Abstract
Mammalian cleavage factor I (CF Im) is composed of two polypeptides of 25 kDa and either a 59 or 68 kDa subunit (CF Im25, CF Im59, CF Im68). It is part of the cleavage and polyadenylation complex responsible for processing the 3′ ends of messenger RNA precursors. To investigate post-translational modifications in factors of the 3′ processing complex, we systematically searched for enzymes that modify arginines by the addition of methyl groups. Protein arginine methyltransferases (PRMTs) are such enzymes that transfer methyl groups from S-adenosyl methionine to arginine residues within polypeptide chains resulting in mono- or dimethylated arginines. We found that CF Im68 and the nuclear poly(A) binding protein 1 (PABPN1) were methylated by HeLa cell extracts in vitro. By fractionation of these extracts followed by mass spectral analysis, we could demonstrate that the catalytic subunit PRMT5, together with its cofactor WD45, could symmetrically dimethylate CF Im68, whereas pICln, the third polypeptide of the complex, was stimulatory. As sites of methylation in CF Im68 we could exclusively identify arginines in a GGRGRGRF or “GAR” motif that is conserved in vertebrates. Further in vitro assays revealed a second methyltransferase, PRMT1, which modifies CF Im68 by asymmetric dimethylation of the GAR motif and also weakly methylates the C-termini of both CF Im59 and CF Im68. The results suggest that native—as compared with recombinant—protein substrates may contain additional determinants for methylation by specific PRMTs. A possible involvement of CF Im methylation in the context of RNA export is discussed.
Keywords: protein arginine methyltransferase, PRMT1, PRMT2, PRMT5, mammalian cleavage factor I, CF Im
INTRODUCTION
Messenger RNA precursors (pre-mRNAs) in eukaryotes are processed extensively before they serve as templates for protein synthesis. pre-mRNAs are cleaved at specific sites downstream from the coding region by the cleavage and polyadenylation complex (also called the 3′ processing complex), followed by the addition of a poly(A) extension to the upstream cleavage fragment. A recent proteomic analysis of a purified mammalian 3′ processing apparatus identified about 85 polypeptides (Shi et al. 2009). The core factors of the complex are the cleavage and polyadenylation specificity factor (CPSF) consisting of eight polypeptides: CPSF-30, CPSF-73, CPSF-100, and CPSF-160, as well as FIP1 (Kaufmann et al. 2004), WDR33, Rbbp6, and PP1. Other members of the core complex are the cleavage stimulation factor (CstF; 50, 64, and 77 kDa), cleavage factors I and II (CF Im and CF IIm; two subunits each), and a poly(A) polymerase (PAP), which is responsible for the addition of the poly(A) tail and is assisted by nuclear poly(A) binding protein 1 (PABPN1) for poly(A) length control (for reviews, see Wahle and Keller 1996; Zhao et al. 1999; Martin and Keller 2007; Kühn et al. 2009; Shi et al. 2009).
CF Im is a complex that contains one polypeptide of either 59 or 68 kDa (CF Im59, CF Im68) and a 25 kDa homodimer (CF Im25) (Rüegsegger et al. 1996; Coseno et al. 2008). Splice variants of both factors were also described: CF Im59_S1 as a short form of CF Im59 and CF Im68_L1 as a long form of CF Im68 with a size of 72 kDa (previously called CF Im72 kDa) (see Fig. 1A). CF Im59/59_S1 and CF Im68/68_L1 are encoded by two different genes (Rüegsegger et al. 1996), yet share a high degree of homology (Fig. 1). Both contain an RNA recognition motif (RRM) fold at the N-terminus, a proline-rich region in the center, and an RS-like domain at the C-terminus. The CF Im25 dimer together with one of the two larger subunits is sufficient to stimulate cleavage, although it is not known in what function the large subunits differ (Rüegsegger et al. 1996, 1998). Analysis of the subnuclear localization of CF Im68 has identified this protein in speckles and paraspeckles and its location varies during the cell cycle (Cardinale et al. 2007). Moreover, sequence-specific RNA binding of the CF Im68 subunit to motifs within the 3′ UTR of its own pre-mRNA has been reported to suppress cleavage (Brown and Gilmartin 2003). CF IIm is composed of the subunits hClp1 and hPcf11 (De Vries et al. 2000). Results of several investigations indicate an extensive network of interactions between the factors of the 3′ processing complex and other RNA processing events such as splicing (Gunderson et al. 1994, 1998; Vagner et al. 2000; Kyburz et al. 2006) and alternative splicing (Graveley 2000).
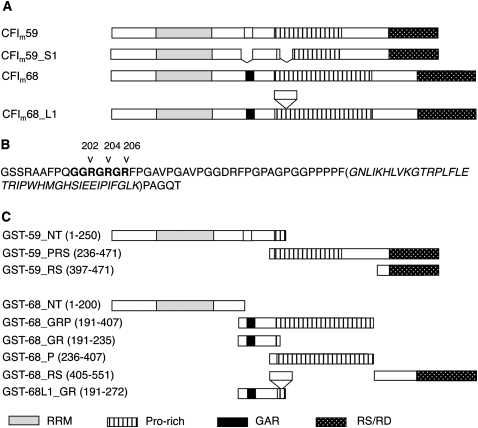
FIGURE 1.
Graphic map of CF Im59 and CF Im68 proteins. (A) Domains are indicated by different patterns (see code under C). RRM, RNA recognition motif; Pro-rich, proline–rich domain; GAR, glycine-arginine motif (the box of potential or cryptic GAR is white in CF Im59 because no methylation was identified); and RS/RD signifies an RS-like domain. Also indicated as insets are included and skipped exons in alternatively spliced forms of CF Im. (B) The protein sequence of the CF Im68 fragment in the GST-68_GR clone and with the exon included (in parentheses) in GST-68L1_GR. (C) Fragments of CF Im59 and CF Im68 used in the experiments. Protein segments are not drawn to exact scale.
Post-translational modifications are present in many factors of the mammalian pre-mRNA 3′ end formation complex (for review, see Ryan and Bauer 2008). They include phosphorylation, as found in CF Im59, Cstf-64, CstF-77, Pcf11, CPSF-100, CPSF-160 (Olsen et al. 2006), and PAP (Colgan et al. 1996), and lysine acetylation, which regulates the interaction between CF Im25 and PAP (Shimazu et al. 2007). In addition, CF Im68 was shown to recruit the CBP acetyltransferase to modulate the localization of PAP through an acetylation–deacetylation cycle (Shimazu et al. 2007). Other examples are sumoylation of PAP (Vethantham et al. 2008) and arginine methylation of CPSF-100, CF Im25, CF Im68, and PABPC2, as found in a screen with the sym10 antibody (Boisvert et al. 2003). All these modifications are thought to be essential for proper functioning and regulation of 3′ end processing.
Arginine methylation is a widespread post-translational modification of proteins in eukaryotes. The enzymes responsible for this activity belong to a family of protein arginine methyltransferases (PRMTs) of which 11 different members have been characterized (for review, see Bedford et al. 2000; Bedford and Richard 2005; Lee and Stallcup 2009; Nicholson et al. 2009; Wolf 2009). PRMTs catalyze the transfer of one or two methyl groups from the methyl donor S-adenosyl methionine (SAM) to arginine guanidino nitrogen atoms. Intermediates with single methyl additions are ω-NG monomethyl arginines. There are two classes of PRMTs that differ in the mode of attachment of the second methyl group. Type I PRMTs produce asymmetric ω-NG,NG-dimethylarginine and include PRMT1 (Lin et al. 1996), PRMT2 (Qi et al. 2002; Lakowski and Frankel 2009), PRMT3 (Tang et al. 1998), PRMT4/CARM1 (Chen et al. 1999), PRMT6 (Frankel et al. 2002), and PRMT8 (Lee et al. 2005a).
Type II methyltransferases PRMT5 (Branscombe et al. 2001), PRMT7 (Miranda et al. 2004; Lee et al. 2005b), and PRMT9 (Cook et al. 2006) promote the formation of symmetric ω-NG,N′G-dimethylarginines. In addition, PRMT10 and PRMT11 were predicted to represent type II PRMTs, but have not been further characterized (for review, see Wolf 2009, and references therein).
The cellular functions of PRMT activities are very diverse and can involve transcriptional regulation by histone arginine methylation, RNA processing, signal transduction, DNA repair, and protein–protein interactions, to name just a few (for review, see Pahlich et al. 2006). In pre-mRNA processing, mRNA transport, and translational control, several proteins have been described to be modified by PRMTs. An example is PABPN1, which is modified by PRMT1, PRMT3, and PRMT6 (Fronz et al. 2008). There, arginine methylation does not prevent self-association of PABPN1 and it is unclear whether arginine methylation has any effect on PABPN1 aggregation, which causes oculopharyngeal muscular dystrophy (Fronz et al. 2008). Another substrate of PRMT1 is heterogeneous nuclear ribonucleoprotein K (hnRNP K), an activator of the tyrosine kinase c-Src. Asymmetric dimethylation of five distinct arginine residues in hnRNP K reduces the interaction with and activation of c-Src and thereby inhibits the c-Src-dependent phosphorylation of hnRNP K (Ostareck-Lederer et al. 2006). PRMT1 appears to be the main PRMT in human cells with the largest spectrum of substrates (Pahlich et al. 2006).
PRMT5, another well-documented methyltransferase, acts in a complex (also called methylosome) with WD45 (also named MEP50) and pICln on many different protein substrates (Meister et al. 2001; Friesen et al. 2002). The PRMT5 complex is also involved in the methylation and subsequent assembly of the spliceosomal U snRNPs, a process that is assisted by a second complex containing the survival of motor neuron protein SMN (Meister et al. 2002; Neuenkirchen et al. 2008).
In this study, we systematically searched for enzymes that modify factors of the 3′ processing complex by adding methyl groups to arginine residues. We demonstrate that CF Im68 and PABPN1 are methylated by factors in nuclear and cytoplasmic HeLa cell extracts. Purification of the activity revealed that the PRMT5/WD45 complex and PRMT1 were responsible for these activities. In addition, the RS-like domains of CF Im59 and CF Im68 could be methylated weakly by recombinant PRMT1. Western blots with antisera, which recognizes symmetrically or asymmetrically dimethylated arginine residues, confirmed that CF Im68 is symmetrically dimethylated and asymmetric dimethylation is detectable only to a minor extent, whereas CF Im59 is dimethylated asymmetrically. Possible roles of arginine methylation in CF Im are discussed.
RESULTS
Systematic search for methyltransferase activity acting on 3′ processing factors
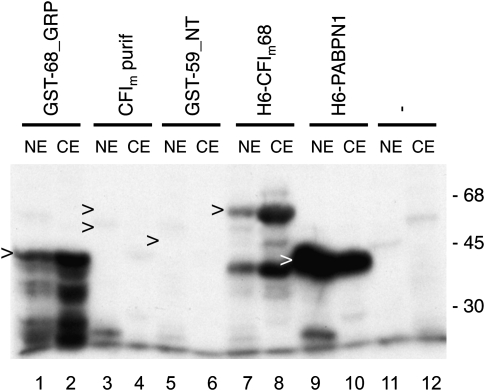
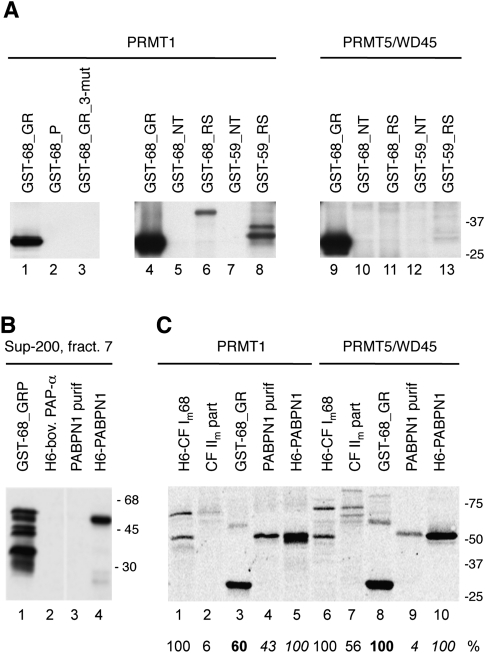
We initiated our study with a systematic search for methyltransferase activity toward characterized factors of the mammalian 3′ processing complex available as recombinant proteins expressed in Escherichia coli or insect cells or as proteins purified from HeLa cells. Methylation reactions typically contained HeLa cell nuclear or cytoplasmic extract, [3H]SAM, and purified proteins as substrates. Some results are shown in Figure 2, and a list of all of the protein substrates tested is presented in Supplemental Table 1. Methylation was only found when CF Im68 or PABPN1 was used as a substrate. The shortest fragment of CF Im68 used (GST-68_GRP) encompassed a region between amino acids 191 and 407 located downstream from the RRM and included the proline-rich domain (Fig. 2, lanes 1,2). We also found methylation of CF Im68_L1, a CF Im68 splice variant (see below; Fig. 3). Interestingly, with both HeLa cell nuclear and cytoplasmic extracts no methylation of the recombinant CF Im59 protein or its fragments was detected (Fig. 2, lanes 5,6; Supplemental Table 1). Methylation of PABPN1 was found to be stronger with nuclear compared with cytoplasmic extract, whereas CF Im68_GR was more intensely methylated by cytoplasmic extract (Fig. 2). CF Im25 was not found to be methylated in several assays (Supplemental Table 1; results not shown). In a proteomics screen CF Im25 was pulled down with an antibody against asymmetrically dimethylated arginines (Boisvert et al. 2003), most likely as a complex with CF Im68.
FIGURE 2.
Methylation activity of nuclear and cytoplasmic extracts on recombinant and purified cleavage and polyadenylation factors. Arrowheads indicate locations of corresponding proteins on Coomassie stained SDS gels (0.5–1 μg were loaded). NE, HeLa nuclear extract; CE; HeLa cytoplasmic extract.
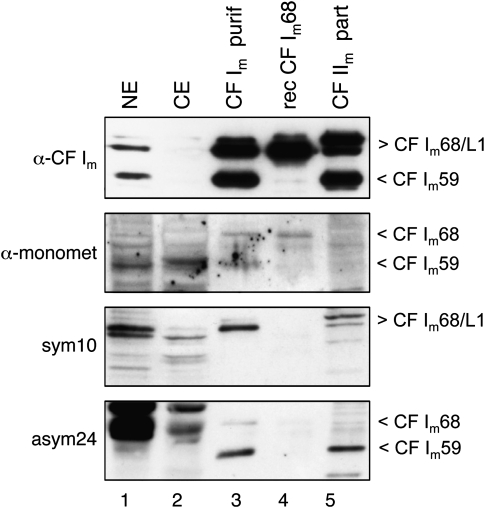
FIGURE 3.
CF Im59 and CF Im68 subunits are differentially dimethylated. Nuclear extract (NE) or cytoplasmic (CE) extract, purified (CF Im purif) recombinant P. pastoris expressed CF Im (rec CF Im) or partially purified CF IIm (CF IIm part) fractions separated on SDS gels and blotted to NC filters were probed with an α-CF Im antibody (reacts with both CF Im59 and CF Im68 subunits) or α-monomethyl, sym10, or asym24 antibody. The label CF Im68/L1 points to CF Im68 (lower band) and CF Im68_L1 (upper band).
Identification of differential methylation in purified CF Im factors
Commercially available antibodies were used to test recombinant and purified CF Im59 and CF Im68 proteins for the presence of symmetric or asymmetric dimethylated arginine residues (Fig. 3). In addition to CF Im68 expressed in Picchia pastoris and partially purified CF IIm (which contains a large proportion of CF Im), fractions of CF Im purified from HeLa cells (Rüegsegger et al. 1996) were separated on SDS gels and probed with polyclonal antibodies specific for monomethylated (α-monomet), and symmetrically (sym10) and asymmetrically dimethylated arginines (asym24) (Boisvert et al. 2003). Both purified and insect cell expressed CF Im proteins are weakly monomethylated (α-monomet panel). Purified CF Im68 was found to be mainly symmetrically dimethylated, suggesting that CF Im68 is a substrate for type II PRMTs (Fig. 3, sym10 panel, lane 3). Also, the longer form of CF Im68, CF Im68_L1 seems to be symmetrically dimethylated (Fig. 3, sym10 panel, lane 5); the signal is weaker because this form of the protein is present in smaller amounts in this fraction (Rüegsegger et al. 1996). A weaker signal with CF Im68 is also seen in Figure 3, lane 3 in the asym24 panel, indicating some asymmetric dimethylation in this protein. CF Im59 only reacted with antibodies against asymmetric dimethylation in the blot, indicating this subunit to be a substrate for type I PRMTs. Later during this work we noticed that antibody sym10 is partially unspecific (see below).
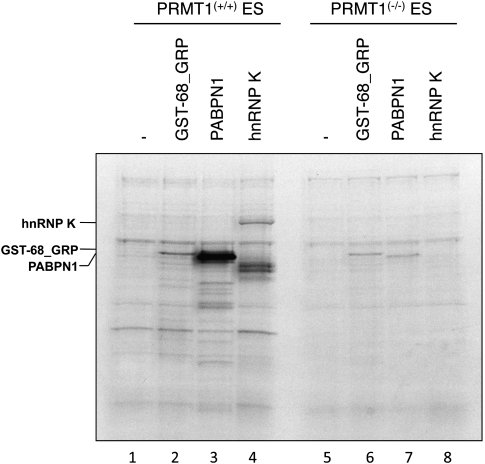
Tests for PRMT1 activity on CF Im68 with extracts from PRMT1 deletion cell lines
PRMT1 was found to methylate factors involved in mRNA translational control, such as hnRNP K (Ostareck-Lederer et al. 2006). Because PRMT1 appeared as a likely candidate for asymmetric methylation of CF Im, we tested methyl transfer to the GST-68_GRP substrate with extracts of the PRMT1 deletion embryonic stem (ES) cell line PRMT1(−/−) (Pawlak et al. 2000). We found GST-68_GRP to be methylated by both the PRMT1(+/+) and the PRMT1(−/−) extracts (Fig. 4, lanes 2,6), although there is a slight reduction of the signal in the PRMT1(−/−) lane. Thus, PRMT1 could be responsible for the asymmetric part of the CF Im68-specific methylation. The control substrate hnRNP K was not methylated by the PRMT1(−/−) extract (Fig. 4, lane 8), consistent with the finding that PRMT1 is the only enzyme that methylates hnRNP K in vitro and in vivo (Ostareck-Lederer et al. 2006). When PABPN1 was incubated with PRMT1(−/−) extract a reduced signal could be detected that most likely occurred from PRMT3 and PRMT6 (Fig. 4, lane 7), which are PRMTs that were shown to act on PABPN1 in addition to PRMT1 (Fronz et al. 2008). Methylation of PABPN1 has been extensively characterized recently (Smith et al. 1999; Kühn et al. 2003) and the protein was found to be asymmetrically dimethylated by PRMT1, PRMT3, and PRMT6 (Fronz et al. 2008). We therefore concentrated on the proteins CF Im59 and CF Im68 in this study.
FIGURE 4.
Activity of PRMT1-deficient cell extracts on protein substrates. Cell extracts from PRMT1(+/+) and PRMT1(−/−) ES cells were incubated with [14C]SAM and recombinant GST-68_GRP, PABPN1, or hnRNP K (10 pmol each). Reactions were resolved on 12% SDS PAGE and methylation was detected by autoradiography.
Purification of methyltransferase activity from HeLa cells and identification of methyltransferase by mass spectrometry
In order to identify CF Im68-specific methyltransferases we first purified the activity in HeLa extracts by column chromatography and used mass spectral analysis to detect potential candidate PRMTs among the purified polypeptides. HeLa nuclear and cytoplasmic fractions were prepared as described (Rüegsegger et al. 1996). Cytoplasmic extract was used for the purification of the activity because we found higher methyltransferase activity specific for CF Im68 in this fraction compared with the nuclear extracts (Fig. 2). Also, the prospective CF Im methyltransferase PRMT1 was reported to be mainly concentrated in the cytoplasm (Herrmann et al. 2005). Starting with 20 mL cytoplasmic extract, the CF Im68-specific PRMT activity was purified as described in Materials and Methods with GST-68_GRP as a test substrate. All affinity columns were low pressure type (except for the final Phenyl Superose and MonoQ columns) and all were run on an FPLC system (from GE Healthcare; see Materials and Methods).
In these experiments, very weak activity specific for CF Im59 was detected with fractionated cell extracts and was consistently lost during the subsequent purification steps (data not shown). Fractions from the gel filtration column with peak activity acting on the GST-68_GRP test substrate were then submitted to mass spectral analysis. These fractions we considered “enriched” with respect to the concentration of active PRMTs, which we estimate in the 1%–5% range. Consistently, PRMT5 and WD45 appeared in the list of identified peptides at positions with high ranks in the mass spectral data (Supplemental Table 2). When the methylation activity was purified over six columns (for details, see Materials and Methods), PRMT5 and WD45 were always present with several peptides. In addition, when column fractions were separated by SDS PAGE and gel slices in the range between 68 and 75 kDa (the predicted molecular weight of PRMT5 is 73 kDa) were analyzed, the number of peptides originating from PRMT5 with high scores was further increased. No peptides of PRMT1 or other members of the PRMT family except for PRMT5 appeared in the mass spectra. In addition, the third subunit of the methylosome, pICln, was never found in the mass spectral data. This was the first indication that WD45 is essential and that the PRMT5/WD45 complex is sufficient for methyltransferase activity acting on CF Im68 (see below). Apparently, pICln is loosely associated with PRMT5 and dissociated during purification under our conditions. In support of these findings, pICln has recently been reported to be an assembly chaperone specific for spliceosomal Sm proteins and it appears to be only transiently associated with PRMT5/WD45 (Chari et al. 2008). Furthermore, fraction 7 of the gel filtration column (results of mass spectral analysis for this fraction are listed in Supplemental Table 2) was used for methylation assays (see below) shown in Figure 8B, below, with substrates PABPN1 and GST-68_GRP.
FIGURE 8.
In vitro methylation of CF Im59 and CF Im68 subfragments. (A) The gels depicted on the middle and right-hand autoradiograms are exposed about 10-fold longer than the autoradiogram to the left. GST-68_GR_3-mut is GST-68_GR with all three arginines (202, 204, and 206) in the GAR motif mutated to alanines. (B) The [3H]SAM activity test with fraction 7 of Superdex-200 column. (C) The [3H]SAM activity test on the native and recombinant substrates. For an explanation of substrate names above the gel see Figure 1 and Supplemental Table 1. Assays were done in the presence of 1.3 pmol of H6-CF Im68, 1.2 pmol of native CF Im68, 4.4 pmol of H6-PABPN1, and 4 pmol of native PABPN1. Also included are 4.8 pmol of PRMT1 and PRMT5/WD45. The signal strength normalized to equal amounts of the substrate are indicated as a percent below the gel and the corresponding numbers are indicated by different fonts (normal, bold, and italic).
These results suggested that the GST-68_GRP substrate was methylated by the PRMT5/WD45 complex and that this fragment should have a GAR motif to be a true substrate. In fact, a predicted GAR motif (GGRGRGRF) is located at residues 201–207 of CF Im68 (Fig. 1B). This is consistent with the results of a proteomic analysis of arginine methylated protein complexes in which CF Im68 was identified as one of the symmetrically dimethylated proteins that is modified at this motif (Boisvert et al. 2003).
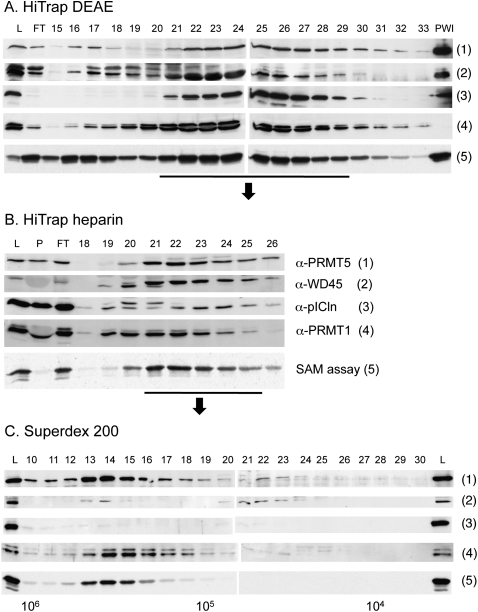
To confirm these results we repeated the fractionation of the HeLa cytoplasmic extract by a different scheme, first using a HiTrap DEAE column, followed by HiTrap heparin and Superdex-200 (see Materials and Methods), and we probed fractions over the entire elution profile with antibodies against PRMT5, WD45, pICln, and PRMT1. In addition, we tested the fractions in a methylation assay (Fig. 5). As a substrate in these assays we used a recombinant polypeptide comprising only the GAR motif (GST-68_GR) with few flanking amino acids. The results indicate that methylation activity coelutes with PRMT5 and WD45 and that pICln does not contribute to activity. Moreover, the Western profile for PRMT1 only partially overlaps with the activity profile, but this does not exclude that PRMT1 contributes to the methylation of GST-68_GR.
FIGURE 5.
Fractionation of the HeLa cytoplasmic extract on the HiTrap DEAE (A), HiTrap heparin (B), and Superdex-200 (C) gel filtration columns. Fractions were probed on Western blots with antibodies against PRMT5, WD45, pICln, and PRMT1 and an activity assay was performed with substrates GST-68_GR and [3H]SAM. L, load; P, pellet; FT, flow through; and PWI indicates 900 ng of the PRMT5/WD45/pICln complex. The numbers are the column fraction numbers.
Generation of recombinant methylosome subunits
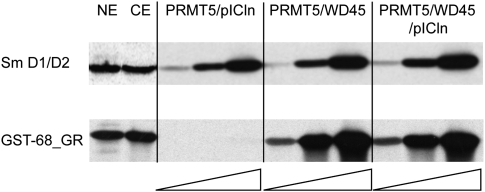
To further analyze the methylation by the PRMT5 complex we made several attempts to reconstitute a complex with recombinant proteins. Expression of the three subunits in E. coli resulted in efficient expression of PRMT5 and pICln but with insoluble WD45. Surprisingly, the resulting PRMT5/pICln complex was highly active in methylating the Sm protein D1, but inactive on the GST-68_GRP substrate (Fig. 6). Finally, a dicistronic vector construct with PRMT5 and WD45 could successfully be expressed in insect cells, resulting in soluble proteins. The complex was fully reconstituted with bacterially expressed pICln if required. As expected from the mass spectrometry data, the PRMT5/WD45 complex showed increasing levels of activity with the GST-68_GR protein substrate (Fig. 6), and addition of pICln did not further stimulate this activity.
FIGURE 6.
The PRMT5/WD45 complex is needed for methylation of the CF Im68 GAR motif. The control substrate Sm proteins D1/D2 (1 μg) and GST-68_GR (800 ng) were incubated in standard methylation reactions with 1 μL nuclear extract (NE) or cytoplasmic extract (CE) or with increasing amounts of the PRMT5 complexes as indicated. Amounts of enzyme present in the reactions were (from left to right) PRMT5/pICln: 10, 100, and 1000 ng; PRMT5/WD45 and PRMT5/WD45/pICln: 1, 10, and 100 ng.
Because the reactions in these tests went to completion, differences in activity with the various subcomplexes are not obvious and kinetic studies were therefore needed to visualize the dependence on substrate concentrations. The results of steady-state kinetics tests clarified the contribution of pICln for CF Im68 methylation (Table 1). A protein-substrate-dependent Michaelis constant KM was several-fold lower when pICln was included in the reaction in addition to PRMT5 and WD45. This suggests that the complete methylosome with all three polypeptides is responsible for the methylation of CF Im68 in vivo. Also, Vmax was more than four times higher with the ternary complex, indicating that only the complete methylosome acts at the full catalytic rate. Moreover, the results of a time course experiment indicate a stimulation of the second methyl addition to monomethylated arginines when the full methylosome complex was present in the reaction (results not shown).
TABLE 1.
GST-68_GR-dependent steady-state kinetics parameters defining methyl transfer by two different PRMT5 complexes
Identification of methylated arginines in CF Im68 by deletion analysis and site directed mutagenesis
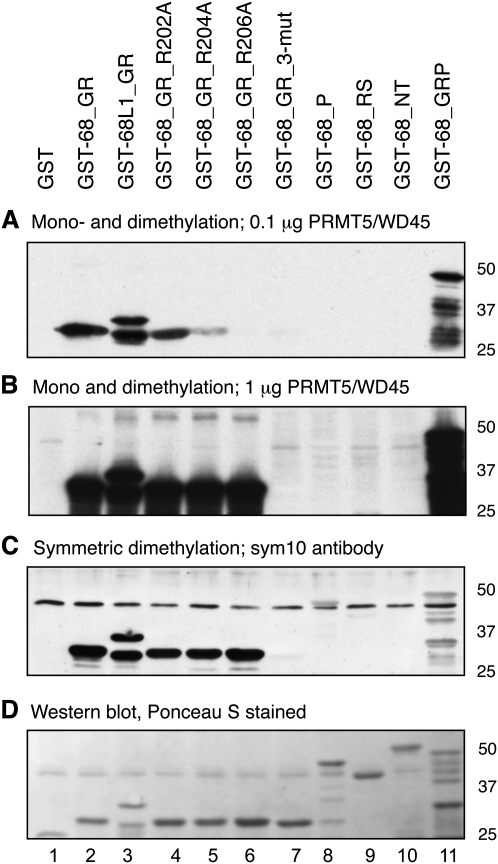
To determine the substrate arginines for PRMT5 within CF Im68, further subfragments of the CF Im68 polypeptide were generated and, in addition, the three arginines within the GAR motif were mutated to alanines (Fig. 1C). Methylation was found exclusively in the recombinant protein fragment containing the GAR motif (GST-68_GR) (Fig. 7A–D, lanes 2–6), whereas the proline-rich domain (Fig. 7A–D, GST-68_P, lane 8) that contains four single RG or GR motifs between amino acids 375 and 400 was not methylated. In addition, fragments representing the RS domain downstream from residue 405 and comprising the N-terminal RRM domain were not methylated in the in vitro assays (Fig. 7A–D, lanes 9,10).
FIGURE 7.
Only arginines within the GGRGRGRF motif are methylated in CF Im68. Site directed point mutations within the GAR motif in the GST-68_GR construct and fragments of CF Im68 were subjected to methylation in vitro by the recombinant PRMT5/WD45 complex. GST-68_GR_3-mut is GST-68_GR with all three arginines (202, 204, and 206) in the GAR motif mutated to alanines. (A) One microgram of the recombinant substrate protein was incubated with 100 ng of PRMT5/WD45 and [3H]SAM. (B) The same as A except that 1 μg of PRMT5/WD45 was used. (C) The same as B except that the reaction contained cold SAM and the blot was probed with the sym10 polyclonal antibody against the SDM arginines. (D) Blot from C stained with Ponceau-S.
Mutation of single arginines to alanine within the GAR motif of CF Im68 resulted in variable effects on methylation activity when 100 ng of recombinant PRMT5/WD45 was present in the assay (Fig. 7A). Mutation of R206 or of all three arginines (R202A, R204A, and R206A) resulted in a complete loss of methylation, whereas the mutation of R202 and R204 had less of an effect (Fig. 7A, lanes 4–6). When the concentration of the PRMT5/WD45 complex in the reaction was increased to 1 μg, the methylation activity was completely lost only when all three arginines were mutated (Fig. 7B, lane 7). And obviously, when the products were probed for symmetric dimethylation by sym10 antibodies, mutation of all three arginines was required to abolish activity (Fig. 7C, lane 7).
In vitro assays with recombinant PRMT1 and PRMT5/WD45
To further analyze the different domains in CF Im59 and CF Im68 for methylation by the recombinant PRMTs, we incubated recombinant subfragments of the CF Im subunits with PRMT1 and PRMT5/WD45. The results indicate that PRMT1 also acts on the same GAR motif in CF Im68 as PRMT5 does, because the fully mutated motif (GST-68_GR_3-mut: all three arginines mutated to alanine) is not methylated by PRMT1 (Fig. 8A, lane 3). In addition, PRMT1 does not methylate the proline-rich and the RRM domain of CF Im68 (Fig. 8A, lanes 2,5). However, after longer exposure, methylation of the RS-like domains of CF Im59 and CF Im68 by PRMT1 is clearly visible (Fig. 8A, lanes 6,8), whereas PRMT5/WD45 does not have such an activity.
In initial tests we noticed that fractions from the Superdex-200 column used for purification of the methylating activity of HeLa extracts methylated recombinant but not purified native, CF Im68 and PABPN1 proteins (Fig. 2, lanes 3,4; Fig. 8, lanes 3,4; Supplemental Table 1). We therefore did another experiment with high concentrations of recombinant PRMT1 and PRMT5/WD45 to see whether the low activity with native proteins in initial tests was just the result of the low concentration of PRMTs present in the HeLa cell extracts, in addition to the low substrate concentrations used (Fig. 8C). It indeed seems that high concentrations of PRMT1 and PRMT5 can also methylate the native CF Im68 and PABPN1 proteins. However, we observe now that PRMT1, as the major PRMT for PABPN1 (Smith et al. 1999; Fronz et al. 2008), adds about half of the SAM to the native compared with the recombinant PABPN1 (Fig. 8C, lanes 4,5). In contrast, PRMT5/WD45 is active on both native and recombinant CF Im68 substrates (Fig. 8C, lanes 6,7), consistent with our observations that PRMT5 is the main PRMT for CF Im68. It is thus possible that native proteins contain additional information to become more specific substrates for a particular PRMT. Therefore, the use of truncated substrates (such as GST-68_GR for CF Im68) always raises the question of physiologically relevant specificity.
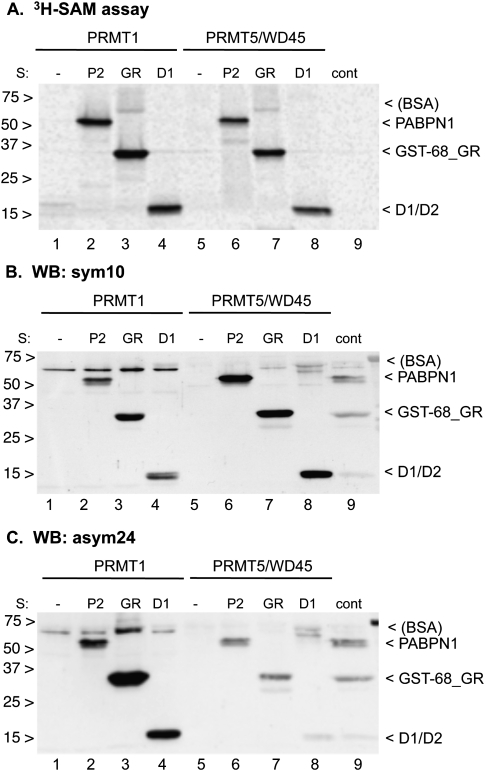
In order to test and assess the activity of PRMT1 and PRMT5 on the substrates GST-68_GR, PABPN1, and D1/D2, we compared the total methyl transfer with tritium labeled SAM. In addition, we determined the fractions of symmetrical and asymmetrical dimethylation with the antibodies sym10 and asym24. Results of the [3H]SAM assay indicate that PRMT1 is slightly more active on PABPN1 when compared with PRMT5/WD45 (Fig. 9A, lanes 2,6). In contrast, on average in several repeats of this assay, we found PRMT5 to be 30%–50% more active on the GST-68_GR and D1/D2 substrates when PABPN1 was normalized.
FIGURE 9.
In vitro activities of recombinant PRMT1 and the PRMT5/WD45 complex. Assays were done in the presence of 2 pmol each of substrates PABPN1, GST-68_GR, or D1/D2 and the reactions were incubated for 2.5 h with 0.32 μM PRMT1 (P1) or the 0.32 μM PRMT5/WD45 complex (P5W) as described in Materials and Methods. (A) For the [3H]SAM assay, 1.04 μCi (or 1.02 μM) [3H]SAM was complemented with an unlabeled SAM to a total concentration of 20 μM. Protein gels were blotted to nitrocellulose and exposed to PhosphorImager screens. (B,C) For Western blots, an unlabeled SAM (20 μM) was present in each reaction. The blots were probed with the sym10 and asym24 antibodies (1:2000 dilution).
For Western blots we incubated 0.14 μM protein substrates PABPN1, GST-68_GR, and D1/D2 with 20 μM SAM with and without 1.5 μM PRMT1 or 5 μM PRMT5/WD45, and probed the proteins blotted to nitrocellulose with the sym10 and asym24 antibodies (Fig. 9B,C). As expected, the PRMT5/WD45 complex indeed appears to add a higher rate of symmetric dimethyl groups to the substrates than PRMT1 (Fig. 9B, lanes 6–8) after subtracting the signal of the control (Fig. 9B, lane 9). In contrast, in the blot that was probed with the asym24 antibody, only PRMT1 strongly adds asymmetric dimethyl groups to all substrates (Fig. 9C, lanes 2–4), whereas no such methylation is detected with PRMT5/WD45 beyond the level seen in the control (Fig. 9C, lane 9).
These results indicated that at least the sym10 antibody is partially nonspecific because PRMT1 is not known to have symmetric dimethylation activity. Also, both sym10 and asym24 antibodies react weakly with the unmethylated substrates in the control lane (Fig. 9C, lane 9). We believe that this cross-reactivity occurs by the binding of the two antibodies to the unmodified GAR motifs because no binding is seen to bovine serum albumin (BSA) in the blot, which is present at about the same concentration as the three protein substrates [indicated by the label < (BSA) next to the gel panels in Fig. 9].
In summary, these results show that in vitro, PRMT1 and PRMT5/WD45 mono- and dimethylate the substrates PABPN1, GST-68_GR, and D1/D2. In addition, we conclude that the results obtained with dimethylation-specific antibodies, in particular sym10, must be regarded with caution.
Mass spectral analysis of an in vitro methylated recombinant CF Im68_GR fragment
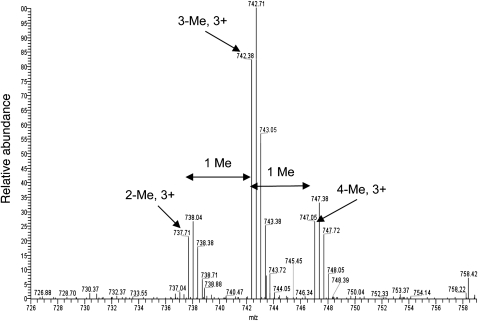
To further characterize the GAR motif with respect to its methylation pattern, the recombinant GST-68_GR fusion protein was methylated with the recombinant PRMT5/WD45/pICln complex. Products containing symmetrically dimethylated arginines were immunoprecipitated with sym10 antibodies, separated by SDS gel electrophoresis, and bands corresponding to the GST-68_GR protein were excised and submitted to mass spectral analysis (see Materials and Methods for details). The purpose of the gel was to separate the GST-68_GR protein from IgG and the methylosome components.
One tryptic peptide was detected with the GAR motif in three peaks of additional masses, accounting for the addition of two, three, or four methyl groups (Fig. 10). This indicates that one of the arginines must be dimethylated; only this would allow immunoprecipitation by the sym10 antibody. The two other species may have either one (for triple methylation) or two additional monomethylations or a second dimethyl group, resulting in quadruple methylation. The arginine at the C-terminus of the peptide is assumed to be unmethylated because trypsin does not cleave the C-terminal to methylated arginines. Apparently, the presence of one doubly methylated arginine in the GGRGRGR sequence seems to prevent cleavage or leads to inefficient cleavage at the two other arginines within the motif.
FIGURE 10.
Mass spectral analysis of the methylated GAR motif. Sum of the spectra from the LC/MS run of di-, tri-, and quadruply methylated AAFPQGGRGRGRFPGAVPGGDR-peptide with a triple charge.
We made several attempts to identify peptides containing methylated arginines from the GAR motif in HeLa cell CF Im68 by mass spectrometry, but did not detect any modified peptides with sufficient probability scores. As the source we used either CF Im68/59/25 purified from HeLa cells or immunprecipitated with anti-CF Im68 or the sym10 antibody and digested with trypsin and chymotrypsin. However, we also could not identify unmethylated peptides from the GAR region in the mass spectrum.
Proof that a dimethylated form of CF Im68 exists in HeLa cells comes from Western blots (Fig. 3, lanes 3,4), where we show that the sym10 antibody reacts with CF Im68 purified from HeLa cells, but not with CF Im68 expressed in insect cells. In addition, when we probed proteins previously immunoprecipitated with anti-CF Im antibody with anti-CF Im or sym10 antibodies in Western blots both signals were comparable in intensity (data not shown).
DISCUSSION
To investigate the potential post-translational modifications in 3′ processing factors we systematically tested the addition of methyl groups to purified and recombinant proteins by methyltransferases present in nuclear and cytoplasmic extracts. A first in vitro methylation screen identified two proteins as substrates, PABPN1 and CF Im68. Chromatographic purification of methylation activity on CF Im68 in cytoplasmic extracts led us to the mass spectral identification of the arginine methyltransferase PRMT5 and a cofactor WD45 strongly suggesting that PRMT5 is the major enzyme responsible for CF Im68 methylation. We could subsequently confirm that PRMT5 and WD45 were essential for methylation of the GAR motif in CF Im68 by use of a reconstituted recombinant complex of the two factors, and we found pICln to be stimulatory to this activity.
The site of methyl addition in CF Im68 could be tracked down to a single GAR motif containing three characteristic tandem GR motifs. We could partially dimethylate the GAR motif arginines in vitro by use of a reconstituted recombinant methylosome. The two subunits PRMT5 and WD45 were essential and apparently sufficient for activity on CF Im68. Steady-state kinetics tests indicated the catalytic rate with the full PRMT5/WD45/pICln complex was higher for the GST-68_GR substrate than for the PRMT5/WD45 complex, but also the KM value was lower, resulting in a higher catalytic efficiency, expressed as kcat/Km, for the ternary complex (Table 1). This suggests a function for WD45 as a specificity factor in the methylation of CF Im68. In contrast, with a spliceosomal Sm D1/D2 heterodimer we found both the PRMT5/pICln and the PRMT5/WD45 subcomplexes to be active. pICln was recently identified as an assembly chaperone that promotes spliceosomal snRNP formation together with the SMN complex (Chari et al. 2008). The stimulating effect of pICln on activity with CF Im68 is most likely caused by the stabilization of the complex. Interestingly, no methylation of the substrate GST-68_GR was detected when pICln was added as a potential cofactor to recombinant PRMT5 in the assay (Fig. 6). It remains to be shown whether the WD45 subunit is a specificity factor for proteins other than CF Im68.
We had indications that purified CF Im68 also contains asymmetrically dimethylated arginines, modifications that would require type I PRMTs. One obvious candidate for such an activity is PRMT1. We could confirm this in experiments with a PRMT1 deletion cell line (Fig. 4) and also by in vitro assays with recombinant PRMT1 (Figs. 5, 8, 9). The fraction of asymmetric dimethylation within the total arginine methylation in CF Im68 is rather small and there could be competition between the PRMTs for a given substrate. In our tests, methylation of the C-termini of CF Im59 and CF Im68 by PRMT1 was weak and it cannot be excluded that this activity is nonspecific considering the large number of arginines in the RS-like domains present in both factors. In addition, it is possible that other type I PRMTs are involved in methylating these proteins. Moreover, we should point out that physiological relevance has not been demonstrated by linking the information obtained on the truncated substrate GST-68_GR with the native CF Im68. The results depicted in Figure 8C confirm these concerns and suggest that specificity for PRMTs can be different in native compared with recombinant protein substrates.
In order to search for factors interacting with CF Im59 and CF Im68, we also carried out a yeast two-hybrid screen (results not shown; see Materials and Methods for details). We found an interaction between the SH3 domain of the protein arginine methyltransferase PRMT2 and CF Im59, whereas no interaction was found of any PRMT with CF Im68. PRMT2 was shown to have asymmetric dimethylation activity on histone H4 (Lakowski and Frankel 2009). An interaction between CF Im59 and PRMT2 was also described in a proteome-scale screen (Rual et al. 2005). However, we could not detect any methylation activity with recombinant PRMT2 (results not shown).
We made several attempts to identify a function for arginine methylation in CF Im. Such functions could include regulation of interactions with either other protein factors or with an RNA substrate, where CF Im could also have a role as a specialized adaptor in a pre-mRNA transcription and processing interactome. Methylation of the GAR motif could regulate interactions shown to exist with the SR proteins 9G8, Srp20, and hTra2b (Dettwiler et al. 2004). Also, interactions could be regulated by WW proteins that bind to proline-rich motifs in their interactors (Ingham et al. 2005). However, in extensive tests we could not find evidence for an effect of methylated versus unmethylated CF Im68 on its interaction with its partner CF Im25 or with various RNA molecules (results not shown).
Another function of methylation within the CF Im68 GAR motif could be the regulation of RNA export carried out by CF Im68, as recently reported by Ruepp et al. (2009), who demonstrated that CF Im68 shuttles between the nucleus and the cytoplasm in a transcription-dependent manner and, at the same time, interacts with the mRNA export receptor NXF1/TAP. In addition, it was postulated that CF Im is a functional analog of the 3′ end processing factor Hrp1p (Nab4) in the yeast Saccharomyces cerevisiae (Zhao et al. 1999). Indeed, both CF Im and Hrp1p are involved in poly(A) site recognition and 3′ processing complex assembly and function in poly(A) site cleavage and polyadenylation (Venkataraman et al. 2005). Moreover, both Hrp1p (Shen et al. 1998) and CF Im (Boisvert et al. 2003; this work) can be modified by arginine methylation. We are currently investigating a possible role of the methylation state of CF Im on shuttling and RNA transport by testing effects of PRMT knockdown or mutagenesis in the CF Im68 GAR motif.
Furthermore, to confirm that PRMT5 is responsible for methylation of CF Im68 we used RNAi to knock down PRMT5 expression in HEK293 cells. The results indicated that PRMT5 is a very stable protein in the cells. Although we were able to dramatically reduce the level of PRMT5 after 3–5 d by siRNA transfection, the level of symmetric dimethylation in immunoprecipitated CF Im68 or CF Im68_L1 was not reduced (results not shown). This indicates that not only the CF Im proteins, but also their modifications are very stable. Also, our observation that dimethylation in CF Im and of PABPN1 purified from HeLa cells is stable and is not lost during the purification process, and in addition cannot be further dimethylated in vitro (Fig. 2 A,B), indicates that these modifications are extremely stable and may not be reversible.
In summary, this work was intended to shed some light on the pattern of arginine methylation in factors involved in pre-mRNA 3′ end processing with a focus on CF Im. We have found two arginine methyltransferases that add methyl groups either symmetrically or asymmetrically to arginines within one particular motif in CF Im68. Our results should form a basis for further research to elucidate a function of these modifications within the complex mechanism of pre-mRNA 3′ end processing.
MATERIALS AND METHODS
Proteins and antibodies
Nuclear and cytoplasmic extracts were prepared with minor modifications as described previously (Dignam et al. 1983). For the cloning of CF Im59, peptide sequencing was carried out from the same pool of human CF Im and according to the same procedure as described previously for CF Im68 (Rüegsegger et al. 1998). The sequence of CF Im59 was submitted to the GenBank database (accession AJ275970).
Because both CF Im59 and CF Im68 could not be expressed as full-length proteins in bacteria, they were produced in insect cells or Picchia pastoris (used for Western analysis) (Fig. 3, lane 4). For baculovirus/insect cell expression of proteins in the Bac-To-Bac expression system (Invitrogen), coding regions or fragments of CF Im were inserted into the transfer vector p-FASTBAC-Hta, and generation of recombinant proteins was done as described previously (Dettwiler et al. 2004). Reconstituted recombinant CF Im25 and CF Im68 subunits expressed in P. pastoris were described previously (Rüegsegger et al. 1998). For the Western blot in Figure 3 (lane 4), 2 μL of fraction 25 from the original Phenyl Superose column was used.
Purified CF Im from HeLa cells was described previously (Rüegsegger et al. 1996). Fraction 40 (5 μL) of the final MonoQ eluate was used for Western blots depicted in Figure 3 (CF Im purif, lane 3). Partially purified CF I/IIm was a gift from Henk de Vries and was described previously (De Vries et al. 2000). Fraction 43 (10 μL) of Phenyl Superose column 4 was used for Western blots (Fig. 3; CF IIm part, lane 5).
Recombinant PRMT1 was expressed and purified as described previously (Ostareck-Lederer et al. 2006). PRMT5 and WD45 were coexpressed in Sf21 (Spodoptera frugiperda) insect cells with the MultiBac system (Berger et al. 2004). PRMT5 contained an N-terminal His-tag. Harvested cells were resuspended in 20 mM HEPES-NaOH (pH 8), 0.5 M NaCl, and 10 mM imidazole, lysed by sonication, and the lysate clarified by centrifugation at 40,000 rpm in a 45Ti rotor. The PRMT5/WD45 heterodimer was purified from the cleared lysate by Ni-NTA chromatography (Qiagen), followed by dialysis and anion exchange chromatography on a 1 mL HiTrap Q column (GE Healthcare). The final purification step was gel filtration on a Superdex 200 column (GE Healthcare) run in 20 mM HEPES-NaOH (pH 7.5), 0.2 M NaCl, and 5 mM DTT.
The PRMT5/pICln heterodimer was expressed in BL21(DE3) cells induced with 1 mM IPTG at 25°C for 5 h from a tricistronic expression plasmid encoding PRMT5, WD45, and pICln with an N-terminal His-tag on pICln. WD45 was insoluble when expressed with this construct. Harvested cells were resuspended in 20 mM HEPES-NaOH (pH 8), 0.5 M NaCl, and 10 mM imidazole, lysed by sonication, and the lysate clarified by centrifugation at 40,000 rpm in a 45Ti rotor. The PRMT5/pICln heterodimer was purified from the cleared lysate by NiNTA chromatography (Qiagen). The final purification step was gel filtration on a Superdex 200 column (GE Healthcare) run in 20 mM HEPES-NaOH (pH 7.5), 0.2 M NaCl, and 5 mM DTT.
Details for the bacterial expression and purification of pICln have been described elsewhere (Chari et al. 2008). For reconstitution of the heterotrimeric PRMT5/WD45/pICln complex, the PRMT5/WD45 heterodimer was incubated with an equimolar amount of bacterially expressed pICln at 4°C overnight. The reconstitution mixture was then purified by gel filtration chromatography on a Superdex 200 column (GE Healthcare) run in 20 mM HEPES-NaOH (pH 7.5), 0.2 M NaCl, and 5 mM DTT. The Sm proteins D1/D2 were purified as described previously (Chari et al. 2008).
The anti-PRMT5, anti-symmetric (sym10), and anti-asymmetric arginine (asym24) antibodies were from Upstate and the anti-monomethyl arginine antibodies (ab414) were from Abcam. The anti-PRMT1 antibodies (MAT-B12) were from Santa Cruz. Anti-CPSF-100 and CPSF-160 antibodies were described previously (Jenny and Keller 1995), and the anti-CF Im antibodies were also described previously (Rüegsegger et al. 1998).
In vitro protein methylation assay
In a typical methylation assay, the reaction mixture contained between 400 and 1000 ng of a protein substrate, between 0.5 and 1 μCi S-adenosyl-L-[methyl-3H]-methionine (82 Ci/mmol from GE Healthcare or 70.8 Ci/mmol from NEN/Perkin Elmer), and methyltransferase preparations. The reaction was done in 10 or 15 μL total volume and incubated at 37°C for 2 h or up to overnight. Ten microliters of 2× SDS PAGE loading buffer was added and the products were separated on 10% or 12.5% SDS PAGE gels depending on the size of the substrate. The proteins were then blotted by a semidry blotting system (NOVEX from Invitrogen) onto nitrocellulose (Protran BA85, Whatman) and the filters were exposed to a PhosphorImager Tritium screen (GE Healthcare) for quantification or to a Kodak BioMax MS film with a BioMax TRANSCREEN LE intensifying screen (from Kodak) for 4 h or overnight.
PRMT1(+/+) and PRMT1(−/−) ES cell extracts and methylation assays
Methylation assays with ES cell extracts were done as described in Ostareck-Lederer et al. (2006).
Fractionation of protein methyltransferase activity
The following buffers were used. Buffer A contained 25 mM Tris-HCl (pH 7.9), 10% glycerol, 0.2 mM EDTA (pH 8.0), 0.02% NP-40, 0.5 mM dithiotreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.4 μg/mL leupeptin hemisulfate, and 0.7 μg/mL pepstatin. Buffer B was buffer A with 1 M KCl, and buffer C was buffer A with 0.1 M KCl. All columns were run on an FPLC system (GE Healthcare) inside a cooling cabinet.
The HeLa cytoplasmic extract was prepared as described previously (Dignam et al. 1983), except that in all buffers, Hepes-KOH was replaced by Tris-HCl (pH 7.9). Cytoplasmic extract corresponds to the S-100 fraction of Dignam (Dignam et al. 1983). Typically, 20 mL of dialyzed cytoplasmic extract was spun to remove insoluble materials and applied to three 5 mL HiTrap-DEAE-FF columns connected in series and equilibrated in buffer C. The column was washed with 1.5 column volumes of buffer C and protein was eluted with a linear gradient of buffers A and B from 0.1 to 1 M KCl. Fractions were assayed with 500 ng of GST-68-GRP and [3H]SAM as substrates. The active fractions were pooled, dialyzed against buffer A with Hepes (pH 8.0) and 50 mM KCl, loaded onto a 5 mL HiTrap SP column (GE Healthcare), and eluted with a 20–1000-mM gradient; activity was in the flow through. The flow through was directly loaded on a 1 mL HiTrap heparin column (GE Healthcare) and eluted with a gradient of 50–1000 mM KCl (protease inhibitors were omitted from here on). Active fractions were concentrated in a Centricon-30 device and separated on a Superdex-200 gel filtration column (GE Healthcare) in buffer A with 150 mM KCl. The active fractions were subjected to ammonium sulfate precipitation (35%) by direct addition of solid ammonium sulfate. The precipitate was collected by centrifugation at 10,000 rpm for 30 min and the pellet was resuspended in buffer A with 500 mM ammonium sulfate and loaded on a 1 mL Phenyl-Superose column (GE Healthcare). A decreasing gradient of 500–20 mM ammonium sulfate was applied. The active fractions were dialyzed against buffer A containing 20 mM KCl, loaded on a MonoQ column (GE Healthcare), and eluted with a 20–800 mM KCl gradient.
Yeast two-hybrid analysis of CF Im59
The coding sequence for the full-length FLJ12529 protein (GenBank accession number gi: 22450805) was PCR-amplified and cloned into pB27 as a C-terminal fusion to LexA (N-LexA-FLJ12529-C) and sequenced. Yeast two-hybrid screening was performed by Hybrigenics, S.A. (http://www.hybrigenics.com). Details of the procedure can be obtained by request from the corresponding author.
Mass spectral analysis
The protein fractions enriched for methyltransferase activity were ethanol precipitated before tryptic digestion. For determination of methylarginine in the GAR motif protein GST-68_GR (4 μg) was methylated with 1 μg PRMT5/WD45/pICln methylosome, 0.1 mM SAM, and a buffer consisting of 40 mM Tris-HCl (pH 7.9), 200 mM NaCl, and 5 mM DTT overnight at 30°C. Dimethylated products were immunoprecipitated with sym10 antibody coupled to protein A Sepharose beads, 25 μL nonreducing SDS loading buffer was added to washed beads, loaded on a 10% SDS gel, stained with colloidal Coomassie blue (Novex kit from Invitrogen), gel slices containing the expected protein were isolated, and proteins digested in gel with trypsin. Peptides were subjected to electrospray mass spectral analysis.
RNAi
The double-stranded siRNA oligos PRMT5-1 (sense sequence: 5′-GUAUGAGUGGGCUGUGACA[dT][dT]), PRMT5-2 (sense sequence: 5′-CACAGUACUACAUGGCUUU[dT][dT], and CF Im68 (sense sequence: 5′-GACCGAGAUUACAUGGAUA[dT][dT]) were from SIGMA. For reverse transfection according to the Lipofectamine RNAiMAX manual (Invitrogen), 104 HEK293 cells per well were seeded in 24-well plates with 20 nM siRNA and Lipofectamine-RNAiMAX (Invitrogen) in 0.6 mL OptiMEM medium (Invitrogen), and left to grow for 3–5 d at 37°C with 5% CO2.
Steady-state kinetics
The parameters for the sum of mono- and dimethylation were determined by titration of the substrate GST-68_GR in reactions with [3H]SAM and either the PRMT5/WD45 or PRMT5/WD45/pICln complex. After incubation, reactions were precipitated with 10% TCA and counted in a scintillation counter. The results were analyzed on Lineweaver–Burk plots.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Christiane Rammelt for help with RNAi, Suzette Moes and Paul Jenö for the mass spectral analysis, Isabelle Kaufmann for the purified CPSF, Henk de Vries for the purified CF Im/CF IIm, Uwe Kühn for the recombinant PABPN1 plasmid, and Elmar Wahle for the purified PABPN1. U.F. was supported by grant no. FOR855 from the German Research Council. A.O.-L. was supported by DFG grant nos. OS290/2-1 and OS290/2-2 and Heisenberg-Fellowship OS290/1-2. W.K. was supported by the University of Basel and the Swiss National Science Fund (Grant no. 3100A0-102132/2).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2164210.
REFERENCES
- Bedford MT, Richard S 2005. Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem 275: 16030–16036 [DOI] [PubMed] [Google Scholar]
- Berger I, Fitzgerald DJ, Richmond TJ 2004. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 22: 1583–1587 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Richard S 2003. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics 2: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S 2001. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem 276: 32971–32976 [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 12: 1467–1476 [DOI] [PubMed] [Google Scholar]
- Cardinale S, Cisterna B, Bonetti P, Aringhieri C, Biggiogera M, Barabino SM 2007. Subnuclear localization and dynamics of the pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol Biol Cell 18: 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U 2008. An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell 135: 497–509 [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR 1999. Regulation of transcription by a protein methyltransferase. Science 284: 2174–2177 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL 1996. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature 384: 282–285 [DOI] [PubMed] [Google Scholar]
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S 2006. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun 342: 472–481 [DOI] [PubMed] [Google Scholar]
- Coseno M, Martin G, Berger C, Gilmartin G, Keller W, Doublie S 2008. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res 36: 3474–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries H, Rüegsegger U, Hübner W, Friedlein A, Langen H, Keller W 2000. Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J 19: 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein–protein interactions, and subcellular localization. J Biol Chem 279: 35788–35797 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT 2002. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem 277: 3537–3543 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem 277: 8243–8247 [DOI] [PubMed] [Google Scholar]
- Fronz K, Otto S, Kolbel K, Kühn U, Friedrich H, Schierhorn A, Beck-Sickinger AG, Ostareck-Lederer A, Wahle E 2008. Promiscuous modification of the nuclear poly(A)-binding protein by multiple protein-arginine methyltransferases does not affect the aggregation behavior. J Biol Chem 283: 20408–20420 [DOI] [PubMed] [Google Scholar]
- Graveley BR 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW 1994. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76: 531–541 [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264 [DOI] [PubMed] [Google Scholar]
- Herrmann F, Lee J, Bedford MT, Fackelmayer FO 2005. Dynamics of human protein arginine methyltransferase 1(PRMT1) in vivo. J Biol Chem 280: 38005–38010 [DOI] [PubMed] [Google Scholar]
- Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. 2005. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 25: 7092–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Keller W 1995. Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res 23: 2629–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Nemeth A, Meyer S, Wahle E 2003. The RNA binding domains of the nuclear poly(A)-binding protein. J Biol Chem 278: 16916–16925 [DOI] [PubMed] [Google Scholar]
- Kühn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 284: 22803–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205 [DOI] [PubMed] [Google Scholar]
- Lakowski TM, Frankel A 2009. Kinetic analysis of human protein arginine N-methyltransferase 2: Formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem J 421: 253–261 [DOI] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR 2009. Minireview: Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol 23: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT 2005a. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem 280: 32890–32896 [DOI] [PubMed] [Google Scholar]
- Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S 2005b. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem 280: 3656–3664 [DOI] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein–arginine N-methyltransferase. J Biol Chem 271: 15034–15044 [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W 2007. RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U 2001. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 11: 1990–1994 [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U 2002. SMN-mediated assembly of RNPs: A complex story. Trends Cell Biol 12: 472–478 [DOI] [PubMed] [Google Scholar]
- Miranda TB, Miranda M, Frankel A, Clarke S 2004. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem 279: 22902–22907 [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U 2008. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 582: 1997–2003 [DOI] [PubMed] [Google Scholar]
- Nicholson TB, Chen T, Richard S 2009. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol Res 60: 466–474 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Rucknagel KP, Schierhorn A, Moritz B, Huttelmaier S, Flach N, Handoko L, Wahle E 2006. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J Biol Chem 281: 11115–11125 [DOI] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H 2006. Protein arginine methylation: Cellular functions and methods of analysis. Biochim Biophys Acta 1764: 1890–1903 [DOI] [PubMed] [Google Scholar]
- Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE 2000. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol 20: 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ 2002. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem 277: 28624–28630 [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Rüegsegger U, Beyer K, Keller W 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113 [DOI] [PubMed] [Google Scholar]
- Rüegsegger U, Blank D, Keller W 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1: 243–253 [DOI] [PubMed] [Google Scholar]
- Ruepp MD, Aringhieri C, Vivarelli S, Cardinale S, Paro S, Schümperli D, Barabino SM 2009. Mammalian pre-mRNA 3′ end processing factor CF Im68 functions in mRNA export. Mol Biol Cell 20: 5211–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Bauer DL 2008. Finishing touches: Post-translational modification of protein factors involved in mammalian pre-mRNA 3′ end formation. Int J Biochem Cell Biol 40: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev 12: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Horinouchi S, Yoshida M 2007. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J Biol Chem 282: 4470–4478 [DOI] [PubMed] [Google Scholar]
- Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E 1999. Unusual sites of arginine methylation in poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem 274: 13229–13234 [DOI] [PubMed] [Google Scholar]
- Tang J, Gary JD, Clarke S, Herschman HR 1998. PRMT3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem 273: 16935–16945 [DOI] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM 2005. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL 2008. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev 22: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Keller W 1996. The biochemistry of polyadenylation. Trends Biochem Sci 21: 247–250 [PubMed] [Google Scholar]
- Wolf SS 2009. The protein arginine methyltransferase family: An update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci 66: 2109–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]