leading edge stop. The leading edge (+20/21) stop was weak under the current toeprinting conditions. However, when present, its intensity correlated well with the +2–5 stop across mutants, and no mutant ever showed a stronger leading edge toeprint than wild type. Thus, we utilized the stronger +2–5 toeprint in this study to characterize the ability of pseudoknot mutants to place the AUG in the mRNA binding cleft. An additional difference between the toeprints observed here, and in several previous reports, is that the leading edge stop falls at +20/21 relative to the AUG, rather than +14–15 or +16–18 (Pestova et al. 1998; Kolupaeva et al. 2000; Otto and Puglisi 2004; Locker et al. 2007). As with the +2–5 P-site stop, the leading edge stop at +20/21 has been consistently observed with the present HeLa 40S/FF luciferase reporter system (Fraser et al. 2009). The appearance of a strong stop at +2–5 and the leading edge stop being at +20/21 rather than the previously observed +16–18 may be due to the use of HeLa rather than rabbit 40S subunits, the FF luc reporter construct, the particular reverse transcription primer, or reverse transcription conditions.
The primer extension patterns of IRESs with mutated SII base pairs showed a weakening of the SII stop in the absence of 40S subunits, although this effect was less pronounced with the bottom base pairs, as compared with the middle and top base pairs (Fig. 2C,D). This SII stop was significantly strengthened in the compensatory mutants for all sets of base pairs, suggesting that the base-pairing mutations had the expected structural effects. The P-site toeprint was clearly reduced in the presence of 40S subunits for the disrupted base pairs of SII that resulted in the strongest translation defects (middle and bottom). Toeprinting was restored by compensatory base-pairing mutations (Fig. 2C,D), as was translation activity (Fig. 1C; Table 1). The P-site toeprint with 40S subunits had near-WT strength for the disrupted top base pairs of SII that caused the smallest defect in translation activity, despite destabilization of SII in the absence of 40S subunits (Fig. 2C).
Primer extension on previously published Ent SII mutants (Kieft et al. 2001; Fig 1B) showed that SII was indeed destabilized by disruptive mutations and restored by compensatory mutations (Fig. 2E, −40S). Nevertheless, very little P-site toeprint was seen for the compensatory mutant, consistent with the reported <10% translation activity (Kieft et al. 2001). Closer examination of the primer extension patterns in the absence of 40S subunits revealed that despite SII formation in the compensatory mutants, there were other structural problems with these mutants as evidenced by the lack of a top SI stop. These structural defects may account for the lack of toeprints and translation activity by these IRESs. Mutation of the bottom SII base pairs on the 5′ side (with respect to the primary sequence) led to a similar loss of the top SI stop in the absence of 40S subunits that was not restored by compensatory mutations (Fig. 2D). This UG sequence may therefore participate in a tertiary interaction with SI.
Based on a previous analysis of mutations in SII (Kieft et al. 2001), we hypothesized that the present mutants would be defective in translation initiation downstream from 40S binding. Strong changes in the primer extension pattern were observed for all mutants in the presence of 150 nM 40S subunits (Fig. 2C–E), suggesting that mutant IRESs were still capable of binding to 40S subunits, even if their toeprints were weak. Accordingly, a dose–response toeprinting experiment of the mutant SII mid 5′× RNA showed a gradual increase in the top SI stop as a function of 40S subunit concentration (Fig. 2F). This top SI stop is strengthened for nearly all mutants examined in this study, and likely represents a direct impediment posed to reverse transcriptase elongation by the binding of the 40S subunit. While a concurrent loss in the IIIe stop is observed, this is likely a direct consequence of the strengthening of the downstream top SI stop, as there are no additional strong stops observed upstream of the IIIe stop in the presence of 40S subunits.
Global pseudoknot structure contributes to translation and toeprint activity
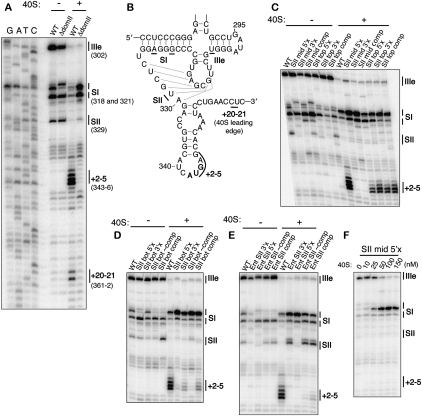
To further probe the overall structure of the pseudoknot, we systematically shortened or lengthened each of the predicted stems and loops of the pseudoknot. Based on their positions within the tertiary structure of the pseudoknot, we envisioned that some elements would be more tolerant of such changes than others. Stem II was more tolerant of shortening than lengthening by 1 base pair (bp) (80% versus 16% translation activity) and SI/J was very sensitive to lengthening and shortening, especially by 2 bp (Fig. 3A; Table 1). SI was more tolerant of changes than the other stems, with the IRES showing 87% activity when SI was lengthened by 2 bp. Shortening of SI by 1 bp was highly tolerated (90% activity) whereas shortening by 2 bp led to an inactive IRES (3% activity). This translation defect from the shortened SI was not due to insufficient space between domains II and III, as the deletion of 2 nt from the poly(C) linker between these domains still yielded 83% activity. Rather, reverse transcription of the SI del 2 mutant (Fig. 3B) showed that the entire pseudoknot structure was highly destabilized in this mutant, suggesting that the global structure cannot tolerate the deletion of 2 bp from SI, at least not the deletion of the two GC base pairs specifically tested here. Notably, most mutants examined in this study maintained strong aspects of the WT primer extension pattern, suggesting that the vast majority of mutants did not compromise the global pseudoknot structure (Figs. 2, 3; data not shown).
FIGURE 3.
Tolerance of the pseudoknot structure to stem and loop length. (A) Mutations of the pseudoknot stem lengths. Mutated nucleotides are shown in lowercase and the mutants are boxed in colors according to their translation activity, as defined in the legend. Deletions of nucleotides are shown as Δ. (B) Primer extension inhibition and toeprinting of stem length mutants. The positions of the major stops are indicated at the right of each gel and are defined in Figure 2, A and B. (C) Mutants of the pseudoknot loop lengths and base pairing at the terminus of SI, with the mutations and the activities represented as in A. (D) Primer extension inhibition and toeprinting of SI terminus base pair mutants, labeled as in B.
With respect to the loops of the pseudoknot, L2 had the most stringent length requirement with deletion of the single uridine or lengthening by two uridines leading to 29% and 11% translation activity, respectively (Fig. 3C; Table 1). L1 had an intermediate tolerance, with elimination of the loop preferred over its lengthening by 2 nt (70% versus 36%, respectively). L3, on the other hand, was highly tolerant of either insertions or deletions, with the elimination of L3 yielding a surprising 102% activity. The observation that two of the three predicted pseudoknot loops can be eliminated without deleterious effects suggests that if the pseudoknot stems are to resemble coaxially stacked helices, which require spanning loop sequences, the termini of predicted stems might actually melt to allow for longer loops.
To test the ability of the terminus of stem I to melt in the pseudoknot, each side of the first two dom II-proximal base pairs was mutated to its complement, or both to their complements to allow for compensatory base pairing. While disruption of the base pairs either from the 5′ or 3′ side in the primary sequence did lead to a reduced translation activity (37% and 32%, respectively), the compensatory base-pair mutation showed minimal restoration of activity (50%) (Fig. 3C; Table 1). This suggests that the sequence of the dom II-proximal terminus of SI is more important than base pairing within the stem. Mutation of the 3′ side of the SI terminus had a deleterious effect on the SI stops in the primer extension reactions, even in the compensatory mutant (Fig. 3D, −40S), suggesting that this sequence may be involved in a tertiary interaction with other pseudoknot elements.
Strong correlation between toeprint strength and translation activity
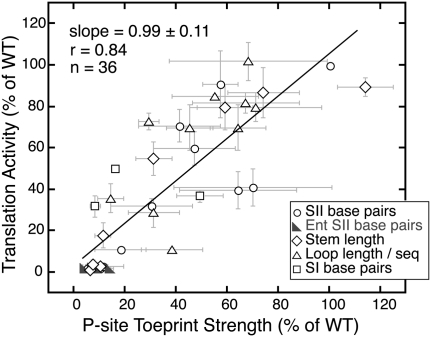
Given this diverse set of mutations of the pseudoknot sequence, we wished to determine how well 40S toeprint strength would correlate with IRES-mediated translation activity. Reproducible changes in the primer extension pattern were observed for all mutants in the presence of 150 nM 40S subunits (Fig. 3B,D; data not shown), suggesting that mutant IRESs were all capable of binding to 40S subunits. The P-site (+2–5) toeprint stop from each mutant was quantified and normalized to the WT IRES from the same experiment. Each mutant's translation activity was then plotted against this normalized toeprint strength (Fig. 4). Regression analysis of translation activity on toeprint strength produced a slope coefficient of 0.99 ± 0.11 and a correlation (r) of 0.84, which is significant at the 0.0001 level using a Student's t-test. The slope coefficient near 1.0 shows that translation activity is proportional to toeprint strength, and is consistent with the IRES pseudoknot structure principally functioning to position the initiation codon. Although the correlation between translation activity and toeprint strength is strong, the relationship is not perfect, indicating that the pseudoknot structure may affect other aspects of translation initiation beyond positioning the start codon. The dispersion of observations from the regression line may also be due in part to the large amount of error inherent in a dynamic assay such as toeprinting, seen in the horizontal error bars in Figure 4.
FIGURE 4.
Correlation between toeprint strength and translation activity. Translation activity (as quantified in Table 1) plotted against toeprint stop intensity, quantified by densitometry, and normalized to the toeprint of the WT IRES. Plotted points reflect the mean value of activity across four in vitro translations from two independent transcriptions, and the mean value of the toeprint strength from two independent transcriptions. The error bars around the points extend 1 SD above and below the mean. Mutants are grouped into categories based on what structural features they disrupt, as indicated by the legend, but linear regression was conducted on the full set of 36 mutants together. The estimated slope with a standard error is 0.99 ± 0.11; the correlation is 0.84, which is significantly different from zero at the 0.0001 level.
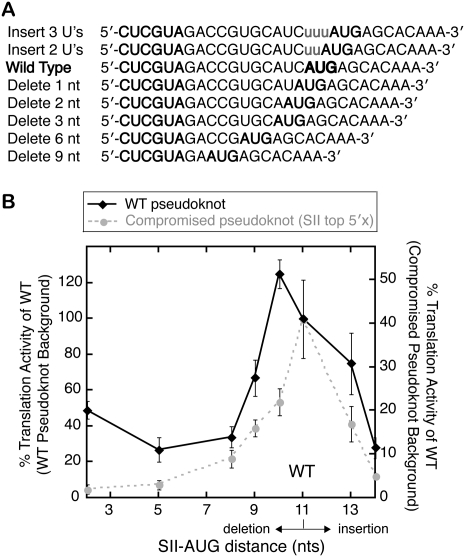
Inhibition of translation by varying pseudoknot–AUG distance
Since the pseudoknot structure affects AUG and ORF positioning in the mRNA binding cleft of the 40S subunit, we tested the dependence of translation activity on the distance between the pseudoknot and the AUG start codon. The WT spacing is 11 nt for the HCV IRES and is well conserved between most related IRESs at 9–13 nt between the end of SII and the AUG (Hellen and de Breyne 2007). Uridines were inserted or nucleotides were deleted directly before the AUG to vary the SII–AUG distance within the IRES (Fig. 5A). Either deletion or insertion of nucleotides between the pseudoknot and start codon impaired translation activity, with the exception of the deletion of a single nucleotide (Fig. 5B, black line). Thus, the WT IRES sequence has nearly the ideal distance between the pseudoknot and start codon. Primer extension inhibition analysis showed that these mutants retained a strong P-site toeprint with 40S subunits, even as the sequence present in the P-site was varied (data not shown).
FIGURE 5.
Inhibition of IRES translation activity by varying the pseudoknot–AUG distance. (A) Sequences of mutations made to either lengthen or shorten the pseudoknot–AUG distance. Inserted nucleotides are shown in lowercase letters, and the start codon and SII are in bold. (B) Translation activity plotted against the pseudoknot–AUG distance with deletions plotted to the left of the WT AUG position and insertions to the right. Translation activities from the WT IRES background are plotted on the left vertical axis and those from a compromised pseudoknot (SII top 5′× mutant) on the right vertical axis, with 100% activity on the left axis scaled to 40% starting activity on the right. Plotted points represent the mean activities of four translation reactions from two independent transcriptions and the error bars extend 1 SD above and below the mean.
Cryo-electron microscopy (cryo-EM) reconstructions of the HCV IRES bound to the 40S ribosomal subunit show that the pseudoknot is thought to bind to the back of the 40S subunit platform, ∼50–70 Å away from the P-site (Spahn et al. 2001). One model for pseudoknot function is that the global structure of the pseudoknot orients dom IV toward the P-site. This hypothesis predicts that a compromised pseudoknot would have a smaller window of tolerance for pseudoknot–AUG distance changes since the orientation of the ORF would be imperfect. To test this possibility, nucleotides were inserted or deleted between the pseudoknot and the AUG of the SII top 5′× mutant IRES. This mutant possesses a compromised pseudoknot but still retains 40% activity and a P-site toeprint. The compromised pseudoknot mutant indeed showed a more stringent requirement for the correct pseudoknot–AUG distance when compared with a WT pseudoknot (Fig. 5B, cf. gray and black lines).
Inhibition of HCV IRES translation with pseudoknot-targeted 2′-OMe oligonucleotides
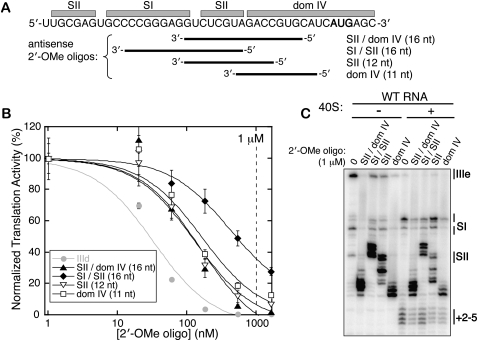
The role of the HCV IRES pseudoknot in orienting the ORF toward the 40S mRNA binding cleft suggests that the pseudoknot may be an attractive drug target. It is much easier to imagine a small molecule perturbing the conformation of the RNA structure in the pseudoknot region than disrupting the binding affinity between the 40S subunit or eIF3, given the large surfaces of these interaction areas. As a proof-of-principle demonstration that the pseudoknot structure can be disrupted in trans, several 2′-OMe oligonucleotides were designed with complementarity to different regions of the pseudoknot and dom IV (Fig. 6A). These oligonucleotides were titrated into in vitro translations of a WT IRES-FF luc reporter in salt-adjusted RRL. As a positive control, a previously established inhibitory 2′-OMe oligonucleotide, which hybridizes to the IIId loop of the IRES (“IIId”) (Tallet-Lopez et al. 2003), was tested and had an IC50 in our system of ∼30 nM (Fig. 6B, gray). The reverse complement of this sequence served as a negative control and showed no inhibition of IRES-FF luc signal through concentrations of 5 μM (data not shown). Three of the four 2′-OMe oligonucleotides targeted against the pseudoknot and dom IV showed potent inhibition, with IC50's around ∼140 nM (Fig. 6B). The 2′-OMe oligonucleotide that in part targeted SI of the pseudoknot had a considerably weaker IC50 of ∼400 nM, likely due to a lack of access to this kinetically stable stem. These oligonucleotides introduced strong new stops in primer extension inhibition reactions, but the P-site toeprint of IRES-40S subunit complexes was downstream from these new stops (Fig. 6C). This toeprint was reduced by the pseudoknot-targeted oligonucleotides in a manner proportional to their inhibitory activity (Fig. 6C).
FIGURE 6.
Inhibition of HCV IRES translation with pseudoknot-targeted 2′-OMe oligonucleotides. (A) The diagram shows to which regions of the IRES each of the four 2′-OMe oligonucleotides is complementary, with the start site AUG shown in bold. (B) Titrations of the 2′-OMe oligonucleotides in WT IRES-FF luciferase in vitro translation reactions, fit to sigmoidal inhibition curves. Plotted points represent the mean activities of four translation reactions from two independent transcriptions and the error bars extend 1 SD above and below the mean. The 1 μM concentration utilized in toeprinting experiments in C is indicated with a vertical dashed line. (C) Primer extension inhibition and toeprinting on the WT IRES-FF luc mRNA in the presence of 1 μM 2′-OMe oligonucleotides.
DISCUSSION
We set out to establish whether the complete pseudoknot structure forms in the HCV IRES and to probe its structural tolerance and functional contributions to translation initiation. Our results show that the full pseudoknot structure, including SII base pairs, does form in the HCV IRES and that pseudoknot formation leads to correct positioning of the AUG start codon in the downstream mRNA on the 40S ribosomal subunit. We also find that disruption of the pseudoknot structure using antisense 2′-OMe oligonucleotides blocks IRES-mediated translation, providing evidence that the pseudoknot may be an appealing target for therapeutic intervention.
SII base pairing contributes to IRES function
Conservative disruptions and compensatory base changes in the proposed SII base-pairing interactions of the HCV IRES show that the SII structure is required for efficient IRES-mediated translation initiation. The bottom and middle base pairs contribute more to translation activity than the top base pair (Fig. 1C; Table 1), perhaps due to their proximity to the AUG start codon or to preferential 40S subunit contacts with this portion of the stem. Although compensatory base pairs throughout the stem restore translation activity, the middle and bottom pairs only rescue activity to 60% and 71% of WT levels, respectively. This observation likely reflects some contribution of sequence identity to activity. Mutation of the entire SII stem to its complementary sequence results in an inactive IRES (Kieft et al. 2001). Here we conclude that this inactivity is not due to L1 and L2 sequences being changed from uridines to adenosines. Instead, numerous simultaneous mutations cause global misfolding of the pseudoknot, as evidenced by the lack of an SI stop in compensatory mutants (Fig. 2E).
An intriguing possibility is that SII may be a dynamic part of the pseudoknot structure, as suggested from RNase probing experiments showing sensitivity to both single- and double-stranded RNases (Wang et al. 1995; Kolupaeva et al. 2000; Fletcher et al. 2002). Comparison of the primer extension inhibition patterns of IRES mutants in the absence and presence of 40S ribosomal subunits shows that most mutants (with the exception of the Ent SII series) lose their stable SII stop in the presence of 40S subunits. Given that stops upstream of SII are still strong, this may represent a weakening of SII base pairing upon productive 40S binding. It is still an open question whether the dynamic nature of this stem has a functional role in translation initiation. Mutations in SII that strengthen or weaken the thermodynamic stability of this stem did not show a clear correlation with activity level (data not shown).
The pseudoknot positions the start codon on the ribosome
Comparison of 40S toeprint strength to translation activity among the 35 mutants analyzed in this study reveals a strong linear correlation (Fig. 4); mutations that perturb ORF positioning on the 40S subunit also result in defective translation initiation. This suggests that the primary function of the pseudoknot is to position the initiation codon, and that the entire pseudoknot structure is required for this function. This global structure may extend further into the main stem of dom III, as it has recently been shown that there is a Watson–Crick interaction between a IIIe loop pyrimidine and a conserved purine/purine mismatch in the stem of dom III (Easton et al. 2009). This interaction serves to stabilize SI of the pseudoknot, and may or may not contribute to the P-site toeprint in 48S preinitiation complexes (Otto and Puglisi 2004; Easton et al. 2009).
Cryo-EM analysis maps the position of the pseudoknot to the mRNA exit channel at the back of the platform of the 40S ribosomal subunit (Spahn et al. 2001). This location on the small ribosomal subunit is similar to that of the 16S rRNA segment that binds to the Shine–Dalgarno sequence of bacterial mRNAs. In bacteria, this interaction establishes the path of mRNA through the exit channel and into the mRNA binding cleft (Yusupova et al. 2001; Boehringer et al. 2005). Toeprinting analysis shows that the pseudoknots in HCV and CSFV IRESs contribute to AUG positioning (Pestova et al. 1998; Otto and Puglisi 2004; this study). Notably, many more contacts between the pseudoknot and the platform of the 40S ribosomal subunit are observed by cryo-EM in a structure of the IRES with only the 40S subunit than in a structure with the 80S ribosome (Spahn et al. 2001; Boehringer et al. 2005).
Modulation of the pseudoknot–AUG distance in the WT IRES background shows that the IRES maintains a near-optimal distance between the pseudoknot and the initiation codon. This distance range resembles that between the Shine–Dalgarno sequence and the AUG in bacterial mRNAs, which is most commonly 4–12 nt in Escherichia coli (Ringquist et al. 1992; Ma et al. 2002). The stimulation of translation activity observed upon reducing the pseudoknot–AUG distance by 1 nt may be an effect of destabilizing the stem–loop of dom IV (Honda et al. 1996). The deleterious effect of larger deletions in this spacing likely results from the AUG not being able to reach the P-site. It should be noted that this genotype 1b sequence contains an A at position 350 and does not possess the downstream ACG codon that was proposed to serve as an alternate translation start site in other HCV IRES sequences (Reynolds et al. 1995). The observation that insertions between the pseudoknot and AUG hurt translation activity considerably is consistent with previous conclusions that scanning does not occur in CSFV or HCV IRES-mediated translation initiation (Reynolds et al. 1996; Rijnbrand et al. 1997). It also suggests that AUG is not a “ball on a string” suspended from the pseudoknot, but is more actively guided to the binding cleft, quite possibly through the same path as that observed in bacteria (Yusupova et al. 2001; Boehringer et al. 2005).
We predicted that if the pseudoknot positions the ORF in the mRNA binding cleft, a compromised pseudoknot would tolerate a smaller range of SII–AUG distances than the WT pseudoknot for effective translation initiation. Indeed, this is observed for the SII top 5′× mutant relative to the WT IRES (Fig. 5B), supporting a model in which the pseudoknot structure sets the angle at which dom IV is presented to the interface region of the 40S subunit, orienting it appropriately through the mRNA exit tunnel. It is also possible that the pseudoknot structure allosterically manipulates the mRNA binding cleft by influencing the conformation of the 40S subunit and thus influences the P-site primer extension stops.
Toward the global structure of the pseudoknot
In the absence of other tertiary structural constraints, the most thermodynamically stable way for two helices of a pseudoknot to interact is to coaxially stack to form a continuous helix in three-dimensional space (Pleij et al. 1985; Wyatt et al. 1989; Brierley et al. 2007). The fact that all three predicted loops in the HCV IRES pseudoknot are single nucleotides, and especially the observation that L1 and L3 can be eliminated without deleterious effects (Fig. 3C; Table 1), suggests that this pseudoknot may not assume a classic, coaxially stacked structure. A lack of base pairing at the SI terminus could partially compensate for the short loop sequences. The possibility that SII might coaxially stack with either SI/J or SI was considered, but the mutations made to test these possibilities gave no direct evidence for these arrangements (data not shown). The pseudoknot may adopt a complex globular structure that evades modeling based on mutational studies alone. It will be critical in the future to determine the high-resolution structure of this region of the IRES.
The present mutational study provides new insights into the nature of the pseudoknot structure, including the length tolerance of loops and stems within the pseudoknot. In addition, activity assays with mutant IRESs suggest a potential sequence-specific requirement at the dom II-proximal terminus of SI, as compensatory base-pairing mutations in the final two base pairs of this stem do not show a significant rescue in activity over the mutations of either individual side of the stem (Fig. 3C; Table 1). Primer extension inhibition analysis suggests that mutation of the 3′ side (in the primary sequence) of this stem preferentially disrupts SI stability (Fig. 3D), indicating a potential role of these nucleotides in a tertiary structure. In contrast, previous mutational analysis shows that compensatory mutations fully rescue activity when the second-to-fifth bases in this stem are changed from CUCC to AGAA (Wang et al. 1994), and similar results have been seen for internal positions in SIa of the CSFV IRES (Fletcher and Jackson 2002). The simplest explanation for this difference would be that a sequence-specific requirement occurs in the terminal base pair of SI, rather than in the second internal base pair. Other sequence-specific requirements are observed in this region of the pseudoknot in the related CSFV IRES, where mutating the A-rich L3 to contain more uridines leads to a drop in activity (Fletcher and Jackson 2002).
The pseudoknot as a drug target
The HCV IRES is more than a molecular scaffold for recruiting eIF3 and the 40S ribosomal subunit, as mutations can dramatically reduce translation activity without strongly affecting the affinity of the IRES for these translation factors (Kieft et al. 2001; Otto and Puglisi 2004). The places within the IRES where conformations of the RNA contribute to activity are vulnerable regions at which small molecules may be able to interfere. The pseudoknot is likely to be such a structure. Previous binding affinities measured for severe pseudoknot (Ent SII) mutants suggest that proper pseudoknot structure does not strongly contribute to binding affinity for 40S subunits (Kieft et al. 2001). In addition, for all mutations made throughout the global structure of the pseudoknot, consistent changes are observed between primer extension inhibition patterns for the IRES in the absence and presence of 40S subunits (Figs. 2, 3; data not shown). This suggests that any differences in the binding affinities of mutant IRESs for the 40S subunit are relatively small and not significant enough to cause the observed lack of translation activity in rabbit reticulocyte lysate reactions, which we estimate to contain ∼600 nM 40S subunits (Meyer et al. 1982).
The 2′-OMe oligonucleotides that can base pair with sequences within SII of the pseudoknot show potent inhibition of IRES activity (Fig. 6). A 2′-OMe oligonucleotide targeted to the IIId loop of the IRES (used here as a positive control for inhibition) was dropped after a Phase I trial due to a lack of on-target activity and aminotransferase flares (McHutchinson et al. 2006). Nonetheless, inhibition from these pseudoknot-targeted 2′-OMe oligonucleotides serves as a proof of principle that disrupting the pseudoknot structure would be a viable target for novel HCV therapeutics. Indeed, the fact that point mutations are capable of drastically reducing translation activity suggests that a small molecule might be able to accomplish this goal by perturbing the specific pseudoknot conformation required for AUG positioning. To fully realize the goals of understanding the molecular mechanism of the HCV IRES pseudoknot's function and to incapacitate this function with a small molecule, a critical objective will be the determination of an atomic resolution structure of this region of the IRES.
MATERIALS AND METHODS
Cloning of the IRES reporter construct and mutants
A WT IRES-FF luc reporter was generated from a previous MS2 hairpin-HCV IRES-FF luc construct (Ji et al. 2004) by QuikChange mutagenesis (Stratagene) with the primer 5′-CGGAATTCTAATACGACTCACTATAGCTCCCCTGTGAGGAACTACTG-3′ and its reverse complement. Deletion of three MS2 hairpins from the WT IRES-FF luc construct yielded plasmid pKB84. The IRES sequence in pKB84 is genotype 1b, ultimately derived from the pK1b plasmid (Tsukiyama-Kohara et al. 1992) with the exception of a G357A mutation in the coding sequence of the IRES (downstream from dom IV). All IRES mutations were generated from pKB84 using QuikChange mutagenesis. DNA sequencing was used to check the IRES and FF luciferase sequences of each mutant.
Transcription and quantification of reporter mRNA
Uncapped reporter RNAs were in vitro transcribed using T7 polymerase from HindIII-digested plasmids. After DNase I treatment (RNase-free, Promega, 0.09 U/μL for 15 min at 37°C), RNA was purified from free nucleotides and enzyme using RNA Clean&Concentrator-25 columns (Zymo Research). RNA was eluted in 20 mM Hepes at pH 7.5, distributed into small, single-experiment aliquots, and stored at −80°C until use. The initial quantification of RNA concentration was determined by absorbance at 260 nm. Concentrations were adjusted based on densitometry measurements of the full-length RNA, run on an ethidium bromide-stained 1% agarose TAE gel. Sodium dodecyl sulfate (SDS, 0.025%) was present in the 2X formamide loading dye and was essential for RNAs to run as clean bands in nondenaturing agarose gels. The intensities of full-length RNA bands were normalized to a lane-by-lane loading control of 40 ng linearized dsDNA plasmid. All six RNAs for a given translation experiment were run in duplicate on the same 1% agarose gel immediately before the translation experiment, and requantified as above. In rare cases, RNAs that showed significant smearing or degradation were discarded and transcribed again.
In vitro translation reactions
Rabbit reticulocyte lysate (RRL) in vitro translations were performed using Promega's standard nuclease-treated Rabbit Reticulocyte Lysate System. Each 15 μL reaction contained 56% (v/v) nuclease-treated RRL, 20 μM amino acids, 1.3 U/μL RNasin Plus RNase inhibitor (Promega), Complete Protease Inhibitor Cocktail (Roche: one tablet in 1 mL used as 50X), 2 mM DTT, and 1.8 mM MgCl2, 45 mM KCl, and 26 mM KOAc, to obtain final salt concentrations of 2.2 mM Mg2+, 45 mM KCl, and 90 mM KOAc (Kozak 1990; KE Berry, B Peng, D Koditek, N Pagratis, J Perry, J Parish, W Zhong, JA Doudna, I-h Shih, in prep.). Translation reactions were initiated with the addition of 10 ng reporter mRNA (quantified as described above), incubated at 30°C for 30 min, and stopped with the addition of 7.5 μL 60 μM puromycin. These conditions were in the linear range for signal with respect to both RNA concentration and translation time. Luciferase activity was measured in a Veritas microplate luminometer (Turner BioSystems) using 50 μL of Luciferase Assay Reagent (Promega) for each well, according to the manufacturers' instructions. A mutant's translation activity is reported as the mean of its activity normalized to wild type from each of four independent translation experiments (from two independent transcriptions), and is reported with the standard deviation (SD) between these four values.
For in vitro translation experiments containing 2′-OMe oligonucleotides, serial dilutions of 10X oligonucleotides were made, and the oligonucleotides were mixed with translation extracts prior to the addition of WT IRES-FF luc reporter RNA. The means for duplicate translation reactions at each oligonucleotide concentration were plotted and fit to the inhibition curve %Translation activity = 100/(1+10[log(IC50)-log(inhibitior)]) using Kaleidagraph (Synergy Software). Sequences for the 2′-OMe oligonucleotides were IIId: 5′-ACCCAACACUACUCGGC-3′; SII/dom IV: 5′-UGCACGGUCUACGAGA-3′; SI / SII: 5′-CUACGAGACCUCCCGG-3′; SII: 5′-CGGUCUACGAGA-3′; and dom IV: 5′-GAUGCACGGUC-3′.
Primer extension inhibition (toeprinting) assays
The 40S ribosomal subunits were purified from a HeLa cytoplasmic lysate (a gift from R Tjian, University of California at Berkeley) as described previously (Fraser et al. 2007). Reverse transcription was conducted from a 5′-end-labeled DNA primer that hybridizes to nucleotides 18–37 of the FF luciferase ORF (5′-GCGCCGGGCCTTTCTTTATG-3′) (Fraser et al. 2009). Toeprinting reactions were performed based on previous descriptions (Pestova et al. 1998; Fraser et al. 2009) with minor modifications. Briefly, ∼50 nM IRES-FF luc RNA, 50 nM 5′ end-labeled prime, and 150 nM 40S subunits were mixed in a 10μL reaction in 50 mM Hepes at pH 7.5, 50 mM KCl, 2 mM Mg(OAc)2, and 1 mM DTT. Binding reactions were heated to 37°C for 10 min, and then incubated for 5 min at RT and 5 min on ice. Reverse transcription was initiated with the addition of a 1 μL extension buffer containing 80 mM Mg(OAc)2, 10 mM DTT, 4 mM dNTPs, and 12U/μL Superscript III reverse transcriptase (Invitrogen). Reactions were incubated at 30°C for 45 min and stopped on ice with the addition of 2X formamide loading dye. cDNAs were resolved without further purification on 10% denaturing acrylamide gels alongside dideoxy sequencing reactions and detected by phosporimaging. While primer extension gels were cropped above the IIIe loop (at approximately nucleotide 300) in figures, gels were inspected to ensure the presence of significant stops for each IRES mutant at higher positions on the gel and corresponding to full length RNA, indicating that RNA was not degraded in these reactions.
To quantify toeprint strength, densitometry was performed in ImageQuant TL (Molecular Dynamics) by measuring the intensity within a fixed area around the P-site toeprint for each mutant and normalizing to the toeprint intensity from the WT IRES within the same experiment. Toeprint strength is reported as the mean observation and standard deviation from at least two independent reverse transcription experiments from two independent RNA transcriptions, to control for variability within the assay, as well as variations in lane loading. For each mutant, translation activity was plotted against toeprint strength and ordinary least-squares linear-regression analysis was performed in Kaleidagraph (Synergy Software) to yield an estimated slope and a Pearson correlation coefficient (r).
ACKNOWLEDGMENTS
We thank N. Husain for assistance with cloning, and W. Berry, C. Fraser, and members of the Doudna laboratory for helpful discussions and comments on the manuscript. This work was supported by a Program project grant from the National Institutes of Health and a research gift generously provided by Gilead, Inc. (to J.A.D.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2197210.
REFERENCES
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H 2005. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure 13: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Brierley I, Pennell S, Gilbert RJ 2007. Viral RNA pseudoknots: Versatile motifs in gene expression and replication. Nat Rev Microbiol 5: 598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EA, Zhang H, Ping LH, Lemon SM 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res 20: 5041–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch M, Hadziyannis SJ 2008. Old and emerging therapies in chronic hepatitis C: An update. J Viral Hepat 15: 2–11 [DOI] [PubMed] [Google Scholar]
- Easton LE, Locker N, Lukavsky PJ 2009. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res 37: 5537–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SP, Jackson RJ 2002. Pestivirus internal ribosome entry site (IRES) structure and function: Elements in the 5′ untranslated region important for IRES function. J Virol 76: 5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SP, Ali IK, Kaminski A, Digard P, Jackson RJ 2002. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8: 1558–1571 [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819 [DOI] [PubMed] [Google Scholar]
- Fraser CS, Hershey JW, Doudna JA 2009. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol 16: 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghany MG, Strader DB, Thomas DL, Seeff LB 2009. Diagnosis, management, and treatment of hepatitis C: An update. Hepatology 49: 1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, de Breyne S 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol 81: 5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Brown EA, Lemon SM 1996. Stability of a stem–loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2: 955–968 [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci 101: 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol 292: 513–529 [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU 2000. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1990. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res 18: 2828–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Campbell A, Karlin S 2002. Correlations between Shine–Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol 184: 5733–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHutchinson JG, Patel K, Pockros P, Nyberg L, Pianko S, Yu RZ, Dorr FA, Kwoh TJ 2006. A phase I trial of an antisense inhibitor of hepatitis C virus (ISIS 14803), administered to chronic hepatitis C patients. J Hepatol 44: 88–96 [DOI] [PubMed] [Google Scholar]
- Meyer LJ, Milburn SC, Hershey JW 1982. Immunochemical characterization of mammalian protein synthesis initiation factors. Biochemistry 21: 4206–4212 [DOI] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119: 369–380 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J 27: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij CW, Rietveld K, Bosch L 1985. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res 13: 1717–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J 14: 6010–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ 1996. Internal initiation of translation of hepatitis C virus RNA: The ribosome entry site is at the authentic initiation codon. RNA 2: 867–878 [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand R, van der Straaten T, van Rijn PA, Spaan WJ, Bredenbeek PJ 1997. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol 71: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L 1992. Translation initiation in Escherichia coli: Sequences within the ribosome-binding site. Mol Microbiol 6: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Tallet-Lopez B, Aldaz-Carroll L, Chabas S, Dausse E, Staedel C, Toulmé J 2003. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res 31: 734–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol 15: 836–841 [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A 1992. Internal ribosome entry site within hepatitis C virus RNA. J Virol 66: 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol 67: 3338–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A 1994. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol 68: 7301–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Le SY, Ali N, Siddiqui A 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1: 526–537 [PMC free article] [PubMed] [Google Scholar]
- Wyatt JR, Puglisi JD, Tinoco I Jr 1989. RNA folding: Pseudoknots, loops, and bulges. Bioessays 11: 100–106 [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]