Summary
The Escherichia coli fimbrial adhesive protein, FimH, mediates shear-dependent binding to mannosylated surfaces via force-enhanced allosteric catch bonds, but the underlying structural mechanism was previously unknown. Here we present the crystal structure of FimH incorporated into the multi-protein fimbrial tip, where the anchoring (pilin) domain of FimH interacts with the mannose-binding (lectin) domain and causes a twist in the β-sandwich fold of the latter. This loosens the mannose-binding pocket on the opposite end of lectin domain, resulting in an inactive low-affinity state of the adhesin. The autoinhibition effect of the pilin domain is removed by application of tensile force across the bond, which separates the domains and causes the lectin domain to untwist and clamp tightly around ligand like a finger trap toy. Thus, β-sandwich domains, which are common in multidomain proteins exposed to tensile force in vivo, can undergo drastic allosteric changes and be subjected to mechanical regulation.
Introduction
Adhesive proteins of prokaryotic and eukaryotic cells are generally multidomain in nature, with different domains to bind the ligand on the target cell and to anchor the binding protein to the cell membrane or adhesive organelle. There is growing evidence suggesting that interactions with an anchoring domain can also be an allosteric regulator of ligand binding (Aprikian et al., 2007; Arnaout et al., 2005; Askari et al., 2009; Tchesnokova et al., 2008; Waldron and Springer, 2009). Interdomain allosteric regulation can lead to mechanical reinforcement of adhesive bonds (Alon and Dustin, 2007; Astrof et al., 2006; Friedland et al., 2009; Springer, 2009), or even a catch bond mechanism of adhesion in which the adhesive interaction becomes stronger with increased tensile force (Phan et al., 2006; Yakovenko et al., 2008). However, the structural basis of allosteric catch bonds has not been elucidated.
The allosteric catch bond model has been extensively proposed and experimentally supported for the mannose-specific fimbrial adhesin of Escherichia coli called FimH, which mediates shear-dependent bacterial adhesion. FimH is a 30 kDa protein positioned on the tip of surface filaments called type 1 fimbriae that mediate mannose-specific binding and are the most common adhesive organelles in E. coli and other enterobacteria. FimH consists of two immunoglobulin-like domains: an N-terminal lectin domain that binds mannose ligand and a C-terminal pilin domain that anchors FimH into the fimbrial tip. In the crystal structures obtained previously, no interaction between the binding and anchoring domains were observed because structures were determined either for purified lectin domain (Bouckaert et al., 2005; Wellens et al., 2008) or FimH in complex with molecular chaperone FimC that is wedged between the domains (Choudhury et al., 1999; Hung et al., 2002). In spite of biochemical evidence of allosteric changes in the lectin domain, no structural changes have been directly observed. Furthermore, the FimH mannose-binding domain has a β-sandwich fold that is common in eukaryotic and bacterial matrix and adhesive proteins (Shan et al., 1999; Timpl et al., 2000) (Hashimoto, 2006), and is generally thought to be structurally rigid, thus not allowing for allosteric regulation.
Here we present a crystal structure of FimH integrated into fimbrial tips, i.e. in its native conformation. In this structure, the binding domain of FimH is twisted and compressed by interaction with the anchoring pilin domain that loosens the mannose-binding pocket on the other side of the domain This results in autoinhibition, meaning that FimH is maintained in a low-affinity state by internal contacts. Upon interaction with mannose and/or under tensile force, the domains separate and the binding domain untwists and elongates like a stretched finger trap toy, resulting in a tight mannose-binding pocket that provides for an allosteric regulation of the ligand-receptor interaction by mechanical force.
Results
Crystal structure of type 1 fimbrial tip
Fimbrial tips proteins were expressed, purified, crystallized in the absence of mannose or any other ligand molecule, and the structure determined at 2.7 Å resolution, as described in Experimental Procedures, with crystallographic details in Table 1. The complexes crystallized in space group R32 with two copies in the asymmetric unit. The structure was solved using molecular replacement and manual electron density map fitting with known structures of each subunit. Crystallographic refinement of the structure at 2.7 Å resolution with Refmac5 (Murshudov et al., 1997) (R=0.245, Rfree=0.274) yielded a model deposited in the Protein Data Bank with identifier 3JWN. The two copies of the complex in the asymmetric unit are almost identical - Superposition of 927 Cα atoms common to each complex gives a RMSD of 1.1 Ǻ, mostly due to tiny angular differences between subunits. The two copies are situated in different crystal packing environments and are not related by crystallographic symmetry.
Table 1.
Data Collection and Refinement Statistics
| Data collection statistics | |
| Space group | R32 |
| Unit cell dimensions (a,b,c) (Ǻ) | 216.0, 216.0, 532.1 |
| Complexes per asymmetric unit | 2 |
| Resolution (Ǻ) (last shell) | 50.0–2.69 (2.79–2.69) |
| Unique reflections (last shell) | 119926 (6137) |
| Completeness (last shell) | 90.6% (46.7) |
| Redundancy (last shell) | 9.5 (4.6) |
| <I>/<σ(I)> (last shell) | 14.0 (1.0) |
| Rmerge (last shell) | 0.147 (0.862) |
| Refinement statistics | |
| Resolution (Ǻ) | 50.0–2.7 |
| R factor (working set) | 0.245 |
| Rfree (test set=5% of the overall) | 0.272 |
| # unique reflections | 119854 |
| Number of protein atoms | 13868 |
| Number of solvent atoms | 28 |
| Number of heteroatoms | 12 |
| Wilson B value | 65.5 Å2 |
| Average B value from refinement | 51.0 Å2 |
| Ramachandran quality | 89.0% in most-favored regions |
| 10.2% in additional allowed regions | |
| 0.2% in generously allowed regions | |
| 0.6% in disallowed regions | |
| Rms deviation - bond lengths | 0.006 Å |
| Rms deviation - bond angles | 0.9° |
Each complex contains one FimH subunit, followed by one FimG and two FimF subunits, with the last FimF bound to the FimC chaperone (Figure 1A). The subunits are bound to each other by complementary donor-strand swapping (Choudhury et al., 1999) and the tertiary structures of FimF, FimG and FimC are similar to previous structures of these subunits (Eidam et al., 2008; Gossert et al., 2008; Hung et al., 2002; Puorger et al., 2008), with only 0.5 to 1.3 Å deviation in Cα RMSD. In FimH, the tip-anchoring pilin domain changes only slightly (Cα RMSD=1.3 Å) from previous structures of FimH in complex with FimC. However, the mannose-binding lectin domain (Figure 1B) takes on a conformation that differs significantly - by 3.1 to 3.2 Å RMSD - from those previously obtained for the purified form of lectin domain (Figure 1C) or lectin domain in the FimH/FimC complex. The Cα atoms in the lectin domains of all previously reported structures differ from one another by less than 0.8 Å RMSD.
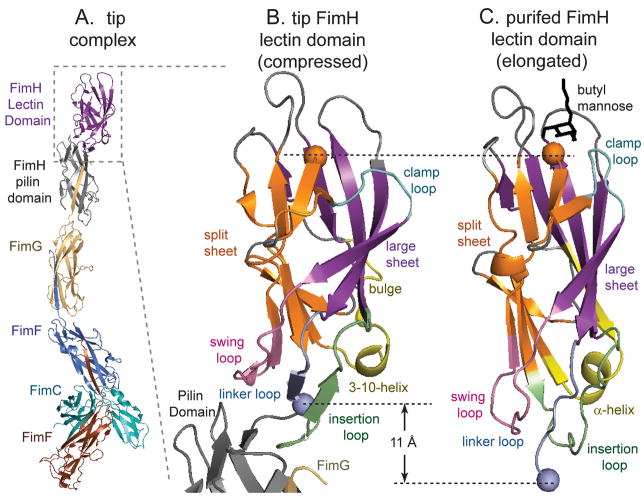
Figure 1. Overall structure.
A. FimH-containing fimbrial tip crystal structure. B. The lectin domain docked to the pilin domain (black) in the fimbrial tips. C. The isolated lectin domain bound to butyl-mannose (black) as previously crystallized (1UWF,(Bouckaert et al., 2005)). In all panels of all figures, the large β-sheet is shown in purple, the split β-sheet in orange, the swing loop in pink (residues 22–35), the linker loop in light blue (151–158), the insertion loop in green (109–124), the clamp loop in cyan (8–16) the 310/α-helix in yellow (59–72), and remaining regions of the lectin domain in gray. The dashed lines indicate the length from the N-terminus (residue 1, orange sphere) to the C-terminus of the lectin domain (residue 158, blue sphere.) These and other structural cartoons were made with Pymol (Delano Scientific LLC.)
Conformational differences in the FimH lectin domain
The lectin domain in the tip-associated adhesin (Figure 1B) has the same general topology as the purified lectin domain (Figure 1C), with a large continuous β-sheet on one side of a β-sandwich (hereafter the large β-sheet, and shown in purple throughout this paper) and a second β-sheet that is split by a small jog in the middle on the other side (split β-sheet, in orange). The mannose-binding site (empty in the tip and with mannose in the purified lectin domain) is at the distal end of the β-sandwich (top in all figures), opposite from the proximal end, where the lectin domain connects to the pilin domain (bottom in all figures). However, the lectin domain in the tip is wider and more compressed (11 Å shorter from Y1 to T158) than the narrower and more elongated purified lectin domain. The compressed (tip-associated domain) and elongated (purified domain) conformations differ mainly in the distal and proximal end loops.
In the proximal end of the tip-associated lectin domain, three loops contact the pilin domain and have different conformations in comparison to ones in the purified form. The loop comprising residues 112 to 125 changes conformation and inserts into the pilin domain, and so is hereafter called the insertion loop (green in figures). The swing loop (residues 23–33; pink), swings out in the compressed tip form. Finally, the linker loop (residues 151 to 158; blue), which links the two domains, is retracted in the compressed form, which is the primary cause of the 11 Å shortening.
At the distal mannose-binding end of the domain, the compressed and elongated conformations differ in the clamp loop (residues 13–17; cyan), which opens outward in the compressed tip form, but is clamped tightly shut in the elongated form of the purified lectin domain.
In addition to these changes in either end, there are also differences in the large β sheet, which has a small bulge formed by residues 58 to 62 and is more tilted along one edge in the compressed conformation (purple).
Notably, these conformational differences are not due to crystal contacts because the two copies of the fimbrial tips in the asymmetric unit are identical in all respects discussed in this paper, as are all copies of the lectin domain in all previous crystal structures (Bouckaert et al., 2005; Choudhury et al., 1999; Hung et al., 2002; Wellens et al., 2008). Three alternative explanations remain for the conformational differences between the tip-associated and purified lectin domains. First, there are differences in primary structure of the lectin domains - all previous crystal structures were obtained from the FimH variant of E. coli strain J96, with V27, N70 and S78, while the tip structure incorporates the FimH variant of E. coli strain F18, which has A27, S70 and N78 instead. Second, the fimbrial tip structure was obtained in the absence of mannose ligand, while the previous structures contained mannose or mannose-like ligand in the binding pocket. Third, in all previous structures, the lectin domain is not interacting with the pilin domain, since the latter is either absent (in the purified lectin domain) or wedged apart by FimC (in the FimH/FimC complex), whereas the two domains form substantial contacts in the tip structure.
Effect of primary sequence on lectin domain conformation
If the three amino acid differences cause the observed conformational differences, then a purified lectin domain derived from FimH variant from E. coli F18 (LDF18) should have similar properties as the tip-associated lectin domain of the same variant. Moreover, both should have properties distinct from those of the purified lectin domain from E. coli J96 (LDJ96), which was previously crystallized in the elongated conformation. If instead the primary structure is not responsible for the conformational change, then purified LDF18 and LDJ96 variants should have similar properties, distinct from those of the lectin domain in the tip.
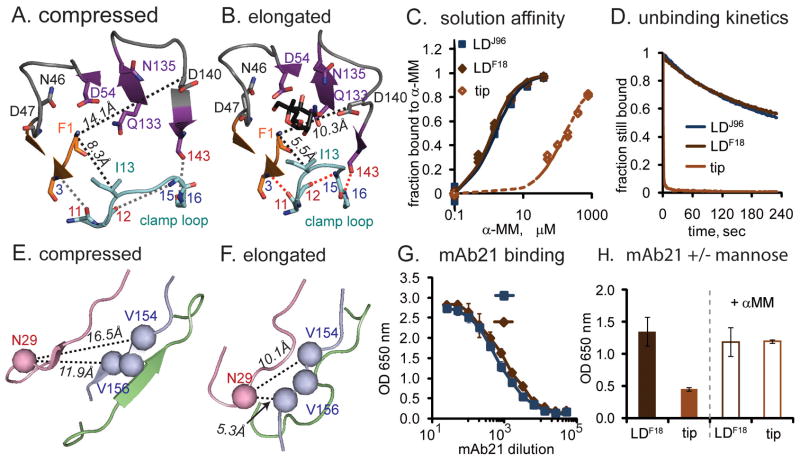
We first considered the differences between the tip and both purified lectin domains in the distal mannose-binding region. The clamp loop forms part of the mannose-binding pocket and is spread wide in the compressed conformation of the tip lectin domain but tightly clamped in the elongated conformation of the purified domain (Figure 2AB). The tight conformation is reinforced by the formation of one backbone hydrogen bond within the clamp loop (nitrogen of G15 to oxygen of P12, or G15:N-P12:O) and two more between the clamp loop and neighboring strands (C3:N-I11:O and G16:N-Q143:O). In the looser tip conformation, two mannose-interacting residues (I13 in the clamp loop and D140 in the large β-sheet) move 3 to 4 Å further away from the other six mannose-binding residues (F1, N46, D47, D54, Q133, N135) (Figure 2AB).
Figure 2. Effect of ligand and primary structure on conformation.
In all structural cartoon panels, co-crystallized butyl mannose is shown in black sticks, while pocket-forming residues are shown as sticks with carbon atoms colored by loop identity (see Figure 1), oxygen atoms in red and nitrogen atoms in blue. Red dotted lines show hydrogen bonds described in the text. Black dotted lines indicate distances described in the text A,B. Top-view of the mannose-binding site in the two conformations. C. Solution affinity curve of tip and purified lectin domains binding to α-MM in Surface Plasmon Resonance (SPR) assays. D. Profiles of F-18 tips and purified lectin domains dissociating from mannosylated bovine serum albumin (man-BSA) in SPR assays. All curves are normalized to the response units at the start of the wash (108 for tip, 79 for LDF18, and 101 for LDJ96) to indicate fraction remaining bound. E,F. Bottom-view of the two conformations of the interdomain regions with the mAb21-binding residues shown as spheres. G, Dilution curve of mAb21 binding to two variants of isolated lectin domain. H. Effect of mannose on mAb21 binding to LDF18 and tip. (Data are represented as mean +/− SEM.)
One would expect that the tightly clamped conformation of the binding pocket has a higher affinity and slower dissociation rate towards mannose than the looser unclamped one. The two purified lectin domains LDF18 and LDJ96 have essentially identical affinities (Figure 2C) and dissociation rates (Figure 2D), indicating that the amino acid differences between them do not affect their pocket structures. In contrast, the tip-incorporated FimH has a 200-fold lower affinity and much faster dissociation rate than both purified lectin domains (Figure 2CD).
To test whether the three amino acid differences affect the conformation of the bottom interdomain region of the lectin domain, we used the mAb21 monoclonal antibody that was obtained against the purified LDJ96 and recognizes a three-dimensional epitope including residues V154-V156 in the linker loop and residue N29 in the swing loop. N29 is positioned over 6 Å closer to residues V154-V156 in the elongated conformation than in the compressed conformation (Figure 2EF), so mAb21 would be expected to bind with different affinity to the two conformations. However, mAb21 recognized LDJ96 and LDF18 similarly (Figure 2G), but bound relatively poorly to the fimbrial tip (Figure 2H, left panel). This indicates that the conformations of the proximal loops of the two purified lectin domain variants are similar and in a different conformation from that of the lectin domain in the tip FimH.
Taken together, these results show that three amino acid differences in primary sequence do not cause the change in conformation of either the proximal or distal ends of the lectin domain that is observed between the tip-associated compressed and purified elongated forms.
Effect of the ligand on lectin domain conformation
If the differential presence of mannose in the crystals is responsible for the different conformations observed in the corresponding X-ray structures, then the purified lectin domain should take on the elongated or compressed conformation depending on whether mannose is present or absent, respectively. However, addition of soluble mannose did not affect the ability of mAb21 to recognize either LDF18 (Figure 2H, right panel) or LDJ96 (not shown), suggesting that the conformation of the proximal loops is not affected by the mannose in the purified lectin domain.
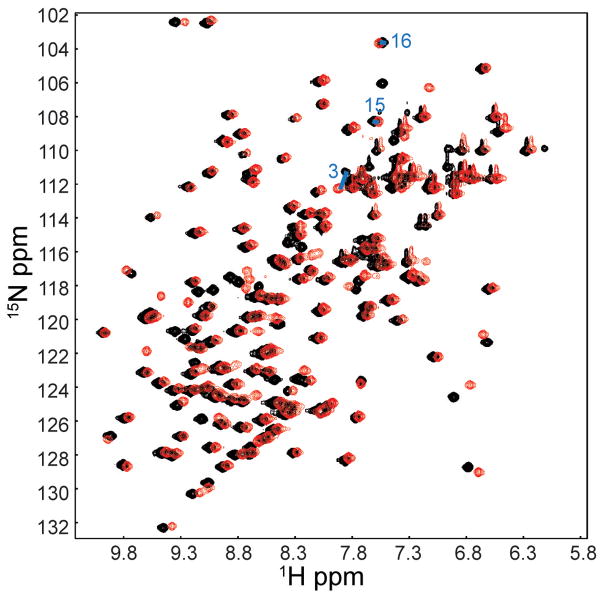
To assess the effect of mannose in a more direct and detailed manner, we collected and assigned NMR spectra of purified LDF18 in the presence and absence of mannose. An overlay of the (1H, 15N)-HSQC spectra is shown in Figure 3, and the residue-by-residue mannose-dependent NMR shift differences are shown in Figure S1. The number and magnitude of differences in the HSQC spectra of mannose-free and mannose-bound LDF18 are too few and much too small to be consistent with the large-scale global conformational differences seen between the elongated and compressed forms of the lectin domain. The only significant chemical shift differences (0.114–0.297 ppm) in the presence of mannose were observed in residues in the mannose binding site that are likely to be due to direct interaction with mannose. At the same time, 31 residues that differentially donate NH groups to hydrogen bonds in the two alternative conformations, including residues C3, G15 and G16 in the clamp loop bonds, showed only insignificant shift differences (≤0.06 ppm). The NMR data thus indicate that the large conformational changes within the lectin domain observed in the two crystal structures, including the clamping of the mannose-binding pocket, are not recapitulated by the presence and absence of mannose in the context of the isolated lectin domain.
Figure 3. NMR spectra of purified 1mM LDF18.
1H-15N HSQC overlay of LDF18 in the mannose-free (black) and mannose-bound (red) forms. The latter contains 1mM α-MM, which is a 1:1 ratio of protein to mannose. Large chemical shift differences are not observed between the two spectra, indicating that binding of mannose induces only small, local conformational changes. See also Figure S1.
While the conformation of the purified lectin domain is not affected by mannose, addition of mannose causes the fimbrial tips to be recognized by mAb21 (Figure 2H, right panel). Thus, the interdomain region appears to switch from the compressed to the elongated conformation when the tip-associated FimH binds mannose. Consistent with the large mannose-induced structural changes of the tip lectin domain, we observed that soaking mannose into fimbrial tip crystals caused their disintegration (not shown). This is consistent with the hypothesis of a reciprocal allosteric connection between the mannose-binding pocket and interdomain region of the lectin domain (Tchesnokova et al, 2008). By this hypothesis, mannose is able to convert the loose conformation of the pocket (as in the compressed tip form) into a tight one (as in the elongated form) that, in turn, causes a structural propagation across the domain to induce the corresponding conformational changes in the loops proximal to the pilin domain.
Together, these data suggest that the purified lectin domain is in the elongated conformation regardless of the presence of mannose, but the fimbrial tip lectin domain is in the compressed conformation without mannose but can switch to the elongated conformation upon binding ligand.
Effect of the pilin domain on the lectin domain conformation
We therefore propose that that, in the absence of mannose, the pilin domain induces the conformational changes observed between the compressed and elongated forms of the lectin domain.
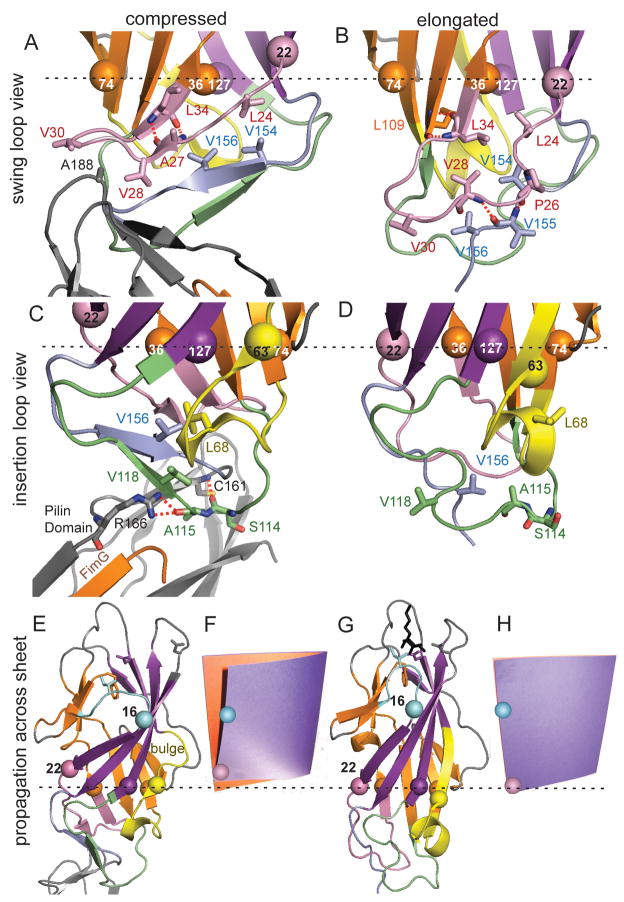
In the compressed conformation, the swing loop residues V28 and V30 move wide and are exposed to interact with residue A188 of the pilin domain (Figure 4A). In the elongated form, V28 faces into the hydrophobic core of the domain and V30 remains close to V156 in the linker loop (Figure 4B). In the compressed form, the insertion loop is extended where S114 and A115 insert deep into a pocket in the pilin domain and form hydrogen bonds with C161 and R166, respectively (Figure 4C.). The insertion would be prevented in the elongated form since the insertion loop reorients, facing A115 inward into the lectin domain (Figure 4D). Moreover, the pilin domain would be pushed away from the proximal loops by the extended linker loop in the elongated form, but not by the compressed linker in the tip.
Figure 4. Structural changes caused by pilin domain interaction.
Colors and representations are the same as in Figures 1 and 3. The Cα atoms are shown as spheres for three β-sheet residues (36, 74, and 127) that are in a plane perpendicular to the field of view, as shown by the dotted black line. Residues 22 and 63 are also shown as spheres to show how they move relative to this plane and the other residues. A, B. View from side of swing loop. C, D. View from side of insertion loop. E, G. Propagation of conformational changes from the proximal region to the distal mannose-binding site. F, H. photograph of a cardboard folder with an orange page representing the split β-sheet and a purple page representing the large β-sheet in an unperturbed state (F) and with the lower left front edge pushed upward (H), to demonstrate the analogy for the page-turning mechanism of β-sandwich allostery.
The conformational changes in the proximal end can be directly connected to the changes in the distal mannose pocket. When the linker loop is retracted to allow close contact of the neighboring loops with the pilin domain, linker residues V154 and V156 move upward in Figure 4AB. This is accommodated by an upward movement of L24 in the swing loop. This in turn is accommodated by an upward movement of residue 22, which anchors the swing loop to the large β-sheet, relative to residue 36, which anchors the swing loop to the split β-sheet (Figure 4AB.) The strand comprising residues 16 to 22 at the edge of the large β-sheet thus slides upwards relative to the opposing residues on the split β-sheet, resulting in the tilt of this strand and the bending and twisting of the large β-sheet (Figure 4EG). This can best be understood by sliding the lower left corner of a page in a book upward as though preparing to turn the page back – the partially opened page (Figure 4F) and the closed page (Figure 4H) resemble the large beta-sheet in the compressed (Figure 4E) and elongated (Figure 4G) conformations of the lectin domain, respectively. Note that the top of the turning page opens outward, just as residue 16 at the top of the tilted edge strand shifts outward in the compressed conformation (Figure 4E–H). Residue 16 anchors the clamp loop to the large β-sheet, and this outward movement breaks the hydrogen bonds in and around the clamp loop, loosening the mannose pocket (as described in Figure 2AB). Thus, interaction with the pilin domain pushes the bottom corner of the large β-sheet upward and loosens the distal mannose-binding pocket through what can be called a page-turning mechanism.
Further inspection reveals a second connection between the pilin domain interaction and tilting of the large β-sheet. The reorientation of the insertion loop in the compressed conformation causes V118 to face inward, while the retraction of linker loop brings V156 upward, creating a hydrophobic pocket inside the loops. This pocket is filled by residue L68, which faces inward in the compressed (Figure 4C) but outward in the elongated conformation (Figure 4D), requiring the 310/α-helix to turn like a screw. As a result, residue A63 at the start of the helix is pushed upward into the large β-sheet relative to residue T74 on the opposing split sheet. Residues 59 to 61 buckle and break away from the neighboring strand, forming the bulge in the large β-sheet seen in Figures 1D and 4E. In the page-turning analogy, this bulge may relieve stress that propagates laterally from the tilted edge, bending the sheet.
Thus, the page-turning mechanism illustrates how conformational changes can propagate across a β-sheet. Most importantly, it explains how docking of the pilin domain to the lectin domain will loosen the mannose pocket while separation of the pilin domain from the lectin domain will clamp the pocket tightly around mannose. Because FimH is allosterically maintained in a low-affinity state by internal contacts rather than by external factors, it can be said to be autoinhibited (Pufall and Graves, 2002). Because the loose binding conformation is more compressed and twisted, the lectin domain also looks and functions much like a finger-trap toy, which is a tube-shaped mesh. This toy, also called a finger prison, has been proposed as a metaphor for force-activated catch bonds (Dembo et al., 1988) because it holds more tightly to fingers inserted on either end when they pull away and it stretches into a more elongated, narrow and less twisted shape. The structural resemblances between the conformational change in the lectin domain and a finger-trap can be noted in the Supplemental Movie 1. Thus, the page turning and finger trap metaphors provide a good general understanding of β-sheet allostery.
Experimental validation of the predicted conformational shift mechanism
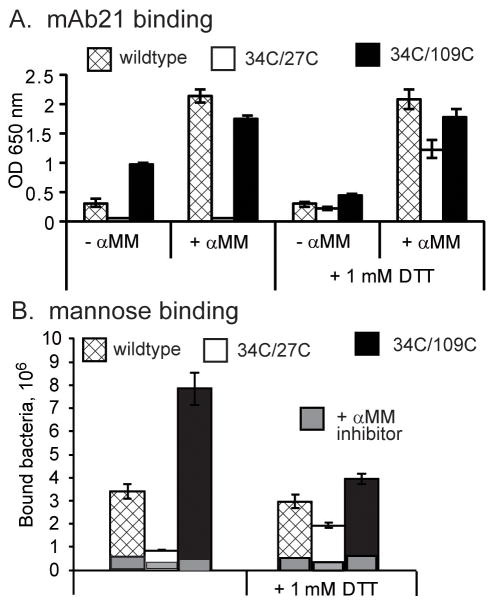
In the compressed conformation, residue L34 faces outward and forms a hydrogen bond with A27 that stabilizes the wide conformation of the swing loop (Figure 4AB). In the elongated form, L34 flips 180° to face inward and bond with L109 instead. We determined with the program MODIP (Dani et al., 2003) that both sets of partners are in position to form disulfide bonds. We thus created a 34C/27C double mutant to lock FimH in the compressed conformation and a 34C/109C double mutant to lock FimH in the elongated conformation. For the mutagenesis studies, fimbriae-incorporated FimH variant from E. coli J96 was used. As predicted, mAb21, which specifically recognizes the elongated conformation, bound strongly to 34C/109C but failed to recognize the 34C/27C fimbriae (Figure 5A). Addition of mannose strongly increased mAb21 binding to wildtype FimH, but failed to enable mAb21 binding to the 34C/27C variant (Figure 5A). Also, 34C/27C bacteria bound much more poorly to mannose-BSA coated surfaces than wildtype, while 34C/109C bacteria bound much more strongly (Figure 5B). The mAb21 and mannose-BSA binding differences between the mutant and wild-type FimH variants were nearly eliminated by addition 1mM DTT, which reduces disulfide bonds. This confirms that both 34C/27C and 34C/109C pairs formed disulfide bonds and locked the lectin domain in compressed low-affinity or elongated high-affinity conformations, respectively, presumably preventing the page-turning mechanism of allosteric propagation described above.
Figure 5. Mutational Regulation.
A. Binding of mAb21 to wildtype or disulfide-locked FimH in fimbriae, with and without α-MM or DTT. B. Number of bacteria expressing wildtype or disulfide-locked FimH binding to man-BSA surface with or without DTT.
Mechanical regulation of allostery
FimH from E. coli F18 is one of the most common structural variants of FimH in E. coli (Sokurenko at al, 2004). On the surface of bacteria, FimH is incorporated into the fimbrial tip and thus is expected to be in the low-affinity compressed conformation. Under in vivo conditions, the elongated conformation can at least sometimes be induced by binding to soluble or surface-bound mannosylated compounds, consistent with the data presented above (Figure 2H) and a previous study (Tchesnokova et al., 2008). However, under equilibrium conditions (in the absence of antibodies or other factors that can affect structural dynamics of the lectin domain), the ligand-induced change is not sustained and is insufficient to mediate strong binding to mannose, as indicated by the reduced affinity and bond lifetimes observed for the tip relative to the purified lectin domain (Figure 2CD).
On the other hand, FimH is known to mediate shear-enhanced adhesion of bacteria to mannosylated cells or surfaces (Thomas et al., 2004), binding weakly at low shear but strongly in high shear conditions. This occurs because FimH forms catch bonds (Thomas et al., 2006; Yakovenko et al., 2008), which are longer-lived under higher tensile force. Thus, it is possible that the high-affinity elongated conformation might be induced and/or sustained by mechanical force. To test this, we probed the fimbrial tips using an Atomic Force Microscope (AFM) with a mannose-BSA-coated AFM cantilever. Unlike a previous study, where force was increased until the bonds ruptured (Yakovenko et al., 2008), here we measure effect of a constant force on lifetimes of single FimH-mannose bonds as described previously for other catch bonds.
At a low force of 20 pN, most of the detected interactions dissociate within one second (Figure 6). However, when tensile force was increased to 30, 50 or 70 pN, up to 70% of the interactions became long-lived. The two dissociation rates indicate that there are two types of interactions. Since experimental conditions are chosen for single bonds (see methods), these results cannot be attributed to multiple bonds. However, two dissociation rates are expected for single receptor-ligand interactions when a receptor has two alternative conformations. Indeed, the fast and slow dissociation rates, which dominate at low (≤ 20 pN) and moderate (30–70 pN) forces respectively, resemble the behavior of the fimbrial tip and lectin domain respectively in SPR assays, where there is no force (Figure 6). This suggests that mechanical force causes FimH in the fimbrial tip to switch from the low-affinity compressed to the high-affinity elongated conformation, presumably by separating the two FimH domains and/or directly favoring the elongated conformation.
Figure 6. Mechanical Regulation.
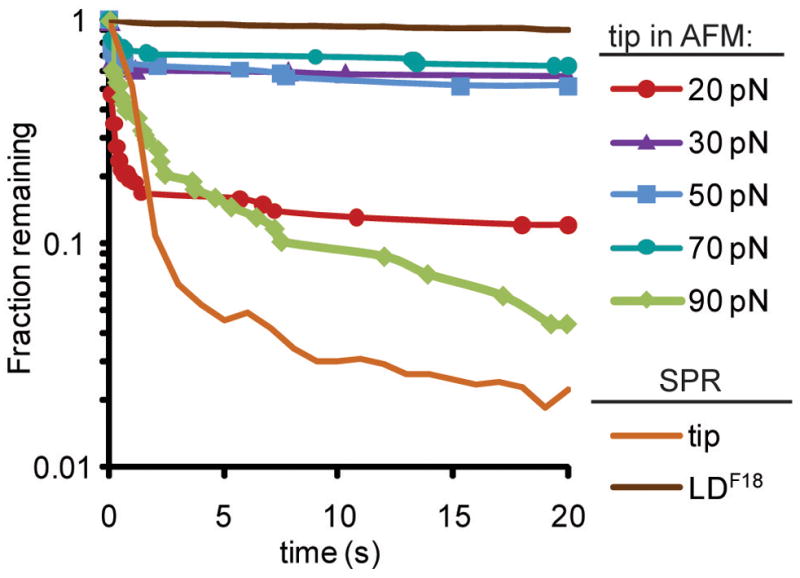
Profiles of tip dissociating from man-BSA at various tensile forces, as measured by AFM. These survival plots show the fraction of interactions that are left as a function of time after force is applied. The BiACore® surface plasmon resonance (SPR) dissociation profiles for the tip and LDF18 are superimposed for comparison.
The exceptionally long lifetime of these bonds at these high molecular forces is consistent with previous studies of these bonds under force (Thomas et al., 2008) and further suggests that FimH is optimized for strong adhesion under force. All bonds, even catch bonds, are overpowered by sufficiently high force (Thomas et al., 2008), and this is seen here for FimH by the shorter lifetimes at 90 pN.
Discussion
Together, these data support the following model of allosteric regulation of FimH. In the absence of mannose, the lectin domain in fimbrial tips is maintained in the compressed conformation with a loose mannose-binding pocket due to allosteric auto-inhibition by the interacting pilin domain. When mannose binds, the pocket can tighten around mannose by forming backbone hydrogen bonds in the clamp loop, which in turn can induce a switch from the twisted compressed to the untwisted elongated conformation and presumably dislodge the pilin domain. However, this ligand-induced change might not occur every time, and when it does occur, the change is transient since the pilin domain can redock and cause the reversion to the compressed conformation. For these reasons, FimH affinity and bond lifetimes remain low. If the compressed conformation of the bond is subjected to mechanical force before mannose dissociates, the pilin domain is pulled away until it is attached only through the linker chain. The bottom region of the lectin domain (proximal to the pilin domain) is switched from the compressed to the elongated conformation by the absence of pilin contacts and possibly also by forced elongation. Similarly, force applied to the elongated conformation can prevent redocking of the pilin domain and reversion to the compressed conformation. Whether force induces or sustains the elongated conformation, the mannose-binding pocket is kept tightly shut, lengthening the bond lifetime under force. In this manner, the conformational changes can be initiated in the binding pocket by mannose or in the interdomain region by force, but in either case propagate across the protein through a distortion of the large β-sheet similar to the distortion in a turning page or in a finger trap toy.
It has been shown that a variety of naturally-occurring point mutations in FimH increase binding to mannose, presumably by favoring the elongated high-affinity conformation. These variants are found mostly in uropathogenic E. coli, and increase bacterial adhesion to mannosylated cells and surfaces under static conditions (Sokurenko et al., 2004). This suggests that these variants benefit from binding strongly even without force in low-flow conditions such as the upper urinary tract. However, evolutionary analysis indicates that the more weakly binding variants, such as FimH from E. coli F18, are evolutionarily predominant (Sokurenko et al., 2004), suggesting that allosteric inhibition and corresponding catch bond behavior are beneficial for transmission or survival. The physiological advantage of allosteric catch-bond adhesion could include resistance to soluble inhibitors (Nilsson et al., 2006; Thomas et al., 2002), and rapid surface colonization (Anderson et al., 2007).
The page-turning mechanism described here can be contrasted to conformational changes in other proteins. Other examples of structural changes in a β-sandwich domain involve various degrees of rearrangement or swapping of the edge β-strands. For example, the adhesion receptors ICAM-1 (Chen et al., 2007), cadherins (Chen et al., 2005; Patel et al., 2006), and a bacterial fibronectin binding protein (Bingham et al., 2008) all rearrange edge β-strands during counter-receptor binding or dimerization. Similarly, many β-sandwich proteins, including the type 1 fimbrial tip subunits, polymerize via β-strand swapping, and many amyloid proteins irreversibly refold into β-sheet structures. In contrast to these rearrangements, we report here that an intact β-sheet can twist and bend to allosterically confer structural information from one end of the β sandwich to the other. This β-sheet-bending mechanism is similar to the tilting of an α helix in allosteric proteins such as integrins (Xiao et al., 2004) and myosin (Holmes et al., 2004), in that the relative rigidity of the helix or sheet allows long-range structural communication.
The structural changes we describe here provide a novel mechanism by which conformational changes can propagate across a β-sandwich domain, raising the question as to whether this occurs in other proteins. Interestingly, β-sandwich domains are present in many adhesive, matrix, and other extracellular proteins in prokaryotes, eukaryotes and even viruses, and are thought to confer stability in the presence of mechanical tension and other harsh conditions. The page-turning mechanism could provide the flexibility needed for allosteric regulation together with stability in such proteins.
Experimental Procedures
Expression and purification
Fimbrial tips were expressed and purified as previously described (Aprikian et al., 2007). Briefly, a plasmid containing genes for FimC (with 6HIS tag at the C terminus), FimF, FimG and FimH was transformed into C43 expression cells. After induction, cells were homogenized and the tip complex purified on Ni-NTA agarose followed by an α-D-mannose-agarose column. Single pick fractions from 10% α-D-mannose elution were concentrated to 12mg/ml and submitted to the Center for High-Throughput Structural Biology (CHTSB) at the Hauptman-Woodward Institute (Buffalo, NY) for screening crystallization conditions (Luft et al., 2003).
Crystallization
The fimbrial tips were crystallized using hanging drop vapor diffusion techniques at room temperature. Crystals were obtained by mixing equal volume drops containing the protein (11 mg/ml in 20mM Hepes pH 7.0; 0.15M NaCl) and a crystallization solution containing 1.6M KCl, 0.1M sodium citrate pH 4.1. Crystals were transferred to a cryosolution containing 30% glycerol before freezing at 100 K in a nitrogen stream for diffraction data collection.
Diffraction data were collected at ALS Beamline 5.0.2 (λ=1.0Å). The space group for the crystals is R32 with two copies of the tip complex in the asymmetric unit. The diffraction data were processed with HKL2000 (Otwinowski and Minor, 1997). Data set statistics are shown in Table 1.
Structure solution and refinement
The crystal structure was obtained using the molecular replacement programs Phaser(McCoy et al., 2007), MOLREP(Vagin and Teplyakov, 1997) and EMPR (Kissinger et al., 1999). Input structures were obtained from previous structures of FimG (Puorger et al., 2008) (PDB ident 3BFQ), FimF (Eidam et al., 2008) (3BWU), and FimC (Eidam et al., 2008) (3BWU). The individual subunit structures are sufficiently different to require separate search models for each. We were unable to locate the FimH domains using a known structure (Hung et al., 2002) (1KLF) and the molecular replacement programs. Instead, the domains were placed into the electron density maps manually and refined using real space refinement as implemented in XtalView (McRee, 1999).
The structural model was refined using REFMAC-5 (Murshudov et al., 1997) in the CCP4 suite (Bailey, 1994). Rfree (Brunger, 1993) was calculated using 5% of the data in the test set. At 2.9 Ǻ resolution, the data set completeness drops below 90% and <I>/<σ(I)> is 2.0. Significant diffraction data were obtained out to 2.7 Ǻ resolution and we chose to use all reflections to that limit for refinement. Sigma A weighted |Fo|−|Fc| and 2|Fo|−|Fc| electron density maps (Read, 1986) were viewed with XtalView (McRee, 1999) and Coot(Emsley and Cowtan, 2004) for graphical evaluation of the model and electron density maps. Table 1 contains refinement statistics for the structure.
Measuring RMSD differences
Cα carbons were used to measure RMSD with N-terminal extensions excluded and the same residues used for alignment and RMSD calculations with VMD (Humphrey et al., 1996). When pdb files had multiple copies of a structure, the closest in RMSD were used.
Surface Plasmon Resonance
SPR experiments with both purified lectin domains and fimbrial tips were performed as described previously on a Biacore2000™ instrument (Biacore Inc.) (Tchesnokova et al., 2008). In brief, man-BSA was immobilized on a CM5 chip via amino coupling and a series of concentrations of tips or lectin domains (analyte) were injected and the average response over 5 seconds at the end of injection was determined to obtain a calibration curve for each tip or lectin domain analyte. Then, 10 μM purified fimbrial tips or 1 μM purified lectin domains were preincubated with αMM at each concentration indicated in Figure 2C, injected for 2 minutes, and washed out with buffer. The average response over 5 seconds at the end of the injection was determined, and the concentration of free analyte calculated using the calibration curve. The fraction of bound analyte is calculated as: bound = (total − free)/total. To measure dissociation profiles in Figure 2D, 10 μM tip, LDF18 or LDJ96 were injected for 2 minutes and then washed out. The curves were normalized to a value of one at the time the wash started, to indicate the fraction remaining bound.
MAb binding
Mouse monoclonal antibody 21 (mAb21; raised against LDJ96) and binding of mAb21 to lectin domain, fimbrial tips, or purified fimbraie was described previously (Tchesnokova et al., 2008). Briefly, proteins were immobilized in 96 well dishes, washed, incubated with mAb21 at a range of dilutions with or without 50 mM αMM, and the latter detected with HRP-conjugated goat anti-mouse antibodies.
NMR
All NMR experiments were collected on a 1 mM, 13C,15N-labeled sample of the isolated FimH lectin domain (LD) in 90% H2O/10% D2O solution containing 50 mM sodium phosphate (pH 7.0) and 100 mM NaCl. NMR spectra were acquired at 27°C on Bruker Avance II 600 and DMX500 spectrometers equipped with triple-resonance, triple-axis gradient probe, with 32 transients per t1 values. The standard suite of three dimensional triple resonance experiments, HNCA, HNCOCA, HNCACB, and CBCACONH were used to assign the mannose-free LD. The assignments of mannose-free LD were then transferred to mannose-bound LD and further confirmed with HNCACB and CBCACONH. Data was processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRView (Johnson, 2004). 134 out of 150 observable, non-proline residues could be assigned. The chemical shift differences between the mannose-free and mannose-bound forms, “Δ(δno mannose − δmannose) ppm” (shown in Figure S1) were computed from the absolute, weighted average of backbone amide proton and nitrogen chemical shift differences as follows: (((Δδ H)2 + (Δδ N/5)2)/2)1/2.
Atomic Force Microscopy
Fimbrial tips were attached to a surface and man-BSA to a cantilever tip as described previously (Yakovenko et al., 2008). 10 ng/ml fimbrial tips were immobilized on polystyrene or glass and 100μg/ml man-BSA on Biolever AFM tips (Olympus; Tokyo, Japan), and blocked with Phosphate Buffered Saline + 0.2% Bovine Serum Albumin (PBS-BSA) overnight. An MFP-3D AFM from Asylum Systems (Santa Barbara USA) was used to probe the forces between the cantilever and surface in PBS-BSA. The tip was pressed to the surface with 30 pN of force for 1 s then withdrawn at 2 μm/s until the indicated tensile force was reached, whereupon it was maintained with a digital feedback loop. Spring constants (nominal 6 pN/nm) were calibrated using the thermal method. Data were taken at 5 kHz and a running average of 100 points (20 ms) used to reduce noise. Each lifetime was measured as the time between reaching within 5 pN of the desired force in the filtered data and the sudden drop to no force, and lifetimes plotted as a survival curve.
Conditions were chosen such that only 10 +/− 5% of pulls show any measurable interaction, so that single bonds could be inferred by Poisson s law of small numbers, the standard for single molecule force spectroscopy (Zhu et al., 2002). This means that no more than 15% of the measured interactions are expected to have multiple bonds, so that the long lifetimes (70% at high force) cannot be due to multiple bonds. When pulls were performed in the presence of 1% soluble αMM, a competitive inhibitor of FimH, interactions were rare and short-lived, so the long lifetimes are not due to nonspecific binding.
Supplementary Material
Acknowledgments
We thank NIH (1RO1 AI50940NIH; Sokurenko P.I. and 5 T32 GM008268) and the AHA Postdoctoral Fellowship #0820107Z for funding this work. We thank Eric Larson for helpful discussions and the Advanced Light Source for beamtime. The Advanced Light, Source is supported by the Director, Office of Science Office of Basic Energy Sciences the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ronald E. Stenkamp, Email: stenkamp@u.washington.edu.
Evgeni V. Sokurenko, Email: evs@u.washington.edu.
Wendy E. Thomas, Email: wendyt@u.washington.edu.
References
- Alon R, Dustin ML. Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity. 2007;26:17–27. doi: 10.1016/j.immuni.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Anderson BN, Ding AM, Nilsson LM, Kusuma K, Tchesnokova V, Vogel V, Sokurenko EV, Thomas WE. Weak rolling adhesion enhances bacterial surface colonization. J Bacteriol. 2007;189:1794–1802. doi: 10.1128/JB.00899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprikian P, Tchesnokova V, Kidd B, Yakovenko O, Yarov-Yarovoy V, Trinchina E, Vogel V, Thomas W, Sokurenko E. Interdomain Interaction in the FimH Adhesin of Escherichia coli Regulates the Affinity to Mannose. J Biol Chem. 2007;282:23437–23446. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- Arnaout MA, Mahalingam B, Xiong JP. Integrin Structure, Allostery, and Bidirectional Signaling. Annu Rev Cell Dev Biol. 2005 doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- Askari JA, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. J Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrof NS, Salas A, Shimaoka M, Chen J, Springer TA. Importance of Force Linkage in Mechanochemistry of Adhesion Receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Bingham RJ, Rudino-Pinera E, Meenan NA, Schwarz-Linek U, Turkenburg JP, Hook M, Garman EF, Potts JR. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc Natl Acad Sci U S A. 2008;105:12254–12258. doi: 10.1073/pnas.0803556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert J, Berglund J, Schembri M, De Genst E, Cools L, Wuhrer M, Hung CS, Pinkner J, Slattegard R, Zavialov A, et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol Microbiol. 2005;55:441–455. doi: 10.1111/j.1365-2958.2004.04415.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Assessment of Phase Accuracy by Cross Validation - the Free R-Value - Methods and Applications. Acta Crystallographica Section D-Biological Crystallography. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kim TD, Carman CV, Mi LZ, Song G, Springer TA. Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization. Proc Natl Acad Sci U S A. 2007;104:15358–15363. doi: 10.1073/pnas.0707406104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- Dani VS, Ramakrishnan C, Varadarajan R. MODIP revisited: reevaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng. 2003;16:187–193. doi: 10.1093/proeng/gzg024. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Eidam O, Dworkowski FS, Glockshuber R, Grutter MG, Capitani G. Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD. FEBS Lett. 2008;582:651–655. doi: 10.1016/j.febslet.2008.01.030. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- Gossert AD, Bettendorff P, Puorger C, Vetsch M, Herrmann T, Glockshuber R, Wuthrich K. NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits. J Mol Biol. 2008;375:752–763. doi: 10.1016/j.jmb.2007.10.059. [DOI] [PubMed] [Google Scholar]
- Hashimoto H. Recent structural studies of carbohydrate-binding modules. Cell Mol Life Sci. 2006;63:2954–2967. doi: 10.1007/s00018-006-6195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Schroder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hung CS, Bouckaert J, Hung D, Pinkner J, Widberg C, DeFusco A, Auguste CG, Strouse R, Langermann S, Waksman G, Hultgren SJ. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. 2002;44:903–915. doi: 10.1046/j.1365-2958.2002.02915.x. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D Biol Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee DE. XtalView Xfit - A versatile program for manipulating atomic coordinates and electron density. Journal of Structural Biology. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nilsson LM, Thomas WE, Sokurenko EV, Vogel V. Elevated shear stress protects Escherichia coli cells adhering to surfaces via catch bonds from detachment by soluble inhibitors. Appl Environ Microbiol. 2006;72:3005–3010. doi: 10.1128/AEM.72.4.3005-3010.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski A, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzym. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Phan UT, Waldron TT, Springer TA. Remodeling of the lectin-EGF-like domain interface in P- and L-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat Immunol. 2006;7:883–889. doi: 10.1038/ni1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- Puorger C, Eidam O, Capitani G, Erilov D, Grutter MG, Glockshuber R. Infinite kinetic stability against dissociation of supramolecular protein complexes through donor strand complementation. Structure. 2008;16:631–642. doi: 10.1016/j.str.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Read RJ. Improved Fourier Coefficients for Maps Using Phases from Partial Structures with Errors. Acta Crystallographica Section A. 1986;42:140–149. [Google Scholar]
- Shan WS, Koch A, Murray J, Colman DR, Shapiro L. The adhesive binding site of cadherins revisited. Biophys Chem. 1999;82:157–163. doi: 10.1016/s0301-4622(99)00115-5. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, Johnson JR, Dykhuizen DE. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004;21:1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- Springer TA. Structural basis for selectin mechanochemistry. Proc Natl Acad Sci U S A. 2009;106:91–96. doi: 10.1073/pnas.0810784105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokova V, Aprikian P, Yakovenko O, Larock C, Kidd B, Vogel V, Thomas W, Sokurenko E. Integrin-like allosteric properties of the catch bond-forming FimH adhesin of Escherichia coli. J Biol Chem. 2008;283:7823–7833. doi: 10.1074/jbc.M707804200. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Forero M, Yakovenko O, Nilsson L, Vicini P, Sokurenko EV, Vogel V. Catch Bond Model Derived from Allostery Explains Force-Activated Bacterial Adhesion. Biophys J. 2006;90:753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WE, Nilsson L, Forero M, Sokurenko EV, Vogel V. ‘Stick-and-roll’ bacterial adhesion mediated by catch-bonds. Molecular Microbiology. 2004;53:1545–1557. doi: 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–317. doi: 10.1016/s0945-053x(00)00072-x. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Waldron TT, Springer TA. Transmission of allostery through the lectin domain in selectin-mediated cell adhesion. Proc Natl Acad Sci U S A. 2009;106:85–90. doi: 10.1073/pnas.0810620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellens A, Garofalo C, Nguyen H, Van Gerven N, Slattegard R, Hernalsteens JP, Wyns L, Oscarson S, De Greve H, Hultgren S, Bouckaert J. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS ONE. 2008;3:e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, Vogel V, Sokurenko E, Thomas W. Fimh forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Long M, Chesla SE, Bongrand P. Measuring receptor/ligand interaction at the single-bond level: experimental and interpretative issues. Ann Biomed Eng. 2002;30:305–314. doi: 10.1114/1.1467923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.