Summary
Quorum sensing (QS) cell–cell communication systems are utilized by bacteria to coordinate their behaviour according to cell density. Several different types of QS signal molecules have been identified, among which acyl-homoserine lactones (AHLs) produced by Proteobacteria have been studied to the greatest extent. Although QS has been studied extensively in cultured microorganisms, little is known about the QS systems of uncultured microorganisms and the roles of these systems in microbial communities. To extend our knowledge of QS systems and to better understand the signalling that takes place in the natural environment, metagenomic libraries constructed using DNA from activated sludge and soil were screened, using an Agrobacterium biosensor strain, for novel QS synthase genes. Three cosmids (QS6-1, QS10-1 and QS10-2) that encode the production of QS signals were identified and DNA sequence analysis revealed that all three clones encode a novel luxI family AHL synthase and a luxR family transcriptional regulator. Thin layer chromatography revealed that these LuxI homologue proteins are able to synthesize multiple AHL signals. Tandem mass spectrometry analysis revealed that LuxIQS6-1 directs the synthesis of at least three AHLs, 3-O-C14:1 HSL, 3-O-C16:1 HSL and 3-O-C14 HSL; LuxIQS10-1 directs the synthesis of at least 3-O-C12 HSL and 3-O-C14 HSL; while LuxIQS10-2 directs the synthesis of at least C8 HSL and C10 HSL. Two possible new AHLs, C14:3 HSL and (?)-hydroxymethyl-3-O-C14 HSL, were also found to be synthesized by LuxIQS6-1.
Introduction
Quorum sensing (QS) is a cell–cell communication system utilized by bacteria to coordinate their behaviour according to cell densities (Fuqua et al., 1996; Miller and Bassler, 2001). Quorum sensing bacteria can produce and secrete small hormone-like chemical molecules into the environment. As the cell density increases, these chemical molecules accumulate, and when a certain threshold concentration is reached, bacteria can respond to these molecules and alter gene expression and thus cell behaviour. Since the first report of a QS system in the bioluminescent marine bacterium Vibrio fischeri (Nealson and Hastings, 1979), QS has been found in many other bacteria and is involved in many aspects of bacterial life, including bioluminescence, virulence, symbiosis, antibiotic production, swarming and swimming motility, biofilm formation, conjugation and growth inhibition. To date, several types of QS signals have been found, including N-acyl-homoserine lactones (AHLs) for Proteobacteria (Whitehead et al., 2001; Bassler and Losick, 2006; Williams, 2007), the oligopeptides for Firmicutes (Mayville et al., 1999; Nakayama et al., 2001), and another family of molecules generically called autoinducer-2 (AI-2), which are produced by both Proteobacteria and Firmicutes, and are presumed to be used for interspecies communication (Xavier and Bassler, 2005). The AHL-based systems have been studied to the greatest extent. These systems usually contain a luxI homologue gene, which is responsible for synthesis of AHLs, and a luxR homologue gene, which is an AHL-dependent transcriptional regulator. AHL-based QS systems have been experimentally identified and studied in more than 40 different bacterial species. In addition, genome sequencing projects have revealed that many more bacteria contain putative luxI/luxR homologues, in some cases multiple systems in single genomes.
The vast majority of microorganisms cannot be cultured (estimated to be more than 99% of the microorganisms in most soils) (Amann et al., 1995; Daniel, 2005). In order to characterize the metabolic capacity of the as-yet-uncultured organisms, the culture-independent methods of metagenomics were introduced. Metagenomic libraries are constructed by directly extracting DNA from environmental samples, and cloning the environmental DNA into vectors maintained in a bacterial host (Handelsman et al., 1998; Rondon et al., 2000; Handelsman, 2004). By direct sequencing or functional screening the genetic properties of those uncultured organisms can be analysed.
Among those uncultured microorganisms, it is possible that the capacity to synthesize a large variety of novel organic molecules that have intercellular signalling activity exists. The isolation and characterization of those novel QS signals will increase our knowledge about the cell–cell communication taking place in the natural community. To date, only a few attempts have been made to identify QS systems in uncultured organisms. In 2005, Williamson and colleagues (2005) reported the isolation of new QS synthase genes from metagenomic libraries constructed using DNA from soil on the floodplain of the Tanana River in Alaska. From this library, they isolated a clone encoding a LuxI family protein, which synthesizes 3-O-C6 HSL. In another study, metagenomic analysis of the gypsy moth gut microbiota led to the identification of a gene that encodes a monooxygenase homologue that mediates production of signal mimics that induce QS (Guan et al., 2007). Recently, screening of the metagenomic libraries generated using DNA from a French pasture soil failed to uncover any clones encoding AHL synthases; however, one clone containing a quorum quenching lactonase was identified (Riaz et al., 2008).
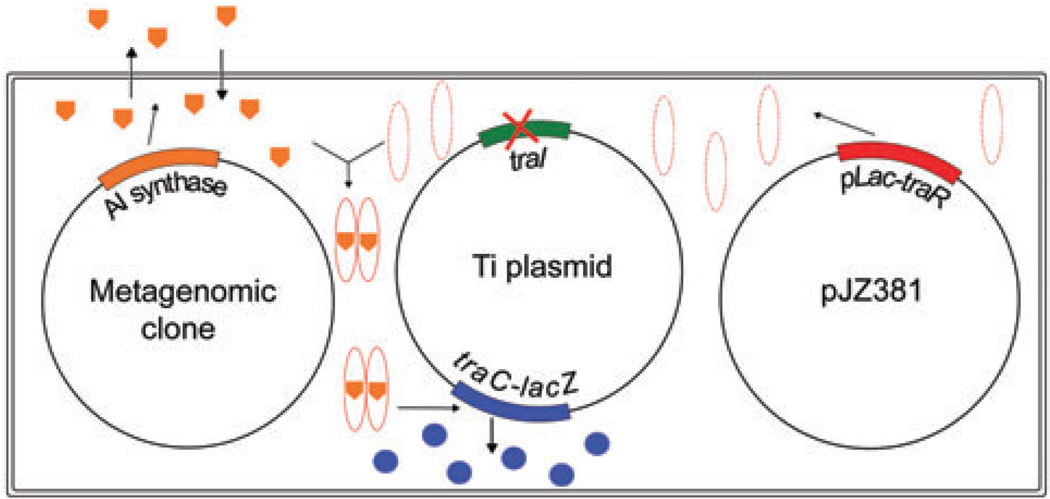
In this study, metagenomic libraries previously constructed in this lab (Wang et al., 2006) were screened for novel QS inducers using the Agrobacterium tumefaciens biosensor strain HC103(pJZ381) (Fig. 1). Isolation and characterization of three novel LuxI/LuxR type QS systems are described below.
Fig. 1.
Screening strategy for quorum sensing inducer synthase gene. The Ti plamid contains a traI non-polar deletion and a traC–lacZ translational fusion. In pJZ381, traR is overexpressed under control of the lac promoter of pBBR1MCS5. Without binding to AHL, the TraR protein is not stable and degrades quickly after synthesis. When metagenomic clones are introduced into the biosensor strain, if quorum sensing inducer synthases are present, the produced active signals bind to TraR and activate transcription of the traC–lacZ fusion, enabling the bacteria to form blue colonies in the presence of X-Gal.
Results
Screening of the metagenomic libraries
In this study, four metagenomic libraries, CX4, CX6, CX9 and CX10, constructed using DNA from activated sludge or soil (Table 1) (Wang et al., 2006) were screened for QS genes. After transferring the metagenomic cosmid clones into the biosensor strain A. tumefaciens HC103(pJZ381) and screening for blue colonies, a number of clones were isolated from each library (Table 1). The five weak blue colonies from library CX4 were not further analysed in this study. The six blue colonies from library CX6 all came from the same clone, QS6-1. The five colonies from library CX9 represent one unique clone that contained an environmental lacZ gene instead of a QS inducer synthase. The six blue colonies from library CX10 were from two unique clones and were designated QS10-1 and QS10-2. The isolated clones showed different levels of β-galactosidase activity. The biosensor strain had a very low level of background activity (3.5 miller units). Of the three isolated clones, QS6-1 showed the lowest activity (27 miller units), QS10-1 showed the highest activity (613 miller units) and QS10-2 showed an intermediate level of activity (255 miller units). When luxIQS6-1 was expressed under the lac promoter in pRK6–1LuxI, the resulting strain HC103(pJZ381)(pRK6–1LuxI) showed a high level of activity (1083 miller units).
Table 1.
Screening of metagenomic library for QS inducer synthases.
| Library | DNA source | No. of clones |
No. of colonies screened |
No. blue colonies | No. unique clones |
|---|---|---|---|---|---|
| CX4 | Pulp | 3 879 | ≈14 000 | 5 (weak blue) | ND (not further analysed) |
| CX6 | Municipal waste | 3 322 | ≈5 600 | 6 | 1 (QS6-1) |
| CX9 | Soil | 22 180 | ≈20 000 | 5 | 1 (environmental lacZ) |
| CX10 | Soil | 8 696 | ≈16 300 | 6 | 2 (QS10-1, QS10-2) |
In vitro transposon insertional mutagenesis and DNA sequence analysis
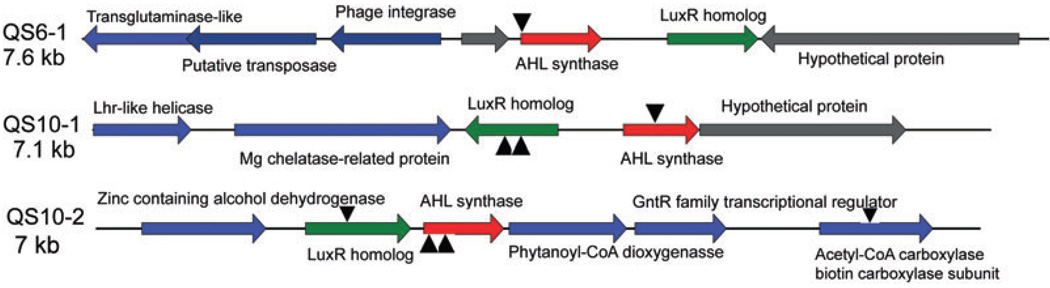
To help localize and sequence the QS signal synthase genes in the cosmid clones, in vitro transposition systems HyperMu™<CHL-1> insertion kit and the EZ-Tn5™ <KAN-2> insertion kit (Epicenter) were used. After random mutagenesis and screening, one white clone for QS6-1, three for QS10-1 and four for QS10-2 were identified. DNA sequences were obtained from the transposon insertion sites and later utilizing a primer walking method. Finally, 7.6, 7.3 and 7.1 kb of sequence information for QS6-1, QS10-1 and QS10-2, respectively, were obtained. The sequences were deposited in the GenBank database under the following accession numbers: QS6-1, FJ041295; QS10-1, FJ041296; QS10-2, FJ041297. Figure 2 shows the gene arrangements in each of the clones as well as the transposon insertion sites. All three clones contain a luxI family AHL synthase gene and a luxR family transcriptional regulator gene adjacent to one another.
Fig. 2.
Gene arrangements on QS6-1, QS10-1 and QS10-2. The luxI homologue AHL synthase genes are highlighted in red, and the luxR homologue genes are highlighted in green. Black triangles indicate the position of transposon insertion sites.
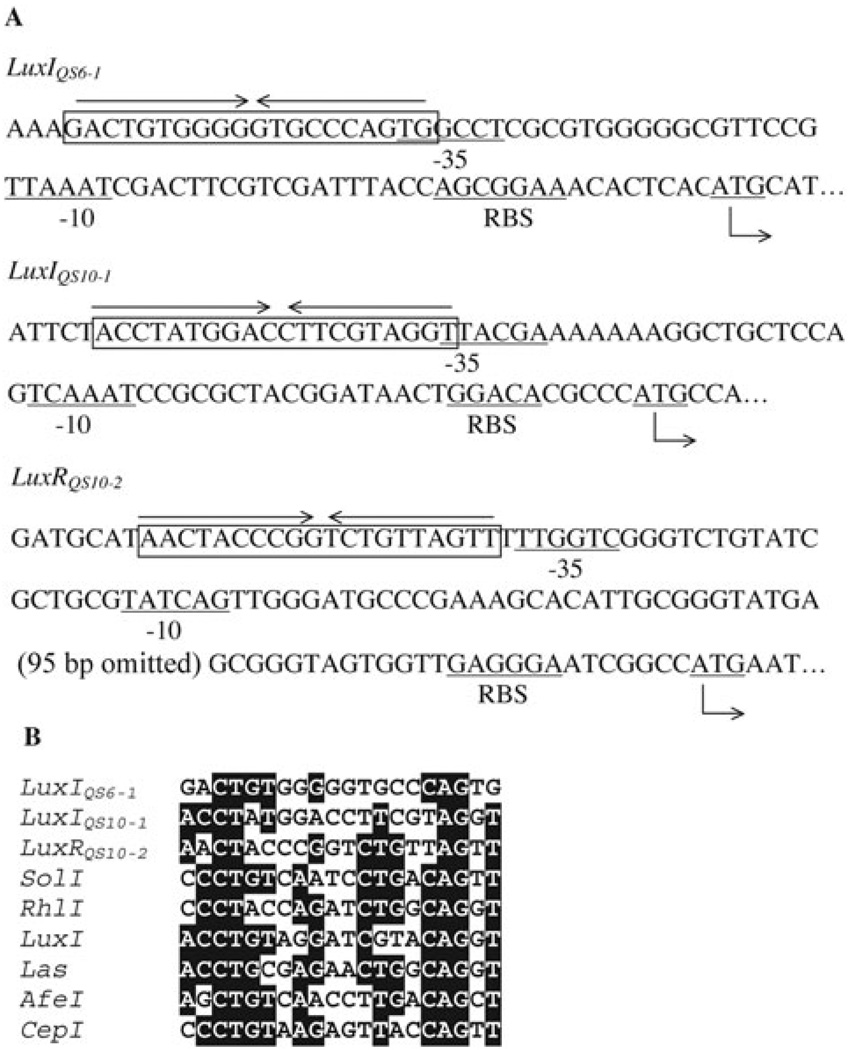
Transposon insertional mutagenesis indicated that not only mutation of the luxI homologues, but also the mutation of the luxR homologues in QS10-1 and QS10-2 (Fig. 2) caused those two clones to lose the ability to induce the biosensor strain to turn blue. A similar phenomenon has been reported for other luxI–luxR family QS systems (Zheng et al., 2006). This may be explained by the fact that in many luxI–luxR type systems the luxI type genes are positively regulated by their cognate R proteins (Waters and Bassler, 2005; Williams, 2007). Mutation of the luxR homologues in QS10-1 and QS10-2 disrupted the positive regulation loop leading to their cognate AHL synthesis genes, so that only very low levels of AHLs were synthesized by the mutants even at high cell density, which could not be detected by the biosensor strain. Since the R protein usually binds to lux box-like sequences, which are typically 20 bp inverted repeat sequences close to or partially overlapping with the −35 box of the σ70 promoter (Devine et al., 1988), the DNA sequences upstream of the luxI homologues were examined. Using promoter prediction software BPROM (Softberry, Mt. Kisco, NY, USA) or SAK (Gordon et al., 2003), a possible σ70 promoter was identified upstream of both the luxIQS6-1 and luxIQS10-1 regions. Possible lux box-like sequences with imperfect dyad symmetry were also identified near the −35 box of both promoters (Fig. 3A). No promoter was predicted upstream of the luxIQS10-2 region. However, a possible σ70 promoter was found upstream of the luxRQS10-2 gene and a putative lux box-like sequence was also found close to the −35 box region (Fig. 3A). Considering that the luxRQS10-2 and the luxIQS10-2 are oriented in the same direction, and there is only 102 bp of intergenic region between them, it is possible that they form an operon. Comparison of the three possible lux box-like elements with known lux box-like sequences indicated conservation at certain residues as well as specificity (Fig. 3B). It was found that interruption of the gene encoding the biotin carboxylase subunit of acyl-CoA carboxylase of QS10-2 also affects the AHL synthesis of that clone.
Fig. 3.
A. Lux box-like elements upstream of luxIQS6-1, luxIQS10-1 and luxRQS10-2. B. Comparison of elements with known lux box-like elements from Ralstonia solanacarum SolI (Flavier et al., 1997), Pseudomonas aeruginosa RhlI and LasI (Latifi et al., 1995), Vibrio fischeri LuxI (Devine et al., 1988), Acidithiobacillus ferrooxidans AfeI (Rivas et al., 2005), Burkholderia cepacia CepI (Lewenza et al., 1999). Sequences with more than 60% identity are shaded.
BLASTP search against NCBI Non-redundant Protein Sequences database and TBLASTN search against NCBI Environmental Samples database were performed, and it was found that the identified LuxI and LuxR homologues showed from 32% to 54% identity to their closest matches (Table 2). BLAST searches were also performed against the CAMERA All Metagenomic ORF Peptides database, the CAMERA All Metagenomic 454 Reads, the CAMERA Non-Identical Peptide Sequences database, and the JGI all IMG genes database; in all cases, no sequence with a higher identity was found.
Table 2.
Closest matches to the identified LuxI and LuxR homologues obtained from BLAST searches.
| Protein | Closest match from NCBI non-redundant protein sequence database (BLASTP search) |
Closest match from NCBI Environmental Samples database (TBLASTN search) |
|---|---|---|
| LuxIQS6-1 | AHL synthase of Nitrosospira multiformis | Freshwater sediment metagenome |
| ATCC 25196 (47% identity) | lwFormaldehyde_BCIB15374_yl (ABSN01034438.1) (53% identity) |
|
| LuxIQS10-1 | Same as above (51% identity) | Same as above (54% identity) |
| LuxIQS10-2 | AHL synthase of Sphingomonas sp. SKA58 (37% identity) | Marine metagenome ctg_1101668694332 (AACY023886981.1) (37% identity) |
| LuxRQS6-1 | LuxR family protein of Nitrococcus mobilis Nb-231 (34% identity) | Marine metagenome 1096626424210 (AACYo20346372.1) (26% identity) |
| LuxRQS10-1 | LuxR family protein of N. multiformis ATCC 25196 (32% identity) | Same as above (31% identity) |
| LuxRQS10-2 | LuxR family protein Sphingomonas sp. SKA58 (37% identity) | Metagenome sequence XZS25054.x1 (AAFX01052353.1) (41% identity) |
To perform phylogenetic analysis, sequences of LuxI and LuxR homologues that have been experimentally determined, together with sequences of the top BLASTP hits against NCBI Non-redundant Protein Sequences database, were used. Forty-four sequences of LuxI homologues with experimental evidence of functionality were combined with the top 12–18 sequences of BLASTP hits of each of the three LuxI homologues. After deleting redundant sequences, in total 70 sequences were used for multiple sequence alignment and phylogenetic analysis. For LuxR homologues, 42 sequences with experimental evidence of functionality were combined with 13–16 top BLASTP hits of each of the three LuxR homologues, in total 71 non-redundant LuxR family sequences were obtained.
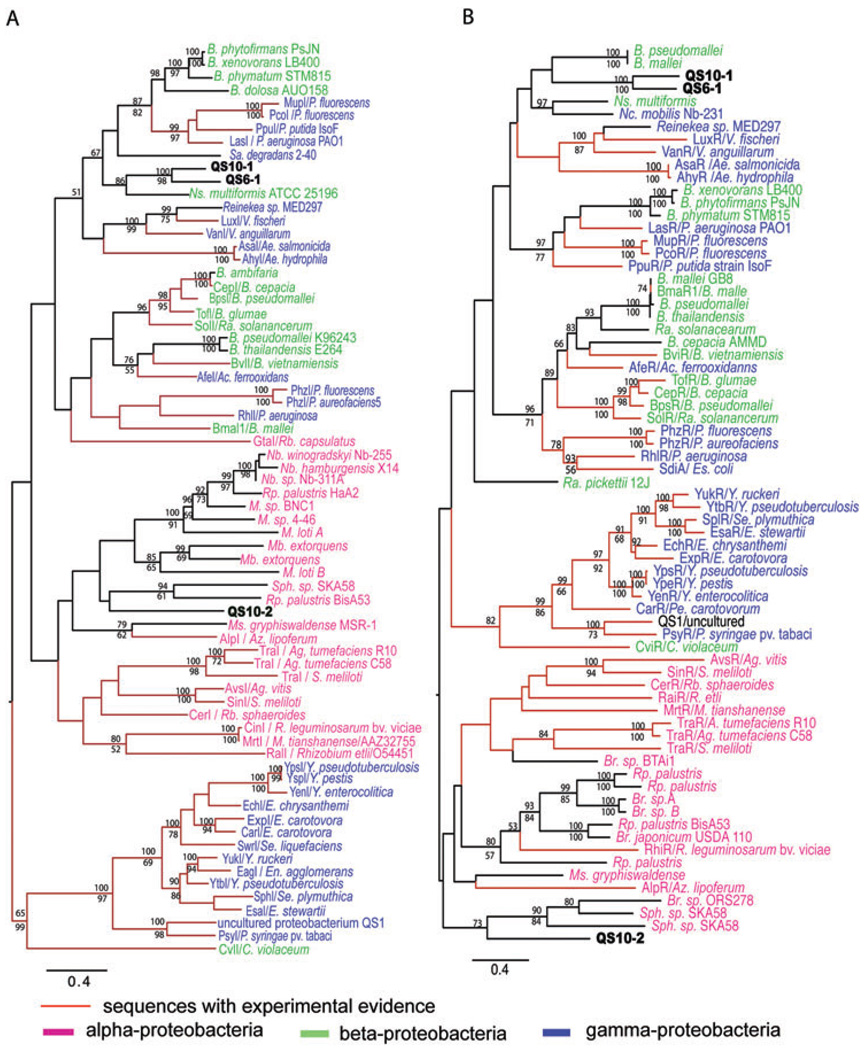
The midpoint rooted maximum likelihood (ML) trees of LuxI and LuxR homologues showed similar topology (Fig. 4). Most sequences from α-Proteobacteria grouped together, while sequences from β-Proteobacteria and γ-Proteobacteria are intermingled, which is different from previously published phylogenetic analysis of LuxI and LuxR homologues (Gray and Garey, 2001; Lerat and Moran, 2004). We note that more sequences from β- and γ-Proteobacteria were used in the present analysis. These results reflect the relatively close relationships of β- and γ-Proteobacteria, and suggest the relatively frequent lateral gene transfer of the luxI and luxR between the two groups. In both trees, QS6-1 and QS10-1 are always grouped together and tend to group with β- or γ-Proteobacteria. In fact, LuxIQS6-1 and LuxIQS10-1 showed 61% identity in amino acid sequence, while LuxRQS6-1 and LuxRQS10-1 showed 53% identity, both of which are higher than the sequence identities they showed to their closest matches from the NCBI BLASTP search.
Fig. 4.
Midpoint-rooted maximum likelihood trees of LuxI (A) and LuxR (B) homologues. Abbreviations for bacterial genus names: Ac, Acidthiobacillus; Ae, Aeromonas; Ag, Agrobacterium; Az, Azospirillium; B, Burkholderia; Br, Bradyrhizobium; C, Chromobacterium; E, Erwinia; En, Enterobacter; Es, Escherichia; M, Mesorhizobium; Mb, Methylobacterium; Ms, Magnetospirillum; Nb, Nitrobacter; Nc, Nitrococcus; Ns, Nitrosospira; P, Pseudomonas; Pe, Pectobacterium; R, Rhizobium; Ra, Ralstoina; Rb, Rhodobacter; Rp, Rhodopseudomonas; S, Sinorhizobium; Sa, Sacharophagus; Se, Serratia; Sph, Spingomonas; V, Vibrio; Y, Yersinia. Tree topology was determined using phyml version 2.4.4 under WAG model. Scale bar indicate the mean number of substitutions per site. Bootstrap values were obtained from 1000 replicates under NJ (upper) and MP (lower) algorithms using PAUP 4.0b program. Only branches with greater than 50% values are shown. The red highlighted branches are sequences with experimental evidence. Purple highlighted sequences are from α-Proteobacteria, green highlighted sequences are from β-Proteobacteria, and blue highlighted ones are from γ-Proteobacteria. Same trees with sequence accession numbers were supplied Fig. S5.
Phylogenetic analyses were also performed for other gene sequences of the three clones. For QS6-1, some of its sequences group with sequences from β-Proteobacteria (Fig. 4, Fig. S1B and C), while some other sequences group with sequences from γ-Proteobacteria or Flavobacteria sp. (Fig. S1 A and B). In agreement with the LuxI and LuxR homologue proteins, phylogenetic analysis of other gene sequences from QS10-1 showed that they grouped with β-Proteobacteria (Fig. S2), and phylogenetic analysis of all other sequences from QS10-2 showed that they grouped with α-Proteobacteria (Figs S3 and S4).
Signal diffusion assay
When streaked on agar plates, QS10-1 and QS10-2 were able to produce signals that can diffuse in agar and induce biosensor strain colonies adjacent to them to turn blue, while diffusion of signals was not observed for QS6-1 (data not shown). When the luxIQS6-1 homologue gene was subcloned into the broad host vector pRK415 and expressed under the pLac promoter, the diffusion of QS signals was observed for the resulting strain HC103(pJZ381)(pRK6–1LuxI).
Elucidation of the structures of the active compounds
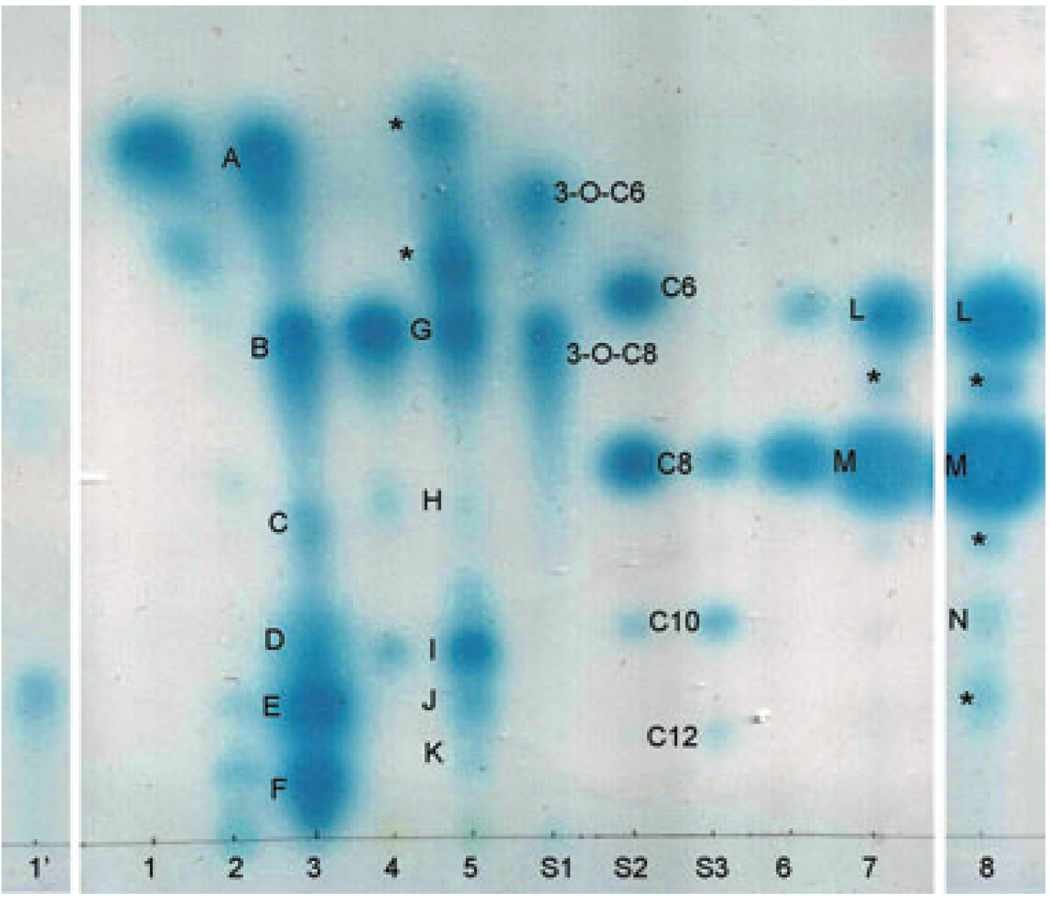
The active compounds produced by each clone were extracted from the culture supernatants and separated using C18 reverse phase Thin Layer Chromatography (TLC) plates (Whatman) (Fig. 5). When expressed in A. tumefaciens under its own promoter, no signal (lane 1) or only very weak signal (lane 1′) was detected for luxIQS6-1. When the luxIQS6-1 was subcloned into pRK415 under the pLac promoter and expressed in A. tumefaciens, or into pET30a+ under the T7 promoter and expressed in Escherichia coli, multiple signals were detected, including both short- and long-chain AHLs (lanes 2 and 3). LuxIQS10-1 was also able to direct synthesis of multiple AHLs when expressed in either A. tumefaciens or E. coli (lanes 4 and 5). Similar to the 3-O-AHL standards, the AHLs synthesized by both LuxIQS6-1 and LuxIQS10-1 tend to form tailed spots on the TLC plate. Extractions from A. tumefaciens HC103(pJZ381)(QS10-2) showed two round spots on the TLC plate with similar shape and migration rate to C6-HSL and C8-HSL (lane 6). When expressed in E. coli, some additional signals were detected with one of them showing similarity to C10-HSL (lane 8).
Fig. 5.
Analytical TLC assay. S1, S2 and S3, synthesized standards (Sigma, Aldrich): S1, 3-O-C6 HSL (2 pmol) and 3-O-C8 HSL(0.1 pmol); S2, C6 HSL(4000 pmol), C8 HSL(100 pmol), C10 HSL(500 pmol); S3, C8 HSL (20 pmol), C10 HSL(1000 pmol) and C12 HSL(9000 pmol). 1′, 1–8, condensed extractions of different strain cultures: 1′, HC103(pJZ381)(pQS6-1) (10 ml); 1, HC103(pJZ381)(pQS6-1) (5 ml); 2, HC103(pJZ381)(pRK6–1LuxI) (5 ml); 3, BL21(DE3)(pET6–1LuxI) (2 ml); 4, HC103(pJZ381)(pQS10-1) (5 ml); 5, BL21(DE3)(pET10–1LuxI) (5 ml); 6, HC103(pJZ381)(pQS10-2) (5 ml); 7, BL21(DE3)(pET10–2LuxI)(3 ml); 8, BL21(DE3)(pET10–2LuxI) (5 ml). The numbers in parentheses are the corresponding volume of culture supernatant extracted. The spots labelled from A to N were purified from preparative TLC plates and further analysed using ESI MS and MS/MS. The spots labelled with an asterisk ‘*’ were unknown compounds and were not analysed using ESI MS and MS/MS.
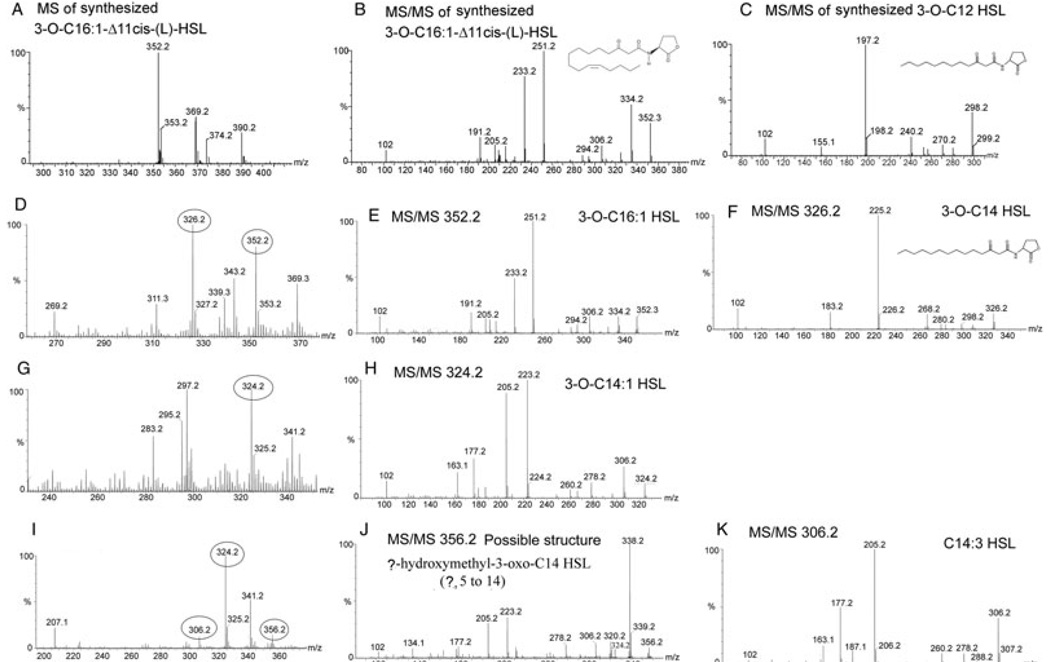
The compounds were partially purified from the silica matrix of a preparative TLC plate and subjected to ESI mass spectrometry (ESI MS) and MS/MS to elucidate their structures. By comparing with the standards and data published in the literature, we were able to identify most of the active compounds synthesized by each LuxI homologue (Table 3). Figure 6 shows the MS and MS/MS results for compounds synthesized by LuxIQS6-1 at the corresponding spots D, E and F on analytical TLC plate (Fig. 5). There are two AHLs identified from spot F (Fig. 6D, E and F). One showed exactly the same MS/MS pattern as synthesized N-3-oxo-hexadec-11Z-enoyl-l-Homoserine lactone [3-O-C16:1-Δ11cis-(l)-HSL] (Cayman chemical) (Fig. 6B and E), which indicates that it is 3-O-C16:1 HSL. The other is 3-oxo-tetradecanoyl-homoserine lactone (3-O-C14 HSL) as it shows similar MS/MS pattern to synthesized N-3-oxo-decanoyl-l-Homoserine lactone (3-O-C12 HSL) (Cayman chemical), except that the molecular weight and the mass of each of the acyl chain-derived fragments are 28 units bigger (corresponding to C2H4) (Fig. 6C and F). Another major AHL was identified from both spot D and E (Fig. 6G and I), and the MS/MS pattern (Fig. 6H) confirms that it is 3-O-C14:1 HSL, as it has a pattern similar to 3-O-C16:1-Δ11cis-(L)-HSL except that the mass is 28 units smaller (Fig. 6B and H). Two minor AHLs were also identified from spot E, one with m/z 306 and another with m/z 356 (Fig. 6I). The MS/MS pattern for the ion at m/z 306 (Fig. 6K) showed a similar pattern to that of the previously published C14:1 HSL and C14:2 HSL (Nieto Penalver et al., 2006) except that the mass is 4 and 2 mass units smaller than C14:1 HSL and C14:2 HSL respectively. This indicates that the AHL at m/z 306 contains three unsaturated bonds on the acyl chain, and is therefore likely to be a novel AHL, C14:3 HSL. When collisionally activated, the ion at m/z 356 gave a major peak at m/z 338 (Fig. 6J), which most likely results from losing a water molecule. According to the relative abundance of the ions at m/z 338 and 356, the molecule most probably contains an OH group. The peak at m/z 324 is 32 mass smaller than the ion 356, which most probably results from losing a methanol moiety. All the other peaks of the MS/MS spectrum are very similar to that of 3-O-C14:1 HSL. So the most probable structure of the compound at m/z 356.24 is (?)-hydroxymethyl-3-oxo-C14 HSL (? could be from 5 to 14). After losing a methanol, the compound will turn into 3-O-C14:1 HSL. Although the synthesized standard 3-O-C16:1 HSL does not give the adduct ion [M+MeOH+H] during MS, it is possible that the compound at m/z 356 is the adduct ion [M+MeOH+H] of 3-O-C14:1 HSL, which is formed during TLC or MS assay. Further experiments are required to determine whether it is a new AHL synthesized by bacteria or just an adduct ion of 3-O-C14:1 HSL and to determine the exact structure.
Table 3.
AHLs synthesized by each LuxI homologue.
| AHL synthase |
Identified AHLs (corresponding position on TLC plate) |
|---|---|
| LuxIQS6-1 | 3-O-C14:1 HSL (spot D and E), 3-O-C16:1 HSL (spot F), 3-O-C14 HSL (spot F), C14:3 HSL (spot E), (?)-hydroxymethyl-3-O-C14 HSL (?, 5–14) (spot E) |
| LuxIQS10-1 | 3-O-C12 HSL (spot I), 3-O-C14 HSL (spot K) |
| LuxIQS10-2 | C8 HSL (spot M), C10 HSL (spot N) |
Fig. 6.
ESI MS and MS/MS results of the active compounds produced by LuxIQS6-1 (at spot D, E and F on Fig. 5).
A. MS results for synthesized 3-O-C16:1-Δ11cis-(L)-HSL.
B. MS/MS for synthesized 3-O-C16:1-Δ11cis-(L)-HSL.
C. MS/MS for synthesized 3-O-C12 HSL.
D. MS for compounds purified from spot F in Fig. 5.
E. MS/MS for the ion at m/z 352.2 in (D).
F. MS/MS for the ion at m/z 326.2 in (D).
G. MS for compounds purified from spot D in Fig. 5.
H. MS/MS for the ion at m/z 324.2 in (G and I).
I. MS for the compound purified at spot E on TLC plate.
J. MS/MS for the ion at m/z 356.2 in (I).
K. MS/MS for the ion at m/z 306.2 in (I).
The MS and MS/MS analysis reveals that the AHL synthesized by LuxIQS10-1 at spot I was 3-O-C12 HSL (Fig. S6), and the AHL at spot K is 3-O-C14 HSL (Fig. S6D). The AHL synthesized by LuxIQS10-1 at spot M exhibits exactly the same spectrum as the synthesized standard C8 HSL (Sigma) (Fig. S7A and B), which confirm that it is C8 HSL, while the AHL at spot N is C10 HSL (Fig. S7C).
According to the analytical TLC assay, the active compounds at spots B and G are similar to 3-O-C8, spot L is similar to C6-HSL, and the compounds at spot A showed a similar shape to 3-O-AHLs, but migrate faster than 3-O-C6 AHL, which could be the product of the recyclization of the open ringed 3-O-C6 AHL (Yates et al., 2002) (Agrobacterium can not detect C4 derivatives). ESI MS and MS/MS were tried twice for those samples, but no suspect AHLs were identified. The attempts to identify the active compounds at spots C and H were also not successful. This may be due to the low abundance of the active compounds, or due to the technical limitations of preparative TLC for purifying AHLs. There is also a possibility that these active compounds could be some signal mimics other than AHLs. The spots marked with an asterisk (*) were not analysed in this study.
Discussion
In this study, three clones containing novel luxI–luxR type QS systems were obtained from environmental samples. Phylogenetic analysis of the DNA sequences showed that, like all other known LuxI-LuxR systems, they most probably come from Proteobacteria, however, they are not closely related to sequences from any known and sequenced species (Fig. 4). Phylogenetic analysis also revealed that α-proteobacterial sequences tend to group together, while sequences from β- and γ-Proteobacteria are intermingled, which likely reflects the relatively frequent horizontal gene transfer (HGT) events between the latter two. The CviI/R of Chromobacterium violacearum, as previously reported (Gray and Garey, 2001), groups with γ-Proteobacteria instead of β-Proteobacteria, which indicates this could result from HGT.
Possible lux box elements were identified in the predicted promoter regions of LuxIQS6-1, LuxIQS10-1 and LuxRQS10-2. No other putative lux box elements were found in the characterized sequences. LuxIQS10-1 seems to form an operon with its downstream gene, predicted to encode a hypothetical protein (QS10-1-5). The N-terminal part of this protein contains a CheY-like signal receiver domain (REC), while the C-terminal part of this protein contains the domain of an uncharacterized conserved protein. According to transposon insertional mutagenesis and the short intergenic distance between them, the four genes in the clone QS10-2, luxRQS10-2, luxIQS10-2, phytanoyl-CoA dioxygenase (phyH) and gntR family transcriptional regulator, seem to form an operon that is QS regulated. The four gene homologues were in the same order and also contain very short intergenic regions among them in the genome of Sphingopyxis alaskensis RB2256 (NCBI genome projects, gene tags: Sala_2588, Sala_2589, Sala_2590 and Sala_2591). PhyH belongs to the subfamily of Fe(II)/α-ketoglutarate-dependent hydroxylases, it catalyses the α-hydroxylation of phytanoyl-CoA. Its role in bacteria is not very well understood, while the deficiency of the PhyH homologue in humans causes Refsum’s disease (Jansen et al., 2000). The GntR family of bacterial regulators are named after the GntR protein of Bacillus subtilis, a repressor of the gluconate operon (Haydon and Guest, 1991). This family of transcriptional regulators contains a conserved N-terminal HTH DNA binding domain, a more heterologous C-terminal effector binding and oligomerization domain, and is involved in different regulatory pathways (Rigali et al., 2002). Transposon insertional mutagensis revealed that mutation of one of the down stream genes, acetyl-CoA carboxylase biotin carboxylase subunit, also affects the production of AHLs. This could be explained by the fact that acetyl-CoA carboxylase (ACC) catalyses the carboxylation of acetyl-CoA to produce malonyl-CoA, which is the first committed step in the biosynthesis of acyl-acyl carrier proteins (acyl-ACPs) used for fatty acid biosynthesis, while the AHL synthesis pathway shares the same pool of acyl-ACPs as fatty acid biosynthesis (Miller and Bassler, 2001).
Each of the LuxI homologues is able to direct synthesis of multiple AHLs. One of the major AHLs produced by LuxIQS6-1 is 3-O-C16:1 HSL, which has been reported to produced by the plant pathogen A. vitis F2/5 strain (Hao and Burr, 2006). Some AHLs with novel structures have been shown to be synthesized by LuxIQS6-1, including an AHL with three unsaturated carbon bonds, C14:3 AHL, and a possible -CH2-OH substituted 3-O-C14 HSL. It was also found that AHLs synthesized by the same luxI homologue in A. tumefaciens and in E. coli were slightly different (Fig. 5). Not only is the relative abundance of each signal different, but also more AHL types were detected in the TLC assay when expressed in E. coli. A similar phenomenon was reported for the LasI protein of P. aeruginosa (Gould et al., 2006). This could be due to the fact that the expression level is much higher in E. coli than in Agrobacterium, so that acyl-ACPs with lower affinities to the AHL synthase proteins could also be used to synthesize AHLs in E. coli. However, there is also a possibility that the expression host could affect the species of AHLs synthesized. Their different growth temperatures and growth rates, the difference in their protein synthesis and matrix transportation mechanisms and different acyl-ACPs pools in the cells could affect the synthesized species of AHLs.
It has been noted that for some LuxR homologues it is very difficult to get soluble protein expression, probably because of the membrane association properties of the proteins (Kaplan and Greenberg, 1987; Smith et al., 2003). In this study, when the three R proteins were overexpressed in E. coli, LuxRQS10-1 could be expressed in the soluble part only in the presence of its cognate signalling AHLs (data not shown), which suggests LuxRQS10-1, like some other R proteins (Zhu and Winans, 2001; Schuster et al., 2004; Urbanowski et al., 2004; Weingart et al., 2005), needs the binding of its cognate signalling ligand for proper protein folding. However, the majority of the LuxRQS6-1 and LuxRQS10-1 remained insoluble even in the presence of cognate AHLs.
The LuxI and LuxR homologues of the three clones were able to synthesize and detect multiple signals with different structures and a wide range of chain lengths. This should make it possible to construct novel biosensor strains that can detect a broad range of AHLs and are useful in the screening and study of luxI–luxR type QS systems for both cultured and uncultured organisms.
While we only isolated three unique QS clones in our study, earlier attempts to isolate QS systems using functional metagenomics approaches were even less successful in isolating novel AHL-based systems (Williamson et al., 2005; Riaz et al., 2008). The specificity of the biosensor systems often requires a cognate luxR to be cloned along with the luxI, as we demonstrated, and this would only occur if the genes are clustered on the same metagenomic library clone. Functional screening also requires that the genes are expressed in the surrogate host. Continuing the use of large-insert libraries, and screening in a number of biosensor strains of diverse genomic backgrounds, should result in isolation of increased numbers of novel QS systems.
Experimental procedures
Bacterial strains, plasmids, metagenomic libraries and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1. Metagenomic libraries were previously constructed by extracting total DNA from activated sludge (CX4 and CX6) or from soil (CX9 and CX10), ligating it into cosmid pRK7813, and maintaining in E. coli HB101 (Table 1) (Wang et al., 2006). 16S rRNA sequence analysis of the soil sample and the CX9 library revealed the presence of members from Proteobacteria (including α, β and Δ), Actino-bacteria, Verrucomicrobia, Fimicutes, Bacteroidetes, etc. (Wang et al., 2006). Agrobacterium strains were cultured in LB (Miller, 1976) medium or ABM (Chilton et al., 1974) minimal medium at 28°C. When required, antibiotics were supplied at the following concentrations: gentamicin (Gm), 50 µg ml−1; kanamycin (Km), 50 µg ml−1; chloramphenicol (Cm), 17 µg ml−1; tetracycline (Tc), 2 µg ml−1. Escherichia coli strains were cultured in LB medium at 37°C. When necessary, antibiotics were supplied at the following concentrations: ampicillin (Ap), 100 µg ml−1; Km, 100 µg ml−1; Cm, 25 µg ml−1; Tc, 20 µg ml−11.
Screening of metagenomic libraries for quorum sensing inducers
The A. tumefaciens biosensor strain HC103(pJZ381) was used to screen the above mentioned libraries for new QS inducers (Fig. 1). Metagenomic clones were transferred to the biosensor strain HC103(pJZ381) by triparental mating using the helper strain E. coli DH5α(pRK600) (Finan et al., 1986). The transconjugants were selected and screened on LB agar containing appropriate antibiotics and 20 µg ml−1 X-Gal. After 48–72 h incubation at 28°C, plates were checked for blue colonies.
The metagenomic plasmids from the identified clones of blue colonies were transferred into E. coli DH5α by triparental mating and then extracted and digested using BamHI. The digestion patterns were analysed and those with the same digestion pattern were deemed to be sibling clones. All unique plasmids were reintroduced into A. tumefaciens HC103(pJZ381) by electroporation to confirm that they form blue colonies in presence of X-gal. Each unique clone was also electroporated into wild-type A. tumefaciens C58, which does not contain a lacZ gene, and incubated at 28°C for 48–72 h. If the colonies turn blue on LB agar containing X-Gal, then the plasmid contains a lacZ gene, otherwise it contains candidate QS genes.
Transposon insertion and sequencing of clones
EZ-Tn5™ <KAN-2> and HyperMu™ <CHL-1> in vitro insertion kits were purchased from Epicentre (Madison,WI) and used for in vitro transposon insertion of the metagenomic clones, according to the manufacturer’s instructions. The mutated metagenomic clones were transferred to the biosensor strain A. tumefaciens HC103(pJZ381) and the transformants were screened for white colonies on plates with appropriate antibiotics and X-gal. The metagenomic clones from the white colonies were transferred to E. coli DH5α by triparental mating. Plasmids were extracted and sequenced using transposon specific primers provided with the kits, MUCHL-1 FP-1 and MUCHL-1 RP-1 (for HyperMu™ <CHL-1>insertions) or KAN-2 FP-1 and KAN-2 RP-1 (for EZ-Tn5™ <KAN-2>insertions). Upstream and downstream regions were sequenced using new primers designed from the sequenced area using a primer walking strategy. All sequencing was performed at MOBIX (McMaster University, Hamilton, ON, Canada) using an ABI 3730 DNA analyser or at York University’s Core Facility for Molecular Biology (Department of Biology, York University, Toronto, ON, Canada) using an ABI 373 A Sequencer.
Phylogenetic analysis
All sequences used were obtained from GenBank or Swiss-Prot. For LuxI and LuxR homologues, sequences with experimental evidence and sequences of top BLASTP (Altschul et al., 1997) hits to NCBI non-redundant protein sequences were used. For other genes, sequences of top BLASTP hits were obtained from GenBank. Multiple sequence alignments were performed using ‘Muscle’ (version 3.6) (Edgar, 2004) and were refined by eye. Some gap-containing areas were removed using BioEdit (Version 7.00) (Hall, 1999). Maximum likelihood trees were constructed using Phyml version 2.4.4 (Guindon and Gascuel, 2003) under WAG model [the best fitting model according to ProTest (Abascal et al., 2005)]. Neighbour-joining and maximum parsimony analysis were performed using PAUP 4.0b (Swofford, 2000). Bootstrap values were obtained from 1000 replicates.
β-Galactosidase activity assay
Agrobacterium tumefaciens strains were grown to stationary phase in ABM minimal medium at 28°C, then subcultured at 28°C in 5 ml ABM with a 1:100 ratio (v/v). For a positive control, 3-O-C8 HSL, which is the cognate AHL of A. tumefaciens R10, was added to a final concentration of 10 nM to HC103(pJZ381) culture. Following 20 h of additional growth, β-galactosidase activity was assayed as described (Miller, 1972).
Subcloning of luxI
Primers were designed to include the start and stop codons of the luxI or the luxR homologue genes (Table S2). KOD hot start DNA polymerase (Novagen, Madison, WI, USA) was used to amplify the fragments. Standard molecular cloning procedures were used to insert the luxI or luxR homologue genes into the BamHI and HindIII sites of pET30a+ or pET30b+ to construct the corresponding vectors, pET6–1LuxI, pET10–1LuxI, pET10–2LuxI (Table S2). The luxIQS6-1 was also inserted into BamHI and HindIII digested pRK415 to yield pRK6–1LuxI.
Extraction, purification and analysis of the active compounds
Agrobacterium tumefaciens strains HC103(pJZ381)(pQS6-1), HC103(pJZ381)(pQS10-1), HC103(pJZ381)(pQS10-2) and HC103(pJZ381)(pRK6–1LuxI) were grown to stationary phase in ABM medium or LB medium at 28°C before extraction. Escherichia coli strains BL21(DE3)(pET6–1LuxI), BL21(DE3)(pET10–1LuxI) and BL21(DE3)(pET10–2LuxI) were grown overnight at 37°C in LB medium with aeration, subcultured using a 1:50 ratio to LB medium and incubated for 1 h at 37°C until the OD600 of the culture reached 0.4–0.5, then IPTG (isopropyl-beta-D-thiogalactopyranoside) was added to a final concentration of 0.5 mM and incubated for 6 h at 28°C before extraction. Extraction of the culture supernatant, analytical TLC (Thin Layer Chromotography) and preparative TLC were performed as described by Shaw and colleagues (1997) with minor modifications. For example, 70% methanol in water was used to develop the reverse phase C18 TLC plates instead of 60%. The biosensor strain A. tumefaciens HC103(pJZ381) was used in the soft agar overlay of the TLC plates.
Extractions from 500 ml of each of the culture supernatants were applied to preparative TLC plates, and were separated and partially purified as described (Shaw et al., 1997). The partially purified compounds were dissolved in 20 µl of 1:1 acetonitrile: water with 0.2% formic acid. Electrospray ionization mass spectrometry (ESI MS) and MS/MS were performed using a micromass Q-TOF ultima global mass spectrometer (Department of Chemistry, University of Waterloo). Argon gas was used as the collision gas, and the collision energy was kept at 15 eV for all experiments.
Supplementary Material
Acknowledgements
We would like to thank Dr Richard Smith from Department of Chemistry, University of Waterloo and Zhenyu Cheng for assistance with the Mass Spectrometry. We are grateful to five anonymous reviewers and Dr Josh Neufeld for providing helpful suggestions on the manuscript. We acknowledge University of Waterloo graduate scholarship to Y.H. Research funding to support this work was provided by Natural Sciences and Engineering Research Council of Canada grants to B.R.G and T.C.C.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3:470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- Devine JH, Countryman C, Baldwin TO. Nucleotide sequence of the luxR and luxI genes and the structure of the primary regulatory region of the lux regulon of Vibrio fischeri ATCC 7744. Biochemistry. 1988;27:837–842. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Kunkel B, De Vos GF, Signer ER. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavier AB, Ganova-Raeva LM, Schell MA, Denny TP. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydroxypalmitic acid methyl ester. J Bacteriol. 1997;179:7089–7097. doi: 10.1128/jb.179.22.7089-7097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- Gordon L, Chervonenkis AY, Gammerman AJ, Shahmuradov IA, Solovyev VV. Sequence alignment kernel for recognition of promoter regions. Bioinformatics. 2003;19:1964–1971. doi: 10.1093/bioinformatics/btg265. [DOI] [PubMed] [Google Scholar]
- Gould TA, Herman J, Krank J, Murphy RC, Churchill ME. Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J Bacteriol. 2006;188:773–783. doi: 10.1128/JB.188.2.773-783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KM, Garey JR. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology. 2001;147:2379–2387. doi: 10.1099/00221287-147-8-2379. [DOI] [PubMed] [Google Scholar]
- Guan C, Ju J, Borlee BR, Williamson LL, Shen B, Raffa KF, Handelsman J. Signal mimics derived from a metagenomic analysis of the gypsy moth gut microbiota. Appl Environ Microbiol. 2007;73:3669–3676. doi: 10.1128/AEM.02617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Hao G, Burr TJ. Regulation of long-chain N-acyl-homoserine lactones in Agrobacterium vitis. J Bacteriol. 2006;188:2173–2183. doi: 10.1128/JB.188.6.2173-2183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon DJ, Guest JR. A new family of bacterial regulatory proteins. FEMS. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- Jansen GA, Hogenhout EM, Ferdinandusse S, Waterham HR, Ofman R, Jakobs C, et al. Human phytanoyl-CoA hydroxylase: resolution of the gene structure and the molecular basis of Refsum’s disease. Hum Mol Genet. 2000;9:1195–1200. doi: 10.1093/hmg/9.8.1195. [DOI] [PubMed] [Google Scholar]
- Kaplan HB, Greenberg EP. Overproduction and purification of the luxR gene product: transcriptional activator of the Vibrio fischeri luminescence system. Proc Natl Acad Sci USA. 1987;84:6639–6643. doi: 10.1073/pnas.84.19.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- Lerat E, Moran NA. The evolutionary history of quorum-sensing systems in bacteria. Mol Biol Evol. 2004;21:903–913. doi: 10.1093/molbev/msh097. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Conway B, Greenberg EP, Sokol PA. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1976. [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Cao Y, Horii T, Sakuda S, Akkermans AD, de Vos WM, Nagasawa H. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol Microbiol. 2001;41:145–154. doi: 10.1046/j.1365-2958.2001.02486.x. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43:496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Penalver CG, Morin D, Cantet F, Saurel O, Milon A, Vorholt JA. Methylobacterium extorquens AM1 produces a novel type of acyl-homoserine lactone with a double unsaturated side chain under methylotrophic growth conditions. FEBS Lett. 2006;580:561–567. doi: 10.1016/j.febslet.2005.12.062. [DOI] [PubMed] [Google Scholar]
- Riaz K, Elmerich C, Moreira D, Raffoux A, Dessaux Y, Faure D. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ Microbiol. 2008;10:560–570. doi: 10.1111/j.1462-2920.2007.01475.x. [DOI] [PubMed] [Google Scholar]
- Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- Rivas M, Seeger M, Holmes DS, Jedlicki E. A Lux-like quorum sensing system in the extreme acidophile Acidithiobacillus ferrooxidans. Biol Res. 2005;38:283–297. doi: 10.4067/s0716-97602005000200018. [DOI] [PubMed] [Google Scholar]
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci USA. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Jr, Rinehart KL, Farrand SK. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Bu Y, Suga H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem Biol. 2003;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA, USA: Sinauer Associates; 2000. [Google Scholar]
- Urbanowski ML, Lostroh CP, Greenberg EP. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol. 2004;186:631–637. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Meek DJ, Panchal P, Boruvka N, Archibald FS, Driscoll BT, Charles TC. Isolation of poly-3-hydroxybutyrate metabolism genes from complex microbial communities by phenotypic complementation of bacterial mutants. Appl Environ Microbiol. 2006;72:384–391. doi: 10.1128/AEM.72.1.384-391.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Weingart CL, White CE, Liu S, Chai Y, Cho H, Tsai CS, et al. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol Microbiol. 2005;57:452–467. doi: 10.1111/j.1365-2958.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- Williamson LL, Borlee BR, Schloss PD, Guan C, Allen HK, Handelsman J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl Environ Microbiol. 2005;71:6335–6344. doi: 10.1128/AEM.71.10.6335-6344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhong Z, Lai X, Chen WX, Li S, Zhu J. A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol. 2006;188:1943–1949. doi: 10.1128/JB.188.5.1943-1949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.