Abstract
The hepatitis C virus internal ribosome entry site (IRES) element contains a three-way junction that is important in the overall RNA conformation, and for its role in the internal initiation of translation. The junction also illustrates some important conformational principles in the folding of three-way helical junctions. It is formally a 3HS4 junction, with the possibility of two alternative stacking conformers. However, in principle, the junction can also undergo two steps of branch migration that would form 2HS1HS3 and 2HS2HS2 junctions. Comparative gel electrophoresis and ensemble fluorescence resonance energy transfer (FRET) studies show that the junction is induced to fold by the presence of Mg2+ ions in low micromolar concentrations, and suggest that the structure adopted is based on coaxial stacking of the two helices that do not terminate in a hairpin loop (i.e., helix IIId). Single-molecule FRET studies confirm this conclusion, and indicate that there is no minor conformer present based on an alternative choice of helical stacking partners. Moreover, analysis of single-molecule FRET data at an 8-msec resolution failed to reveal evidence for structural transitions. It seems probable that this junction adopts a single conformation as a unique and stable fold.
Keywords: RNA structure, RNA junctions, translation, FRET, single-molecule fluorescence
INTRODUCTION
The internal ribosome entry site (IRES) of the hepatitis C virus (HCV) binds eIF3 and the small ribosomal subunit, allowing internal initiation of translation to occur independently of a capped 5′ terminus (Tsukiyama-Kohara et al. 1992; Wang et al. 1993; Sizova et al. 1998). This 341-nucleotide (nt) RNA is found upstream of a gene encoding a large polyprotein. The HCV IRES RNA is highly conserved, and adopts a folded structure in the presence of divalent metal ions (Kieft et al. 1999; Spahn et al. 2001). The RNA has a complex secondary structure (Brown et al. 1992; Reynolds et al. 1995; Rijnbrand et al. 1995; Honda et al. 1996), which includes a number of helical junctions, shown schematically in Figure 1A. Domain III includes a prominent four-way junction. Using the IUB nomenclature of helical junctions (Lilley et al. 1995), this junction is defined as 2HS22HS1, indicating the number of helical (H) and linking segments (S) sequentially around the branchpoint. There is also a less well-defined junction involving helices IIIe and IIIf, and a three-way junction that includes helix IIId (Fig. 1B). Elements of the structure have been determined (Klinck et al. 2000; Lukavsky et al. 2000; Kieft et al. 2002; Lukavsky et al. 2003). We have previously studied the structure and dynamic properties of the 2HS22HS1 four-way junction. The structure of a related construct was solved by X-ray crystallography (Kieft et al. 2002), but we found that this junction was either a stacked cross with axes perpendicular or a dynamic structure oscillating between parallel and anti-parallel stacked structures (Melcher et al. 2003). Interestingly, an anti-parallel conformation was suggested by a single-particle cryo-electron microscopy (cryo-EM) study (Boehringer et al. 2005), whereas a parallel conformation was found in the crystal structure (Kieft et al. 2002).
FIGURE 1.
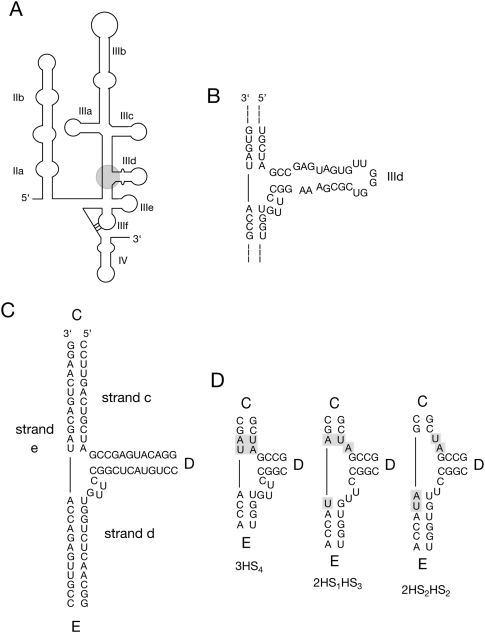
The hepatitis C virus IRES. (A) Schematic of the secondary structure of the IRES. The three-way helical junction studied here is circled. (B) The local sequence around the three-way junction containing helix IIId. (C) The sequence of the three-way junction in the form studied here. The junction comprises three strands (c, d, and e) and the arms are labeled C, D, and E as shown. The loop and non-Watson–Crick pairs have been removed from helix IIId to create a simple helix D for the majority of this study. (D) A form of branch migration creates three possible secondary structures at the junction.
The three-way junction has not been the subject of extensive study, either structurally or functionally. The structure of helix IIId has been studied by nuclear magnetic resonance (NMR) (Klinck et al. 2000; Lukavsky et al. 2000), and the effects of mutations within this helix on IRES activity have been analyzed. To date, there has been no study of the effects of mutations within the core of the junction. However, it has been demonstrated that the apical GGG is critical for the IRES function (Kieft et al. 1999), as it seems to be essential for binding by the 40S ribosomal subunit. Since the conformation of the junction will likely determine the presentation of this loop to the ribosome, we expect that the junction will be critical to IRES function.
Helical junctions are a major structure determinant of complex RNA species (Lilley 2000). Their geometry orients helices relative to one another, allowing long-range tertiary contacts to occur. For example, inspection of the structures of the 16S and 23S rRNA species within the ribosome reveals many helical junctions (Ban et al. 2000; Wimberly et al. 2000). The structure of some autonomously folding smaller RNA species can be almost entirely determined by component junctions (Murchie et al. 1998; Walter et al. 1998), as exemplified by the VS ribozyme where the structural core of the ribozyme comprises five helical segments related by two three-way junctions (Lafontaine et al. 2002; Lipfert et al. 2008). Helical junctions can also generate the catalytic center of ribozymes, such as the hammerhead ribozyme (Martick et al. 2008), and ligand binding pockets of great selectivity, as found in a number of riboswitches (Serganov et al. 2004, 2008; Garst et al. 2008). Three-way junctions are especially common in functional RNA species of all sizes. In the sections that follow we set out a new analysis of the structure and topology of three-way junctions in general, and then follow this with a discussion of the specific sequence of the IRES three-way junction.
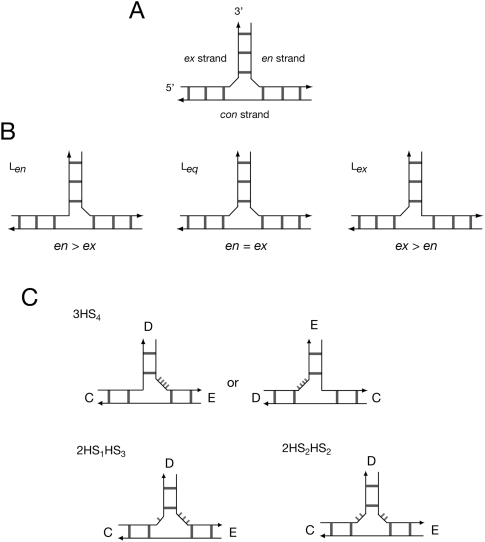
The helical arms of junctions in both DNA and RNA have a strong propensity to undergo coaxial stacking when electrostatic repulsion is minimized by the presence of metal ions (Duckett et al. 1988, 1995; Orr et al. 1998; Walter et al. 1998; Lescoute and Westhof 2006; de la Pena et al. 2009). In the case of three-way junctions, only two arms can be stacked in this way, and the third must remain unstacked. The simplest case is the perfectly base-paired 3H junction, where the three interhelical connections each lack additional nucleotides. However, 3H junctions are rare in natural RNA species, and most have at least one connecting segment comprising one or more unpaired nucleotides, which we term “linking” nucleotides. These probably provide conformational flexibility that is essential to allow the junctions to fold by pairwise coaxial stacking. Examination of the structures of three-way junctions observed in crystal structures of RNA species (Lescoute and Westhof 2006) indicates a strong propensity of junctions fold in a way that minimizes the number of linking nucleotides between the stacked helices. This is a reliable predictor of structure in natural three-way junctions. A junction with two coaxially stacked helices is depicted schematically in Figure 2A. It should be noted that all three component strands are structurally distinct. One strand runs the length of the stacked helices—we call this strand “continuous” (con). The other two pass between the stacked helices and the third helix. One passes out of the stack in a 5′-to-3′ direction—we call this “exit” (ex). The other passes in the opposite direction, and we call this “entry” (en). In a junction that has a single linking segment, the linker (L) can be located on either the ex strand (termed the Lex form), or on the en strand (termed Len) (Fig. 2B). These correspond to the Westhof structural families A and C, respectively (Lescoute and Westhof 2006). It is important to note that because the ex and en strands are inequivalent, the Lex and Len structures are also inequivalent (unlike the two stacking conformers of a four-way junction), and are likely to have different stabilities. Where there are two linkers of equal length we term this “equal” (Leq); this corresponds to the Westhof family B (Lescoute and Westhof 2006).
FIGURE 2.
The structurally distinct strands of three-way junctions with formally unpaired linking segments, and possible structures for the IRES junction. (A) The component strands of a three-way junction with two coaxially stacked helices are distinct. The con strand turns about the shared axis of the stacked helices, while the en and ex strands have their 5′ and 3′ termini in the nonstacked helix. (B) Three possible forms of a junction with unpaired RNA linking the unstacked helix to the coaxially stacked pair of helices. If the linker (or longer linker where there are two) lies on the en or ex strand, we term the structures Len or Lex, respectively, while if there are two linking segments of equal length we call it Leq. (C) In principle the three-way junction of the IRES might adopt alternative secondary structures, so there are a number of structures possible. The 3HS4 secondary structure has a single segment linking the D and E arms, and could therefore adopt either Len (C-on-E stacking) or Lex (D-on-C stacking) structures in principle, or both in equilibrium. The 2HS1HS3 or 2HS2HS2 structures would be expected to fold by C-on-E stacking, forming Len and Leq structures, respectively.
The orientation of the third helix in the plane of the three helices can also be specified with respect to the direction of the con strand. It can be directed toward the 3′ end, so that the axes of the ex strand subtend an acute angle, or it can be directed toward the 5′ end, in which case it is the en strand whose axes make an acute angle. Alternatively, the third helix could be approximately perpendicular to the axis of the stacked helix.
In the present study we have examined the HCV IRES three-way junction that includes helix IIId. Since the terminal loop of helix IIId binds to the 40S ribosomal subunit (Kolupaeva et al. 2000; Kieft et al. 2001; Boehringer et al. 2005), and certain mutations in the loop and internal bulge can abolish HCV IRES binding (Jubin et al. 2000; Klinck et al. 2000; Kieft et al. 2001), the structure of the junction and the resulting trajectory of helix D are likely to be important to the function of the IRES element. For this work we have named the component helices C (the upper arm from which extend helices IIIa, IIIb and IIIc), D (helix IIId), and E (the lower arm that includes IIIe and IIIf) (Fig. 1C). The base pairing is conventionally drawn in the form of a 3HS4 junction. However, we note that the base pairing could be rearranged slightly to create either 2HS1HS3 or 2HS2HS2 junctions (Fig. 1D). In principle, the junction could undergo two steps of branch migration, with the 3HS4 and 2HS2HS2 junctions as the extremes. These three forms are unlikely to be of equal free energy, but in the absence of experimental data, we cannot predict which might be the more stable. However, the secondary structure will have a major influence on the fold adopted by the junction.
The 3HS4 structure has a single linking segment of 4 nt, and two connections with zero length linkers. There are, therefore, two possible structures that might be adopted in which either helices C and E, or helices D and C, are coaxially stacked (Fig. 2C). Note that these are not equivalent, as they are Len and Lex structures, respectively. Indeed, Lescoute and Westhof (2006) speculated that the structure of this junction might fall into either family A or C on the basis of the 3HS4 structure. However, if the junction adopts either the 2HS1HS3 or the 2HS2HS2 secondary structure, there would be two linking segments, and thus only one way of stacking two arms without a linker. In this structure helices C and E are coaxially stacked. The 2HS1HS3 junction is a Len structure, while the 2HS2HS2 form is a Leq structure (Fig. 2C).
From this analysis either secondary structure could adopt the structure based on C upon E coaxial stacking. However, it is likely that the orientation of the D arm (and thus its interaction with the ribosome) would depend on which secondary structure is formed. In this study we have examined the conformation of the IRES junction using a combination of comparative gel electrophoresis and fluorescence resonance energy transfer (FRET) in both ensemble steady-state and single-molecule mode. It emerges that in addition to its functional significance, the IRES junction is an interesting test of the conformational principles of three-way helical branch points, requiring a consideration of both secondary and three-dimensional structure.
RESULTS
Analysis of interhelical angles in the IRES three-way junction
We have studied the structure of the three-way junction shown in Figure 1B. The component helices of the junction are Watson–Crick paired for at least three base pairs extending outward from the point of strand exchange. Helix IIId contains a number of formal non-Watson–Crick pairs, and a small internal loop; however, an NMR structure showed this to be essentially helical without major axial perturbation despite the non-Watson–Crick pairs (Lukavsky et al. 2000). For the major part of this analysis we have therefore retained the central sequence of the junction and extended its arms with perfectly paired helices (Fig. 1C), although we return to the natural sequence later. In this study we have named the helical arms C, D, and E as shown, where helix D is equivalent to helix IIId in the complete IRES. The strands are labeled by lower case letters that correspond to the arm in which the 5′ terminus is located. This junction has been analyzed using comparative gel electrophoresis and FRET.
Analysis of interhelical angles in the IRES three-way junction using comparative gel electrophoresis
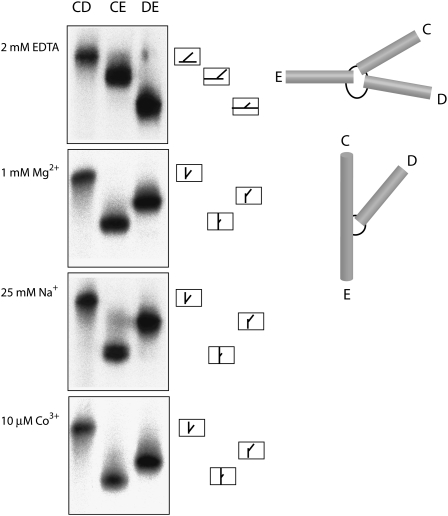
Comparative gel electrophoresis provides a simple yet powerful way to examine the global conformation of a helical junction in RNA under a variety of conditions (Lilley 2008, 2009a). The IRES junction was constructed from three separate strands (each generated by transcription from a DNA template), and prepared in different forms comprising the three possible variants with two long (40 base pairs [bp]) arms and one shortened (10 bp) arm. The species are named according to the long arms; e.g., CD has long C and D arms, and a shortened arm E. The relative electrophoretic mobility in polyacrylamide gels of the three long–short arm species reflects the angle subtended between the two long arms.
The three species of the IRES junction have been electrophoresed in 10% polyacrylamide gels under a variety of ionic conditions (Fig. 3). In the absence of added metal ions (with 2 mM EDTA to chelate any traces of metal ions), the three species migrate with relative rates of DE > CE > CD. This indicates that the largest angle contains the formally unpaired CUUG sequence, suggesting a conformation with an open center. The smallest angle is subtended between the C and D arms. Upon addition of metal ions the pattern of electrophoretic mobility is significantly changed, with relative rates CE > DE > CD. Thus, in common with many helical junctions, the IRES three-way junction undergoes a metal ion-induced change in conformation. The fast mobility of the CE species would be consistent with a predominant species present in a solution whose structure is based upon coaxial stacking of the C and E helices, with the e strand passing continuously between the two helices through the junction. The relative mobilities of the remaining species indicated that helix D is directed into the same quadrant as helix C. The global structure is apparently closely similar in 1 mM Mg2+ and 25 mM Na+ ions, suggesting that specific ion binding may not be required to fold the junction. The same global geometry is also found in the presence of the trivalent ion [Co(NH3)6]3+.
FIGURE 3.
Comparative gel electrophoresis of the conformation adopted by the three-way junction of the HCV IRES in different ionic conditions. The junction studied had a simplified helix D, with the central sequence shown in Figure 1C. Three forms of the junction, each with a different helical arm shortened, are electrophoresed in adjacent tracks of a 10% polyacrylamide gel in the presence of 90 mM Tris-borate (pH 8.3) with the indicated metal ions (or EDTA to chelate metal ions). The long–short-arm junction species are named according to the two long arms. An interpretation of the structures adopted is shown on the right, together with the anticipated structures and mobilities of the long–short-arm junction species. In the presence of EDTA the slow-intermediate-fast pattern is interpreted in terms of the extended structure, while the slow-fast-intermediate pattern observed in the presence of metal ions is interpreted in terms of a structure in which helices C and E are coaxially stacked, with an acute angle subtended between helices C and D.
Analysis of relative end-to-end distances of the arms of the IRES three-way junction using steady-state fluorescence resonance energy transfer
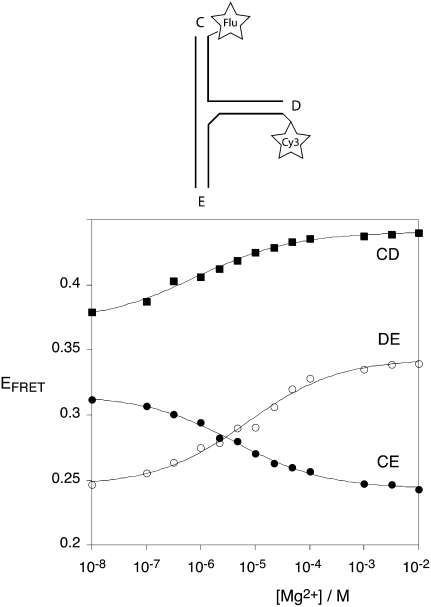
FRET is an alternative way to analyze the global structure of helical junctions in solution, based on energy transfer between donor and acceptor fluorophores attached to the ends of selected pairs of helical arms (Lilley 2009b). Efficiency of the process depends inversely on the sixth power of the distance between the fluorophores (Förster 1948). The IRES junction was constructed with three arms, each comprised of 12 bp (when drawn with 3HS4 secondary structure), labeled with fluorescein donor and Cy3 acceptor attached via the 5′-termini of different strands (Fig. 4). Fluorescein attached at the 5′-terminus is relatively mobile (Norman et al. 2000). This minimizes the complications of fluorophore orientation, so that a simple interpretation based on an inverse relationship between FRET efficiency and distance is unlikely to be misleading. Three such species were constructed, corresponding to the vectors (named according to the arms carrying the donor and acceptor, in that order) CD, CE, and DE.
FIGURE 4.
Analysis of metal ion-induced folding of the IRES three-way junction by steady-state FRET measurements. The junction shown in Figure 1C was prepared with fluorescein (donor) and Cy3 (acceptor) fluorophores attached to the 5′-termini of selected helical arms. These vectors are named by the labeled arms, in the order donor–acceptor, so that the CD vector has fluorescein attached to helix C and Cy3 to helix D. FRET efficiency was measured using the acceptor normalization procedure as a function of Mg2+ ion concentration in 90 mM Tris-borate (pH 8.3), and plotted for the three vectors CD (■), DE (○), and CE (●). The data have been fitted to a two-state model of ion-induced folding (—).
FRET efficiency for the three vectors was measured in the steady state as a function of Mg2+ ion concentration (Fig. 4). This clearly confirms that the structure of the IRES junction depends on the metal ion concentration. The relative interfluorophore distances at low ionic concentration are DE > CE > CD, fully consistent with the global structure deduced from comparative gel electrophoresis. Furthermore, the relative distances in the presence of ≥1 mM Mg2+ (CE > DE > CD) are also consistent with the structure of the predominant species from the electrophoretic experiments in the presence of metal ions. The titrations can be fitted to a two state transition in which the junction folds in response to the binding of Mg2+ ions. Analysis of the data for the CE and DE vectors give half-magnesium concentrations of [Mg2+]1/2 = 4 and 7 μM, respectively. Hill coefficients of 0.52 for both vectors indicate an anti-cooperative process under these conditions, consistent with folding induced by the diffuse interaction of metal ions.
Analysis of the conformation population of the IRES three-way junction using single-molecule FRET
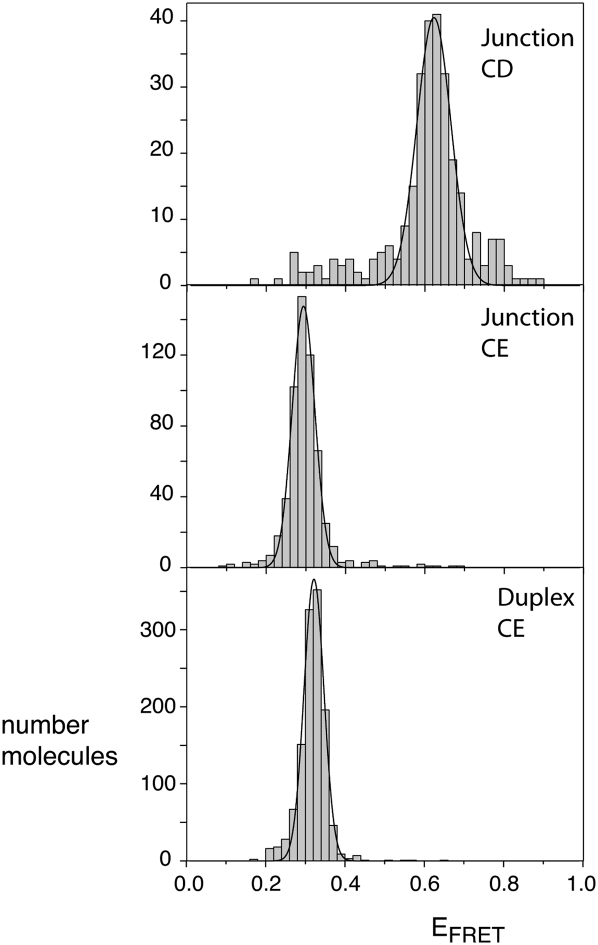
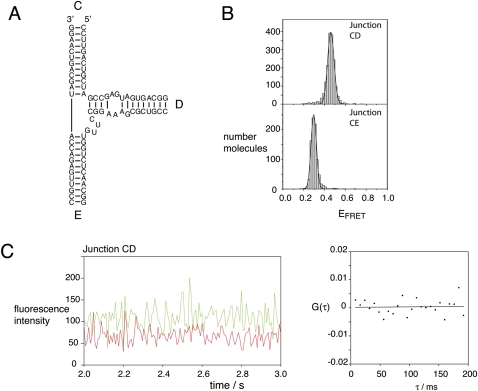
Although electrophoresis and steady-state FRET indicate a predominant conformation that is probably based upon coaxial stacking of C on E arms, these experiments could not exclude a small population of an alternative conformer in equilibrium. We therefore searched for such a conformation using single-molecule FRET experiments. We studied the CE and CD vectors, using Cy3 and Cy5 as donor and acceptor fluorophores, respectively (Fig. 5). In addition, we prepared an identically labeled 24-bp duplex species by hybridization of the e strand to its complement, as a model for C and E arms that are coaxially stacked. All species were studied as single junction molecules encapsulated in phospholipid vesicles in the presence of 10 mM Mg2+ ions, using prism-based total internal reflection fluorescence microscopy. Time traces of donor and acceptor intensities were recorded at time resolutions between 100 and 8 msec, for hundreds of single junctions, and histograms of FRET efficiency were calculated after filtering anomalous data (see Materials and Methods).
FIGURE 5.
Population distributions of FRET efficiencies for Cy3–Cy5-labeled IRES junctions studied as single molecules. Except for the different fluorophores, equivalent junction species as used in the steady-state fluorescence experiments (Fig. 4) were studied. In addition, a Cy3–Cy5-labeled duplex was prepared by hybridization of the junction e strand to its complement, to provide a model for coaxially stacked C and E arms. The different species were encapsulated in phospholipid vesicles in 10 mM Tris-HCl (pH 8.1), 50 mM NaCl, 10 mM MgCl2, and imaging buffer (see Materials and Methods) and studied by total internal reflection fluorescence microscopy. Hundreds of single molecules were studied for each species, and fluorescent intensities at Cy3 and Cy5 emission wavelengths recorded for a number of minutes with a 100-msec resolution. Molecules with aberrant spectral properties were rejected and FRET efficiencies calculated from the remainder. These were plotted as the histograms shown, fitted to Gaussian distributions. Histograms are shown for the CD junction, the CE junction, and the CE duplex. The measured mean EFRET and half-widths for each distribution are presented in Table 1.
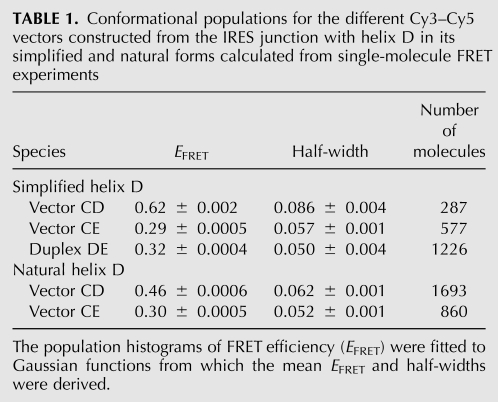
EFRET histograms of data at 100-msec resolution for the junction and duplex species are presented in Figure 5. The CD and CE vectors of the junction give distributions that are well fitted by single Gaussian curves, with mean EFRET values of 0.62 and 0.29, respectively (Table 1). These values are qualitatively in good agreement with the model resulting from the electrophoresis and steady-state FRET data. Moreover, the value for the CE vector is similar to that of the simple CE duplex (EFRET = 0.32), as would be expected if the C and E arms were coaxially stacked in the folded junction. The widths of both distributions for the junction species are not much greater than that for the duplex, indicating that the arms of the junction do not undergo flexural motions of large amplitude. The data for the CD vector has points that are distributed over a wider range of FRET than the Gaussian fit, but these are likely to have arisen from the smaller number of molecules analyzed compared with the CE species.
TABLE 1.
Conformational populations for the different Cy3–Cy5 vectors constructed from the IRES junction with helix D in its simplified and natural forms calculated from single-molecule FRET experiments
Importantly, we see no evidence of additional populations of EFRET, i.e., donor–acceptor distance. In the histogram for the CE vector there is no detectable peak at higher values of EFRET. In the CD vector histogram we observe some points at EFRET = 0.3, but they are randomly distributed rather than forming a distinct peak. Similar histograms were calculated from the data recorded at a faster acquisition time. These data provide no evidence for a minor conformation present in solution under these conditions.
No conformational transitions have been detected in the IRES three-way junction using single-molecule FRET
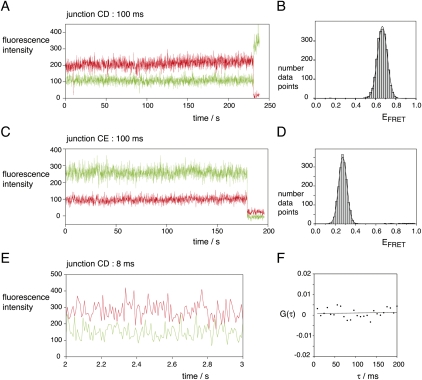
As a further way of seeking alternative conformations, we examined the time records for many single junction molecules, looking for anti-correlated changes in donor and acceptor intensities that would indicate a transition to a different structure. At 100-msec time resolution we can follow donor and acceptor fluorescent intensities over 200 sec or more before photobleaching. Examples of CD and CE vectors are shown in Figure 6, A and C, respectively, with their corresponding FRET efficiency histograms in B and D, respectively. There is no evidence for any structural transition involving a change of FRET efficiency in these traces, and we found very few examples of a transition in the hundreds of time traces examined.
FIGURE 6.
Seeking conformational transitions in single-junction molecules. The CD and CE vectors (the same species used in Figure 5) were separately encapsulated in phospholipid vesicles in 10 mM Tris-HCl (pH 8.1), 50 mM NaCl, 10 mM MgCl2, and imaging buffer, and studied by total internal reflection microscopy. Representative long time records at 100-msec time resolution are shown for the CD and CE vectors (A and C, respectively) up to the point at which fluorophore photobleaching has occurred. Histograms of EFRET for the individual CD and CE molecules are also presented as plots B and D, respectively, for the unbleached sections of the time records. Note that no transitions can be detected over these long time traces. The same preparation of CD vector was also studied at 8-msec time resolution. A 1-sec section of a time record is presented (E). Close examination of this (and many other traces not shown) fails to reveal anti-correlation between Cy3 and Cy5 fluorescence intensity. This is confirmed by performing a cross-correlation analysis on a 4-sec time record (F). The data fit a linear function with no decay, passing through zero.
We also sought short-lived conformational states by examining time traces recorded in the presence of 10 mM MgCl2 at 8-msec time resolution; the highest possible using our EMCCD camera. A 1-sec section of a trace for the CD vector is expanded in Figure 6E. No evidence in this trace (or in many others studied) can be seen for anti-correlation of Cy3 and Cy5 intensities. This is further demonstrated by calculating a cross-correlation function from a randomly chosen 4 sec of data (Fig. 6F). These data show no indication of a decaying anti-correlation, fitting a linear, horizontal function passing through zero. A small subset of junction species (three molecules out of a total of 323) underwent multiple slow transitions between states of different FRET efficiencies; these molecules are clearly of a different character from the majority and are likely to be misfolded or mishybridized junctions.
In summary, using faster acquisition and cross-correlation analysis we have found no evidence for an additional conformation of the junction.
Studying the secondary structure of the IRES three-way junction using in-line probing
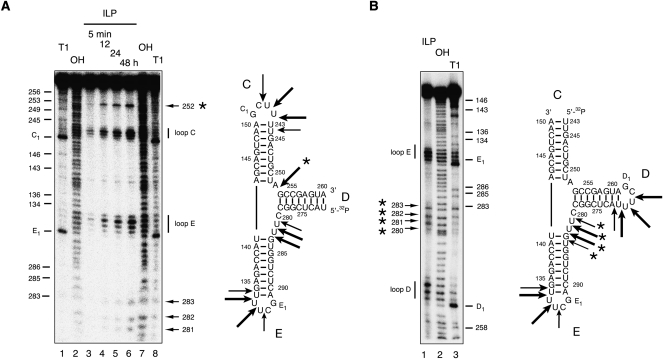
In view of the uncertainty with regard to the secondary structure adopted by the IRES three-way junction, we performed in-line probing experiments in an attempt to identify conformationally flexible nucleotides within the core. In this approach, radioactively [5′-32P]-labeled RNA is subjected to a prolonged incubation in the presence of buffer and Mg2+ ions. Where the backbone is relatively flexible, the RNA can sample a conformation in which the 2′-OH may carry out an in-line nucleophilic attack on the 3′-phosphate, but this is hindered in more rigid parts of the molecule, including duplex regions. Using transcription by T7 RNA polymerase, we prepared two constructs both of which comprised a single strand of RNA, with terminal hairpin loops on either the C and E or the D and E helices (Fig. 7) in order to maximize the resolution of the sequences of interest by gel electrophoresis.
FIGURE 7.
Analysis of the structural flexibility of nucleotides in the IRES three-way junction using in-line probing. Radioactively [5′-32P]-labeled RNA was incubated in 50 mM Tris-HCl (pH 8.3), 100 mM KCl, and 20 mM MgCl2 for various times at 21°C, and analyzed by denaturing gel electrophoresis and phosphorimaging. The junction was studied as a single strand by placing terminal loops onto two helices. In order to maximize the resolution of the sequences of interest, two constructs were studied, each with a single 5′-terminus in either helix D (A) or helix C (B). Bands were assigned by reference to cleavage by ribonuclease T1 (A, lanes 1,8; B, lane 3) and a hydroxide cleavage ladder (A, lanes 2,7; B, lane 2). The junction with open helix D was subjected to in-line probing for 5 min, 12, 24, and 48 h (A, lanes 3–6, respectively), and the open C arm form to a 48-h incubation (B, lane 1). The nucleotides were numbered according to Honda et al. (1999).
As expected, the data reveal that the nucleotides of the loops are flexible. But interestingly, this analysis shows quite clearly that there are two regions of flexibility in the core of the junction. The center of the strand linking the C and E helices (i.e., what would be the e strand if the junction comprised three separate strands) exhibits no detectable reactivity, consistent with the coaxial stacking of the C and E arms. However, the remaining two linking regions do exhibit reactivity, with a single point of cleavage between the C and D arms (equivalent to the c strand), clearly visible in Figure 7A, and four consecutive cleavages between D and E (equivalent to the d strand), seen most clearly in Figure 7B. These data suggest that the predominant form of the junction is 2HS1HS3, although we do not believe that is conclusive. We considered repeating the analysis with junctions in which the secondary structure is locked into place, but this would require changes of sequence at the center of the junction that might well alter the folding significantly. At the present time the most probable conclusion from our data is that the junction is the 2HS1HS3 structure, but it is possible that branch migration occurs that generates a population of the other forms if this did not alter the global folding of the junction.
Restoration of the natural secondary structure of helix D does not significantly perturb the structure of the junction
In the natural form of the junction the secondary structure of helix D contains an internal loop and a number of non-Watson–Crick base pairs after the third base pair from the junction. These were removed from the junction studied above to provide a regular helix to facilitate the analysis. An NMR study of a stem–loop with the sequence of helix D reveals it to adopt the general structure of an A-form helix in broad terms, but in principle, nucleotides from the helix might interact with those of the junction and alter its gross structure or induce transitions between alternative structures. To test this possibility we repeated the single-molecule analysis on a new form of the junction in which the natural structure was restored to arm D. The UUGGGU terminal loop was replaced by four additional base pairs, and the vectors CD and CE were prepared by addition of 5′-linked Cy3 and Cy5 fluorophores as before (Fig. 8A).
FIGURE 8.
Single-molecule FRET analysis of the three-way IRES junction with helix D of the natural sequence. (A) The sequence of the junction studied. This is identical to that of the simplified junction except for the restoration of the base pairing found in the natural junction. However, the terminal loop was removed and the base pairing extended to form a stable helix with a 5′-terminus for fluorophore attachment. The junction is formally depicted in the 3HS4 secondary structure. (B) Population distributions of FRET efficiencies for Cy3–Cy5-labeled vectors encapsulated in phospholipid vesicles in 10 mM Tris-HCl (pH 8.1), 50 mM NaCl, 10 mM MgCl2, and imaging buffer measured at 100-msec time resolution. The total number of molecules included were 1693 and 860 for the CD and CE vectors, respectively. (C) A representative 1-sec time record for the CD vector at 8-msec resolution (left). Four seconds of data were used as input into a cross-correlation analysis (right). These data show no evidence of structural transitions at this time resolution.
The results from the single-molecule FRET analysis are qualitatively similar to those from the junction with a regular helix D. The histograms for the two vectors both reveal a single peak of FRET efficiency, with CD > CE (Fig. 8B). The EFRET value for the CE vector (0.30) (Table 1) is slightly greater than that observed for the simplified junction (0.29), but still very much in the range expected for a coaxial C–E alignment. The half-widths are also similar. The structure of the natural junction is therefore likely to be based on C-on-E helix stacking. However, the EFRET of the CD vector is significantly lower than that for the simplified junction. This might result from a reorientation of helix D arising from new interactions with unpaired bases in that helix. But there are other potential origins for different FRET efficiencies between the two vectors. The D helix in the modified junction is longer than that of the simplified junction by 3 bp, and the Cy5 position is likely to be rotationally different between the two with consequent differences in the relative orientation of the fluorophore transition moments, which can significantly influence FRET efficiency (Iqbal et al. 2008a). Even if orientational effects were unimportant, the FRET efficiency corresponds to a distance that is comparable to the likely Förster length R0, where a relatively small change in distance can result in a significant change in EFRET. As with the simplified junction, we observe no anti-correlated changes in fluorescent intensity between donor and acceptor for either vector (Fig. 8C), providing no evidence for structural transitions in the junction.
DISCUSSION
The three-way junction of the HCV IRES element turns out to be an interesting object lesson in the conformational possibilities of RNA branch points. Like most nucleic acid junctions, it requires the presence of metal ions to fold from an extended form to a form based on pairwise coaxial stacking of helical arms. The transition is induced by Mg2+ ions in low micromolar concentrations, with a Hill coefficient suggestive of nonspecific binding. But the major interest lies in the structure adopted in the presence of metal ions.
Comparative gel electrophoresis and steady-state FRET studies clearly indicate that in the presence of metal ions the junction predominantly adopts a conformation in which arms C and E could be colinear, with an acute angle subtended between arms C and D. The similarity in FRET efficiency between the CE vector of the junction measured in single junction molecules, and a duplex made by hybridizing the e strand to its complement, is consistent with the coaxial stacking of the C and E arms in the folded junction. The half-width of the length distribution for the CE vector of the junction is only a little greater than that of the duplex, indicating that the center of the junction is not very flexible. The mean FRET efficiency is slightly lower for the junction (EFRET = 0.29) compared with the perfect duplex (EFRET = 0.32). While a rotation of the C and E arms hinged about the center would shorten the end-to-end distance and have a relatively small effect on fluorophore orientation, there are two ways in which the efficiency might be reduced compared with the duplex. First, if these arms were unstacked and pulled apart to dislocate the junction, this could increase the end-to-end distance in the junction. Second, a rotation of the helical arms about their common axis might lead to a change in κ2 that gives a lower EFRET. It is known that Cy3 and Cy5 fluorophores may be stacked onto the helical ends of nucleic acid duplexes (Norman et al. 2000; Iqbal et al. 2008b) resulting in significant modulation of energy transfer due to the orientation of their transition dipole moments (Iqbal et al. 2008a). However, the manner of fluorophore attachment to the RNA is not the same as was used in that study, so we cannot be certain about the nature of the RNA–fluorophore interaction in the present case. Both these effects might occur, each contributing to the lower observed EFRET of the junction compared with the perfect duplex.
If the junction significantly populated the 3HS4 secondary structure, it is possible that a subfraction would adopt an alternative conformer in which D and C arms are stacked (Fig. 2C). No minor fraction could be detected using single-molecule FRET, nor were any transitions to such a species observed in the hundreds of single molecules studied. If an alternative conformer exists, it must have a lifetime that is significantly shorter than 8 msec. However, such a fast exchange situation is unlikely. If a small population of the D-on-C stacked conformer was in fast equilibrium with the majority C-on-E form, this would be expected to increase the mean EFRET of the CE vector. But as we have discussed, the measured value is actually lower than the corresponding duplex, not higher. Thus if the junction has the 3HS4 secondary structure, it must be strongly biased to the C-on-E stacking conformer.
By contrast, if the junction has either a 2HS1HS3 or 2HS2HS2 secondary structure the bias to the single C-on-E stacking conformer is immediately explained, because the e strand is the only one lacking a linker. The in-line probing experiment data point toward the 2HS1HS3 structure with the identification of a single flexible nucleotide on the ex strand c, multiple flexible nucleotides on the en strand d, and absence of cleavage on the con strand e. However, this indirect method cannot be regarded as conclusive proof of the structure. If the predominant form is 2HS1HS3, then the structure adopted is quite unusual. It is a Len structure, in which the nonstacked D arm bends into the same quadrant as the C arm. In principle, the nonstacked might bend either way, so that the linker-containing strand turns through an angle that is greater or less than 90°. If we imagine fusing a fourth helix at the position of the linker, these would correspond to parallel or anti-parallel conformations of the resulting four-way junction, respectively. In Len junctions of known structure the linker generally turns through >90° (a pseudoparallel structure, where the ex and en strands cross one another), including the hammerhead ribozyme (Pley et al. 1994; Scott et al. 1995), the VS ribozyme III–IV–V junction (Lafontaine et al. 2002), as well as various cases identified in the ribosome by Lescoute and Westhof (2006). But in the 2HS1HS3 secondary structure, the shorter linker turns through <90°, adopting the pseudo-anti-parallel structure. This seems to be without precedent in the structural database; no 2HS1HS3 three-way junction appears in the RNA junction database (Bindewald et al. 2008). On the other hand, if the junction has a 2HS2HS2 secondary structure the resulting junction is Leq. These tend to fall into the Westhof family B structure, which is pseudoparallel.
In summary, all the evidence indicates that the three-way junction adopts a rather stable fold, in a single stacking conformer, fixing the trajectory of helix IIId (helix D in this study) in the structure. It is possible that it undergoes some limited branch migration that is not detectable in our single-molecule experiments, but no exchange with an alternative stacking conformer has been detected. It is likely that this structurally stable junction has been selected by a requirement for this relatively small RNA to adopt a stable structure that can mediate the interactions with the 40S ribosomal subunit and the translational initiation factor eIF3. While this structure may become distorted upon binding to the ribosome, its stability suggests that it will retain its conformation.
MATERIALS AND METHODS
RNA synthesis and preparation of junctions
RNA oligonucleotides were made by chemical synthesis, either commercially using 2′-ACE chemistry (Dharmacon) or using t-BDMS phosphoramidite chemistry (Beaucage and Caruthers 1981), as described by Wilson et al. (2001). Fluorescein and Cy3 fluorophores were coupled to the oligonucleotides used in the steady-state fluorescence measurements during synthesis. For single-molecule studies all oligonucleotides were made with 5′ aminolinkers, and Cy3 and Cy5 fluorophores were conjugated as N-hydroxysuccinimide esters (Amersham Biosciences). All oligonucleotides were purified by gel electrophoresis in polyacrylamide, and recovered from gel fragments by electroelution or diffusion in buffer followed by ethanol precipitation. Fluorescently labeled RNA was subjected to further purification by reversed-phase HPLC on a C18 column (Waters μ-Bondapak) using an acetonitrile gradient with an aqueous phase of 100 mM triethylammonium acetate at pH 7.0.
Three-way junctions were constructed by mixing one Cy3-labeled strand, one Cy5-labeled strand, and the remaining nonlabeled strand (each 300 pmol) in 90 mM Tris-borate (pH 7.0) and 25 mM NaCl, and annealed by heating at 85°C and cooled slowly to 4°C. The hybridized junctions were purified by electrophoresis in 20% polyacrylamide in 90 mM Tris-borate (pH 8.3) and 25 mM NaCl, and recovered by electroelution and ethanol precipitation.
Transcription of RNA
DNA templates were prepared by 30 cycles of PCR using KOD Hot Start DNA polymerase (Novagen) and 1 μM of each overlapping oligonucleotide, with an annealing temperature of 66°C. RNA was transcribed from 0.15 μM of PCR product in the presence of 40 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 2 mM spermidine, 4 mM of each NTP (Roche), 0.1 U of pyrophosphatase (Sigma), 0.01% Triton X-100 (Sigma), and T7 RNA polymerase for 2.5 h at 37°C (Milligan et al. 1987). The RNA was purified by electrophoresis in a 6% polyacrylamide gel containing 7 M urea. It was visualized by UV shadowing and recovered by electroelution into 8 M ammonium acetate and ethanol precipitation. Purified RNA (6 pmol) was radioactively 5′-32P-labeled using T4 polynucleotide kinase (NEB) and 80 nM of [γ-32P]-ATP (6000 Ci/mmol; Perkin-Elmer) for 1 h at 37°C.
Comparative gel electrophoresis
Radioactively [5′-32P]-labeled species were electrophoresed in 10% polyacrylamide gels (29:1, acrylamide:bis) in 90 mM Tris-borate (pH 8.3), plus added salts or 2 mM EDTA as required. Electrophoresis was performed at 120 V at room temperature, with recirculation of the buffer at >1 l h−1. Gels were dried onto Whatman 3MM paper, exposed to storage phosphor plates, and imaged using a Fuji BAS-1500 PhosphorImager.
The sequences used for the electrophoretic experiments were:
c strand: 5′-GCGCAAGCGACAGGAACCUCGAGGGAUCCGGCGGACUGCUAGCCGAGUACGCGAAGCUUCUCGAGGUUCCUGUCGCUUGCGC-3′;
d strand: 5′-GCGCAAGCGACAGGAACCUCGAGAAGCUUCGCGUACUCGGCCUUGUGGUCUCACGCGUCUAGACUCGAGGUUCCUGUCGCUUGCGC-3′; and
e strand: 5′-GCGCAAGCGACAGGAACCUCGAGUCUAGACGCGUGAGACCAUAGCAGUCCGCCGGAUCCCUCGAGGUUCCUGUCGCUUGCGC-3′.
Further shortened strands were made in order to truncate selected helical arms to 10 bp.
Steady-state fluorescence spectroscopy
Fluorescence spectra were recorded in 90 mM Tris-borate (pH 8.3) at 4°C using an SLM-Aminco 8100 fluorimeter with Phoenix Electronics (ISS). Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of FRET efficiency (EFRET) were measured using the acceptor normalization method (Clegg 1992) implemented in MATLAB. FRET efficiency as a function of metal ion concentration was analyzed on the basis of a model in which the fraction of folded molecules corresponds to a simple two-state model for ion-induced folding, i.e.,
where E0 is the FRET efficiency of the RNA in the absence of added metal ions, ΔEFRET is the increase in FRET efficiency at saturating metal ion concentration, [M] is the prevailing metal ion concentration, KA is the apparent association constant for metal ion binding, and n is a Hill coefficient. Data were fitted to this equation by nonlinear regression. The Mg2+ ion concentration at which the transition is half complete ([Mg2+]1/2) is given by (1/KA)1/n.
The sequences used in the FRET analyses were:
c strand: 5′-CCUUGACUGCUAGCCGAGUACAGG-3′;
d strand: 5′-CCUGUACUCGGCCUUGUGGUCUCAACGG-3′; and
e strand: 5′-CCGUUGAGACCAUAGCAGUCAAGG-3′.
A duplex corresponding to coaxially stacked C and E arms was made by hybridization of Cy5-labeled e strand to its complement (e′ strand) 5′-labeled with Cy3:
e′ strand: 5′-CCUUGACUGCUAUGGUCUCAACGG-3′.
Encapsulation of RNA junctions in phospholipid vesicles for single-molecule studies
A 100:1 molar ratio of unmodified and biotinylated phospholipids was prepared by evaporating a number of aliquots of a mixture of 2.5 mg L-α-phosphatidylcholine and 35 μg 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (Avanti Polar Lipids, Inc.) from chloroform in a stream of argon. The aliquots were hydrated in 250 μL of 50 mM NaCl, 10 mM Tris-HCl (pH 8.1) (TN50 buffer) without (one aliquot, used for surface coating of slides) or with an addition of 200 nM of a given fluorescent RNA junction and 10 mM MgCl2, corresponding to 5 pmol of RNA in a final volume of 250 μL.
The RNA was encapsulated in phospholipid vesicles by repeated extrusion through a polycarbonate membrane containing 200-nm pores using a mini-extruder (both Avanti Polar Lipids, Inc.), creating 200-nm diameter unilamellar vesicles (Boukobza et al. 2001; Okumus et al. 2004). The ratio of RNA to phospholipid resulted in most vesicles being empty, and thus most encapsulations involved a single RNA junction molecule. Slides were prepared by injection of RNA-free phospholipid vesicles into a narrow channel made between a quartz microscope slide and coverslip No 1.5 (VWR international) using double-sided adhesive tape, and left at 4°C for 1 h to allow supported bilayer formation. The sample chamber was then washed with TN50 buffer, treated with 0.2 mg/mL neutravidin (Pierce) for 10 min, and washed. A 1/20th dilution of a chosen encapsulated RNA was injected and allowed to bind to the neutravidin for 15 min. Imaging was performed under the same buffer conditions used for vesicle preparation with an oxygen-scavenging system consisting of 1.6 mg/mL of glucose oxidase, 0.2 mg/mL of catalase, 6% (w/v) glucose, and 1 mM TROLOX (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid).
Total internal reflection single-molecule microscopy
Fluorescence intensities at donor and acceptor wavelengths were acquired from single junction molecules encapsulated in phospholipid vesicles using total internal reflection fluorescence microscopy. The sample was mounted on the stage of an inverted microscope (Olympus IX70) and excited via the evanescent field generated by the total internal reflection of light from a solid state 532-nm laser (Crystalaser) via a quartz prism. Fluorescent emission was collected by a 1.2-numerical aperture 60X water immersion objective lens (Olympus), and separated by a 645-nm dichroic mirror (Chroma Technology) into the donor and the acceptor fluorescence. These were focused side by side into a backilluminated EMCCD camera (iXON, Andor Technology) (Ha 2001). Hundreds of molecules could be recorded simultaneously using an image area of 8.2 × 8.2 mm (512 × 512 active pixels). Data were acquired using software written in Visual C++ (Microsoft), where each frame had a duration of 100 msec (10 frames sec−1) for the population histograms and 8 msec (125 frames sec−1) for the cross-correlation analysis. The highest acquisition rate of the camera is 33 msec, but faster rates can be attained by subdivision of the active number of pixels (16 msec/frame can be achieved dividing the chip by 2 and up to 8 msec/frame using one quarter of the chip).
Measurements were performed at room temperature. Single-molecule FRET efficiency after background correction was approximated by EFRET = IA/(IA+ID), where IA and ID are the fluorescence intensities of the acceptor and donor, respectively. Because the quantum yields and detection efficiencies of Cy3 and Cy5 are very close, EFRET closely matches the true efficiency of energy transfer. However, the spectral overlap separation of Cy3 and Cy5 is not total and the 645-nm separation led to ∼10% of Cy3 leakage into the Cy5 channel. This introduces an apparent EFRET around 0.1 for a single active Cy3 fluorophore in the absence of the Cy5 acceptor.
Data analysis was carried out using laboratory-written analysis routines developed in MATLAB. Single-molecule FRET histograms were obtained using the whole FRET trace, while the population histograms were obtained by averaging frames 11–20 for every individual molecule after manually filtering photobleaching and blinking effects. Filtering involved removal of time traces in which (1) the fluorescence intensity of the acceptor was lower than 50–55 (caused by the absence of Cy5 fluorescence); (2) the total intensity was higher than 550–700 (indicating the presence of multiple fluorophores); (3) absence of anti-correlation between both fluorescence intensities during a blinking, dynamic, or a photobleaching event; (4) the total intensity was irregular over the length of the time-trace; and (5) multiple photobleaching events from the same fluorophore.
Cross-correlation analysis was performed using a program implemented in MATLAB. Records of donor and acceptor intensities as a function of time for single-junction molecules were analyzed using the following equation:
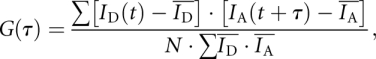 |
where 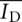 and
and 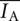 are mean donor and acceptor intensities, respectively, over the whole data record, normalized by the number of points summed (N). The function G(τ) correlates the donor intensity at time t with acceptor intensity at time increment τ later.
are mean donor and acceptor intensities, respectively, over the whole data record, normalized by the number of points summed (N). The function G(τ) correlates the donor intensity at time t with acceptor intensity at time increment τ later.
In-line probing of nucleotide flexibility
Two forms of the IRES junction were generated by transcription of a single RNA species, forming junctions with 5′-termini in either the C or D arms (Fig. 7):
Terminus in C helix: 5′-GGGCAGUCAACUGAUGAGGCCGAAAGGCCGAAACGCGAAAGCGUCUUGACUGCUAGCCGAGUAGCUUUACUCGGCCUUGUGGUCUCAGCUUUGAGACCAUAGCAGUCAA-3′; and
Terminus in D helix: 5′-GGGCCGAGUACUGAUGAGGCCGAAAGGCCGAAACGCGAAAGCGUCUACUCGGCCUUGUGGUCUCAGCUUUGAGACCAUAGCAGUCAAGCUUUUGACUGCUAGCCGAGUA-3′.
During transcription, the hammerhead ribozyme (underlined) underwent cleavage generating a homogenous 5′-end at the IRES junction RNA. These were separately subjected to the in-line probing analysis of Soukup and Breaker (1999). Approximately 0.1 pmol of [5′-32P]-labeled RNA was incubated in 20 μL 50 mM Tris-HCl (pH 8.3), 100 mM KCl, and 20 mM MgCl2 between 1 min and 48 h at 21°C. Aliquots were diluted with 0.5 vol of 100 mM Tris-HCl (pH 7.5) and 20 mM EDTA in formamide (termination buffer). Uniformly cleaved RNA was generated by heating 0.1 pmol [5′-32P]-labeled RNA in 50 mM NaOH at 90°C for 20 sec before addition of six volumes of termination buffer and placing on ice. RNA bands were assigned by comparison with 0.1 pmol [5′-32P]-labeled RNA partial digested with 0.4 U of ribonuclease T1 (which cleaves 3′ to G nucleotides) in the buffer provided by the manufacturer (Ambion). Products of cleavage were separated by electrophoresis in 10% Long Ranger (Lonza) polyacrylamide gels containing 7 M urea, 40% formamide, 90 mM Tris-borate (pH 8.3), and 10 mM EDTA, dried onto Whatman 3MM paper, and visualized by exposure to storage phosphor plates and phosphorimaging (Fuji).
ACKNOWLEDGMENTS
We thank Dr. Tim Wilson for discussions, Dr. Carlos Penedo for providing software, and Cancer Research UK for financial support of these studies.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2158410.
REFERENCES
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Beaucage SL, Caruthers MH 1981. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett 22: 1859–1862 [Google Scholar]
- Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA 2008. RNAJunction: A database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res 36: D392–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H 2005. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure 13: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Boukobza E, Sonnenfeld A, Haran G 2001. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J Phys Chem B 105: 12165–12170 [Google Scholar]
- Brown EA, Zhang H, Ping LH, Lemon SM 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res 20: 5041–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg RM 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 211: 353–388 [DOI] [PubMed] [Google Scholar]
- de la Pena M, Dufour D, Gallego J 2009. Three-way RNA junctions with remote tertiary contacts: A recurrent and highly versatile fold. RNA 15: 1949–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett DR, Murchie AIH, Diekmann S, von Kitzing E, Kemper B, Lilley DMJ 1988. The structure of the Holliday junction and its resolution. Cell 55: 79–89 [DOI] [PubMed] [Google Scholar]
- Duckett DR, Murchie AIH, Lilley DMJ 1995. The global folding of four-way helical junctions in RNA, including that in U1 snRNA. Cell 83: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Förster T 1948. Zwischenmolekulare energiewanderung und fluoreszenz. Ann Phys 2: 55–75 [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T 2001. Single-molecule fluorescence resonance energy transfer. Methods 25: 78–86 [DOI] [PubMed] [Google Scholar]
- Honda M, Ping LH, Rijnbrand RC, Amphlett E, Clarke B, Rowlands D, Lemon SM 1996. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 222: 31–42 [DOI] [PubMed] [Google Scholar]
- Honda M, Beard MR, Ping LH, Lemon SM 1999. A phylogenetically conserved stem–loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol 73: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DMJ 2008a. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci 105: 11176–11181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Wang L, Thompson KC, Lilley DMJ, Norman DG 2008b. The structure of cyanine 5 terminally attached to double-stranded DNA: Implications for FRET studies. Biochemistry 47: 7857–7862 [DOI] [PubMed] [Google Scholar]
- Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM 2000. Hepatitis C virus internal ribosome entry site (IRES) stem–loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol 74: 10430–10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol 292: 513–529 [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol 9: 370–374 [DOI] [PubMed] [Google Scholar]
- Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-ela F 2000. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA 6: 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU 2000. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DMJ 2002. The global structure of the VS ribozyme. EMBO J 21: 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E 2006. Topology of three-way junctions in folded RNAs. RNA 12: 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ 2000. Structures of helical junctions in nucleic acids. Q Rev Biophys 33: 109–159 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ 2008. Analysis of branched nucleic acid structure using comparative gel electrophoresis. Q Rev Biophys 41: 1–39 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ 2009a. Comparative gel electrophoresis analysis of helical junctions in RNA. Methods Enzymol 469: 143–177 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ 2009b. The structure and folding of branched RNA analyzed by fluorescence resonance energy transfer. Methods Enzymol 469: 159–187 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ, Clegg RM, Diekmann S, Seeman NC, von Kitzing E, Hagerman P 1995. Nomenclature Committee of the International Union of Biochemistry: A nomenclature of junctions and branchpoints in nucleic acids. Recommendations 1994. Eur J Biochem 230: 1–2 [DOI] [PubMed] [Google Scholar]
- Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DMJ 2008. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16: 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol 7: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD 2003. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG 2008. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454: 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher SE, Wilson TJ, Lilley DMJ 2003. The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA 9: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherall GW, Uhlenbeck OC 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie AIH, Thomson JB, Walter F, Lilley DMJ 1998. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell 1: 873–881 [DOI] [PubMed] [Google Scholar]
- Norman DG, Grainger RJ, Uhrin D, Lilley DMJ 2000. The location of Cyanine-3 on double-stranded DNA; importance for fluorescence resonance energy transfer studies. Biochemistry 39: 6317–6324 [DOI] [PubMed] [Google Scholar]
- Okumus B, Wilson TJ, Lilley DMJ, Ha T 2004. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J 87: 2798–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr JW, Hagerman PJ, Williamson JR 1998. Protein and Mg2+-induced conformational changes in the S15 binding site of 16 S ribosomal RNA. J Mol Biol 275: 453–464 [DOI] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, McKay DB 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74 [DOI] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J 14: 6010–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W 1995. Almost the entire 5′ nontranslated region of hepatitis C virus is required for cap-independent translation. FEBS Lett 365: 115–119 [DOI] [PubMed] [Google Scholar]
- Scott WG, Finch JT, Klug A 1995. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002 [DOI] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ 2004. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol 11: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2008. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A 1992. Internal ribosome entry site within hepatitis C virus RNA. J Virol 66: 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter F, Murchie AIH, Thomson JB, Lilley DMJ 1998. Structure and activity of the hairpin ribozyme in its natural junction conformation; effect of metal ions. Biochemistry 37: 14195–14203 [DOI] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol 67: 3338–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Zhao Z-Y, Maxwell K, Kontogiannis L, Lilley DMJ 2001. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40: 2291–2302 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]