Abstract
Objectives
NADH cytochrome b5 reductase (b5R) and cytochrome b5 (b5) catalyze the reduction of sulfamethoxazole hydroxylamine (SMX-HA), which can contribute to sulfonamide hypersensitivity, to the parent drug sulfamethoxazole. Variability in hydroxylamine reduction could thus play a role in adverse drug reactions. The aim of this study was to characterize variability in SMX-HA reduction in 111 human livers, and investigate its association with single nucleotide polymorphisms (SNPs) in b5 and b5R cDNA.
Methods
Liver microsomes were assayed for SMX-HA reduction activity, and b5 and b5R expression was semi-quantified by immunoblotting. The coding regions of the b5 (CYB5A) and b5R (CYB5R3) genes were resequenced.
Results
Hepatic SMX-HA reduction displayed a 19-fold range of individual variability (0.06–1.11 nmol/min/mg protein), and a 17-fold range in efficiency (Vmax/Km) among outliers. SMX-HA reduction was positively correlated with b5 and b5R protein content (p < 0.0001, r = 0.42; p = 0.01, r = 0.23, respectively), and expression of both proteins correlated with one another (p < 0.0001; r = 0.74). A novel cSNP in CYB5A (S5A) was associated with very low activity and protein expression. Two novel CYB5R3 SNPs, R59H and R297H, displayed atypical SMX-HA reduction kinetics and decreased SMX-HA reduction efficiency.
Conclusion
These studies indicate that while novel cSNPs in CYB5A and CYB5R3 are associated with significantly altered protein expression and/or hydroxylamine reduction activities, these low frequency cSNPs only appear to minimally impact overall observed phenotypic variability. Work is underway to characterize polymorphisms in other regions of these genes to further account for individual variability in hydroxylamine reduction.
Keywords: sulfamethoxazole, sulfonamide hypersensitivity, cytochrome b5, NADH cytochromeb5 reductase, single nucleotide polymorphism
Introduction
The antimicrobial drug sulfamethoxazole (SMX) is associated with delayed hypersensitivity reactions in 1–3% of the general population [1], with a higher incidence in immunocompromised patients [2,3]. SMX is oxidized to its hydroxylamine (SMX-HA) by CYP2C9 [4] or myeloperoxidase [5]; the hydroxylamine is spontaneously oxidized to an unstable nitroso metabolite (SMX-NO) [6], which is thought to be the proximate immunogen in SMX hypersensitivity reactions [7,8]. SMX-HA and other hydroxylamines are reduced by cytochrome b 5 reductase (b5R) and cytochrome b 5 (b5) in the endoplasmic reticulum [9,10]. This pathway also detoxifies dapsone hydroxylamine [9], as well as hydroxylamines of the carcinogens 4-aminobiphenyl and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) [10]. Thus the microsomal b5/b5R pathway may be clinically and toxicologically important, since it counters the bioactivation of hydroxylamine-forming xenobiotics. Consequently, variability in the activity of this reduction pathway is likely to have direct consequences for the disposition and toxicity of clinically important xenobiotics.
The cytochrome b5 (CYB5A) and the NADH cytochrome b5 reductase (CYB5R3) genes are well characterized. Each gene generates two major splice variants, a soluble cytosolic form found in erythrocytes, and a membrane-bound protein predominantly located in the endoplasmic reticulum of hepatocytes and other tissues [11–13]. However, individual variability in the expression of CYB5R3 and CYB5A has not been well characterized. Two non-synonymous cSNPs in CYB5A, R52K and Y130X, have been reported in the NCBI SNP database, and eight non-synonymous cSNPs have been identified to date for CYB5R3; however, the functional significance, if any, of these and other cSNPs has not been determined.
Although earlier studies have shown significant individual differences in soluble b5/b5R activity (as measured by erythrocyte methemoglobin reduction) [14,15], no genotype-phenotype correlations have been performed for either isoform. Our previous data in microsomes from 27 human livers indicated nearly 10-fold variability in SMX-HA reduction, with a similar range of b5 protein expression but lesser variability in b5R expression [10]. We recently identified a cSNP in CYB5A (T60A) in a screen of leukocyte cDNA from 63 individuals; this cSNP was associated with a 70% decrease in b5 protein expression in vitro [16]. However, the prevalence of this cSNP is not yet clear.
The objectives of this study, therefore, were to characterize the range of hydroxylamine reduction activities in a relatively large sample of normal human livers, and to perform genotype-phenotype correlations between cSNPs in the genes encoding the microsomal forms of b5 and b5R, and hydroxylamine reduction. To meet these objectives, we phenotyped SMX-HA reduction activities in 111 human livers, along with immunoblotting for b5 and b5R protein expression, and resequenced CYB5A and CYB5R3 cDNAs in all livers in order to screen for coding sequence or splice junction polymorphisms.
Methods
Liver samples
Human liver tissue was obtained through the Midwest Division of the Cooperative Human Tissue Network, and through collaboration with Dr. Sharon Weber, from the Division of General Surgery, University of Wisconsin-Madison. Tissue samples were taken from grossly normal margins of excised benign or metastatic liver lesions from adult patients of any race or either gender. Patients with prior intra-abdominal radiation therapy were ineligible. Liver samples were snap frozen in liquid nitrogen and then stored at –80°C until use. All subjects provided informed consent, and all procedures were approved by the Institutional Review Board of the University of Wisconsin-Madison. A sample size of > 100 livers was chosen in order to provide a >95% chance of finding a SNP with an allele frequency of 1% [17].
Subcellular fractionation
Approximately 1 g of liver tissue was homogenized in Dulbecco’s phosphate-buffered saline (PBS), pH 7.4 and centrifuged at 9,000 × g. The resulting nuclear pellet was stored at −80°C for genomic DNA extraction, if needed. The supernatant was further centrifuged at 100,000 × g for 60 minutes to obtain microsomal and cytosolic fractions, which were stored at −80°C. Protein content determination was carried out using the Bradford assay (Bio-Rad, Hercules, CA).
SMX-HA reduction assay and kinetics
SMX-HA (Wilmington PharmaTech, Newark, DE) was dissolved in 50% DMSO with 3mM ascorbic acid, while SMX (Sigma, St.Louis, MO) was dissolved in an aqueous solution of 50% DMSO. Reduction activities were determined by incubating 125 µg microsomal protein with 200 µM SMX-HA and 1mM NADH in PBS, pH 7.4, at 37°C for 15 minutes. Reduced glutathione and ascorbic acid (both at a final concentration of 1 mM) were included to prevent hydroxylamine autooxidation [18]. Negative controls and SMX standards included human serum albumin (125 µg; Sigma) in place of microsomal protein. After reaction termination by the addition of ice-cold methanol, the proteins were pelleted out by centrifugation, and the resulting filtered supernatant was analyzed by gradient HPLC with UV detection at 256 nm. The HPLC conditions were as follows: solvent A consisted of an aqueous solution of 1% acetic acid and 0.05% triethylamine; solvent B was 100% acetonitrile, at a flow rate of 2mL/min. The percentage of solvent B in the mobile phase mixture was initially 10% for 5 minutes, increased to 30% over 7 minutes, kept at 30% for 11 minutes, and finally returned back to 10% over 13 minutes. SMX product formation was quantified by means of peak heights from an SMX standard curve.
SMX-HA reduction kinetics were determined for a subset of human livers (n=18) that had no cSNPs in CYB5A and CYB5R3 and represented a range of SMX-HA reduction activities (high: >90th percentile of activities for the population; intermediate: 45th–55th percentile; and low: <10th percentile). Kinetics were also determined for all livers that were found to have non-synonymous CYB5A or CYB5R3 cSNPs. Kinetic experiments were performed under initial-rate conditions at a concentration range of 50–3000 µM SMX-HA. Data were fit to the Michaelis-Menten equation and apparent kinetic parameters were estimated using GraphPad Prism 5 (GraphPad Software, San Diego, CA). For those livers exhibiting nonlinear kinetics, fitting into several two-site and two-enzyme models resulted in nonsensical estimates of kinetic parameters; for these livers, data from Hanes plots were used in order to derive initial estimates of Km and Vmax for high and low substrate affinity components. An iterative technique was employed to account for the contributions of each component at the substrate range used. The process was repeated until convergence was obtained [19].
b5 and b5R immunoblot analysis
To determine the relationship between b5 and b5R expression and SMX-HA reduction in human liver, rabbit polyclonal antisera to b5 or b5R [9] were used to probe immunoblots prepared from individual human liver microsomal proteins (30 µg). The same blot was probed for both b5 and b5R, along β-actin to control for lane loading. Each blot was probed in the following order: β-actin control, b5, and b5R. Since the β-actin and b5R proteins are similar in size, the PVDF membranes were stripped for 30 minutes at 37°C (Restore Plus Western Blot Stripping Buffer (Pierce Biotechnology, Rockford, IL), prior to b5R immunoblotting. Immunoblotting was performed twice per sample, from separate gels (15% Tris-HCl; Bio-Rad, Hercules, CA). After semi-dry transfer, PVDF membranes (0.45µm; Pierce Biotechnology) were blocked with 5% non-fat dry milk in Dulbecco’s PBS/0.1% Tween-20 for 1 hr and washed with Dulbecco’s PBS/0.1% Tween-20. For loading controls, mouse monoclonal to β-actin, tagged with horseradish peroxidase (Abcam, Cambridge, MA) was diluted 1:5000, and incubated with PVDF membranes for 2 hr at 4°C. Rabbit polyclonal anti-human b5 and b5R antisera, diluted 1:10,000 [9], were each incubated overnight with the PVDF membrane. The secondary antibody, peroxidase-conjugated donkey anti-rabbit IgG (Jackson Immunoresearch Laboratories, West Grove, PA), was diluted 1:10,000 and immunoblotted for 2 hr at 4°C. Bound antibody for all experiments was detected by chemiluminescence (Super Signal® West Pico, Pierce) using a UVP imaging system (UVP, Upland, CA) and quantified by ImageJ software (http://rb.info.nih.gov/ij). Densitometries for bands corresponding to immunoreactive b5 and b5R were each normalized to immunoreactive β-actin.
Resequencing of cDNA for CYB5A and CYB5R3
Total RNA was isolated from 80 µg of individual human liver tissue using the RNAqueous®-4PCR kit (Ambion, Austin, TX). RT-PCR was performed using the RETROscript® KIT (Ambion). Primers were designed to amplify the entire coding sequences of CYB5A and CYB5R3 (Table 1). For amplification of microsomal CYB5A cDNA, the thermocycler parameters were as follows: initial denaturation at 94°C for 2 min, 35 cycles at 94°C for 15 sec, 51°C for 25 sec, and 72°C for 30 sec, followed by a final extension of 72°C for 7 min. For amplification of microsomal CYB5R3 cDNA, a touchdown PCR method was developed: initial denaturation at 95°C for 2 min, then 38 cycles at 95°C for 30 sec, 71–67°C for 30 sec (2 cycles per degree), followed by 28 cycles at the touchdown temperature of 66°C) and 72°C for 1 min 15 sec, followed by a final extension at 72°C for 7 min.
Table 1.
Primers used for cDNA resequencing and SNP confirmation
| Experiment | Orientation | Primer sequence (5′→3′) |
|---|---|---|
| b5 PCR and resequencing |
Forward | GGCCTGGCTCGCGGCGAACCGAG |
| Reverse | CTCCCGTGTCCAAAGCAGGC | |
| b5R PCR | Forward | ACCATGGGGGCCCAGCTCA |
| Reverse | ACTGGGTGAGCGTGAACAG | |
| b5R resequencing | Forward | CTACCTCTCGGCTCGAATTG |
| Reverse | TGTCTCCAATCTGCATGCTC | |
| Novel SNP confirmation resequencing | ||
| b5 S5A | Forward | GTCGGAGGCCCTTCTACTG1 |
| Reverse | TCTTGCTGTGGTTGTGCTTC | |
| b5R R59H | Forward | TGCTCATGAAGCTGTTCCAG |
| b5R R297H | Forward | TTCTGCACGCTTCAAGCT |
anneals to genomic DNA at −159
Four µL aliquots of each PCR reaction were used to cycle sequence the CYB5A and CYB5R3 cDNA amplicons in both directions using BigDye Terminator v3.1 reagents (Applied Biosystems, Foster City, CA). The amplified fragments were submitted to the UW-Madison Biotechnology Core for magnetic bead cleanup and subsequent analysis on an ABI 3700 DNA sequencer. Sequences were aligned using the Staden Package (http://staden.sourceforge.net) and any sequence variants from the published wild-type sequence (NM_148923 for CYB5A, NM_000398 for CYB5R3) were confirmed by repeat sequencing with specific primers (Table 1). The SNPs were also confirmed by direct inspection of sequencing chromatograms, which are available on request from the authors. The first A in the ATG translation initiation codon and the initial methionine residue were designated as +1 for the cDNA and protein respectively, in accordance with the accepted nomenclature (http://www.hgvs.org/mutnomen/recs.html; [20]). The numbering system used for older b5 and b5R studies mentioned in this paper designates the first amino acid of the mature protein as +1. In order to allow comparisons, amino acid changes referred to in previous papers will be identified using the presently recommended nomenclature.
Genomic DNA extraction
Confirmation of one SNP (CYB5A S5A; see below) necessitated designing an appropriate forward primer to anneal to the 5′ untranslated region of exon 1 of CYB5A (Table 1). This required genomic DNA as a template, which was extracted from 12 mg of liver nuclear fraction using the DNeasy Tissue Kit (Qiagen, Valencia, CA).
Data analyses and prediction of SNP impact
SPSS statistical software (SPSS Inc., Chicago, IL) was used to analyze population and outlier activities, kinetics, and expression data. P < 0.05 was considered significant. Pearson correlation, or Spearman rank for non-normally distributed data, were used to investigate relationships among activities, age, and protein expression. Group means were compared via a one-way ANOVA with Tukey’s HSD; for non-normally distributed data, Kruskal-Wallis was used. Analysis of allele frequencies between different races was done using the Fisher’s exact test. Associations among sex and race, and either SMX-HA reduction activities or b5/ b5R protein expression, were investigated by using a general linear model univariate procedure to perform a two-way ANOVA with post-hoc Tukey HSD. Possible interactions between sex and race on activity and protein expression were investigated by estimating the marginal means, which are the group means after controlling for a covariate. Multiple regression analysis was used to verify the association between reduction activities and b5 or b5R expression when controlling for either b5 or b5R expression, with a given β coefficient indicating how much the predicted value of activity changes each time the expression of one protein increases by one unit, while keeping the expression of the other protein constant.
Haplotype analysis, including testing alleles for Hardy-Weinberg equilibrium, testing the significance of D' between two loci, and haplotype association tests with SMX-HA reduction activities were carried out by the Haploview 4.1 software package [21]. Several studies have shown that the impact of amino acid allelic variants on protein structure and function can be predicted by analysis of multiple sequence alignments and protein 3D structures [22–24].The following computational amino acid substitution predictive methods were therefore used: SIFT (http://blocks.fhcrc.org/sift/SIFT.html) [24], PolyPhen (http://genetics.bwh.harvard.edu/pph/) [24], and PMut (http://mmb2.pcb.ub.es:8080/PMut/) [22]. SIFT is based on the premise that important amino acids in a specific protein will be conserved across various species, while PolyPhen predicts the impact of nonsynonymous SNPs on protein 3D structure and function. PMut combines evolutionary information with structural data for prediction of pathological mutations. The neural network servers, Netphos [25] (available at http://www.cbs.dtu.dk/services/NetPhos/) and NetphosK [26] (available at http://www.cbs.dtu.dk/services/NetPhosK/), were used to predict phosphorylation sites for serine and threonine residues affected by SNPs.
Results
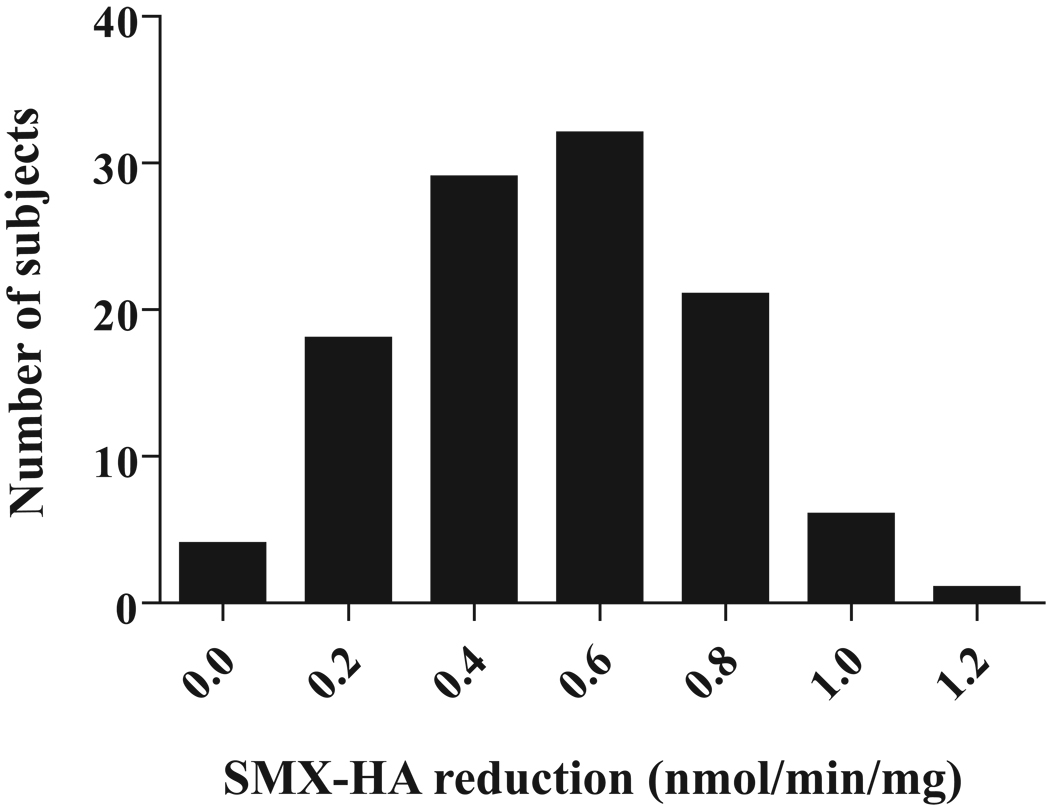
Hepatic SMX-HA reduction activities ranged from 0.06 to 1.11 nmol/min/mg protein (Figure 1), with one sample showing no detectable activity. Mean (± SD) SMX-HA reduction activity was 0.52 ± 0.24 nmol/min/mg microsomal protein (95% confidence interval = 0.47–0.57). SMX-HA reduction activities were not correlated with age (r = −0.145, p = 0.13). However, SMX-HA reduction activities in Caucasian-Americans (CA) (0.55 ± 0.22 nmol/min/mg; n = 93) were significantly higher than in African Americans (AA) (0.32 ± 0.29 nmol/min/mg; n = 15) (p = 0.003; Table 2). In particular, reduction activities in CA females (0.59 ± 0.23 nmol/min/mg; n = 52) were significantly higher than those in AA females (0.28 ± 0.25 nmol/min/mg; n = 9; p = 0.001). Estimated marginal means did not reveal any interaction between sex and race-related differences in activity.
Fig 1.
Distribution of human hepatic sulfamethoxazole-hydroxylamine (SMX-HA) reduction activities in 111 human livers. Mean (± SD) activity was 0.52 ± 0.24 nmol/min/mg microsomal protein in 64 females, 46 males, and one individual of unrecorded sex.
Table 2.
Human hepatic SMX-HA reduction activities, and b5 and b5R protein expression, in 111 human livers, stratified by sex and race.
| Sex | Race | Number of subjects |
Mean age (SD) |
SMX-HA reduction (nmol/min/mg) |
Relative protein expression |
|
|---|---|---|---|---|---|---|
| b5 | b5R | |||||
| All | All | 111 | 58 ± 16 | 0.52 ± 0.24 | 1.3 (0.7–2.2) | 2.4 (1.6–3.9) |
| CA | 93 | 59 ± 16 | 0.55 ± 0.22 | 1.3 (0.8–2.2) | 2.3 (1.7–3.7) | |
| AA | 15 | 53 ± 17 | 0.32 ± 0.29 * | 0.7 (0.5–1.8) | 2.5 (0.7–4.1) | |
| F | CA | 52 | 57 ± 16 | 0.59 ± 0.23 | 1.5 (0.1–4.9) | 2.5 (0.5–8.5) |
| AA | 9 | 53 ± 17 | 0.28 ± 0.25** | 0.6 (0.4–2.0) † | 2.1 (0.4–5.6) | |
| M | CA | 40 | 62 ± 17 | 0.49 ± 0.20 | 1.1 (0.1–5.2) | 2.1 (0.5–7.2) |
| AA | 6 | 53 ± 20 | 0.39 ± 0.35 | 0.8 (0.0–4.4) | 3.4 (0.0–7.2) | |
Mean (± S.D.) SMX-HA reduction activities, and median (interquartile range) b5 and b5R protein expression in immunoreactive units, relative to β-actin.
CA: Caucasian-Americans; AA: African-Americans
Groups were compared via a two-way ANOVA with post hoc Tukey HSD and Tamhane’s T2 tests to explore any possible interactions of sex and race to observed differences in phenotype.
p = 0.003, relative to all CA individuals. Data from 4 livers were excluded from this analysis, including three livers from females of unknown race and one CA liver from an individual of unrecorded sex.
p = 0.001 relative to CA females
Trend p = 0.078 for lower b5 expression in AA females relative to CA females
Immunoreactive b5 and b5R protein expression exhibited five- and nine-fold ranges among individuals, respectively, and were not normally distributed. Immunoreactive b5 and b5R protein were both undetectable in two livers; these livers displayed very low SMX-HA reduction activities (0.06 and 0.07 nmol/min/mg). The single liver sample with no detectable SMX-HA reduction activity exhibited a 30% decrease in both b5 and b5R protein expression compared to median protein expression in the population.
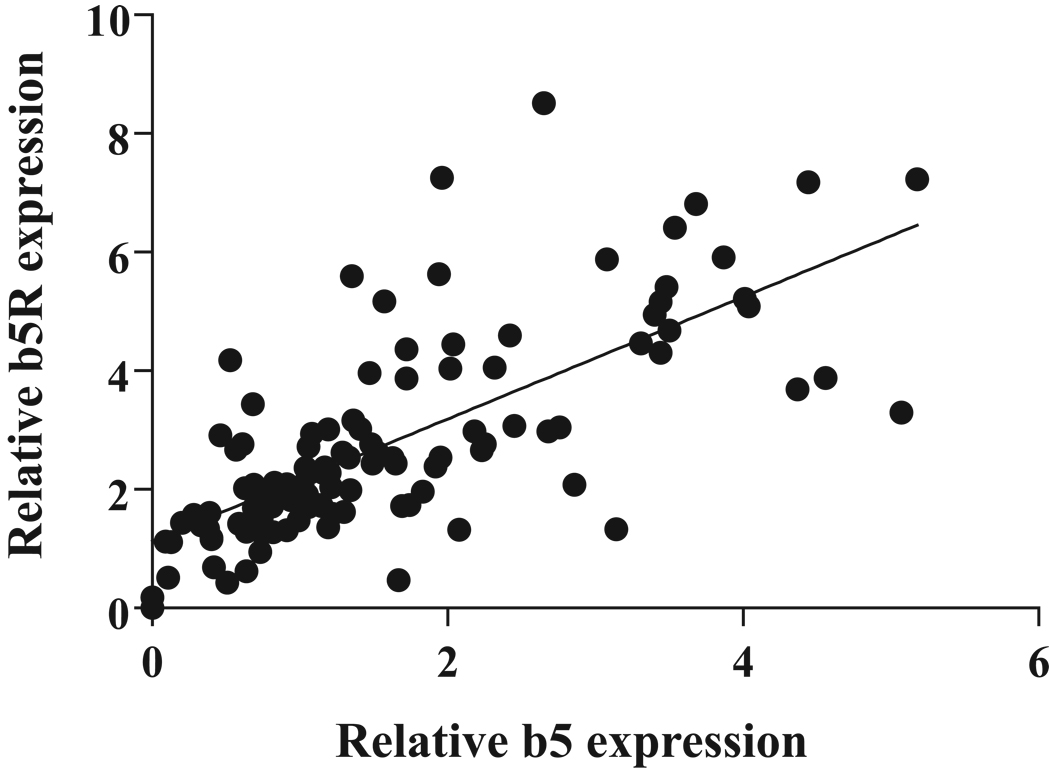
The expression of both b5 and b5R was positively correlated with SMX-HA reduction activities (r = 0.42, p < 0.0001 for b5; r = 0.23, p = 0.01, for b5R). Multiple regression analysis showed that b5 and b5R expression could account for 12.5% of the variability observed in SMX-HA reduction activity in this population, with b5 having more apparent impact than b5R (β coefficient (± S.E.) = 0.07 (± 0.03) for b5, and 0.004 (± 0.02) for b5R). Interestingly, the expression of these two proteins was also significantly correlated with one another (r = 0.74, p < 0.0001) (Fig. 2).
Fig 2.
Relationship between b5 and b5R immunoreactive protein expression, normalized to β-actin, for 111 individual human liver samples. The expression of both proteins was significantly correlated (r = 0.74, p < 0.0001)
Neither b5 nor b5R expression was correlated with age, and there were no significant differences in b5 or b5R expression between AA and CA (p=0.204). The estimated marginal means revealed an interaction between sex and ethnicity, suggesting that CA females had higher b5 expression than CA males, although this apparent interaction did not reach significance (p = 0.063). In AA males and females, expression of b5 and b5R accounted for 95.5 and 82.8%, respectively, of the variability in activities (males: ANOVA F = 15.7, p = 0.03; females: F = 14.4, p = 0.005).
The occurrence of CYB5A and CYB5R3 SNPs in the coding region of the microsomal isoforms was investigated for 111 human livers. Agarose gel electrophoresis of CYB5A and CYB5R3 cDNA did not reveal any amplicon of unexpected size in any subject. All alleles found at each locus were in random association with one other, as expected under Hardy-Weinberg equilibrium conditions. One novel non-synonymous cSNP was found in CYB5A (S5A), while two novel non-synonymous cSNPs were discovered for CYB5R3 (R59H and R297H) (Table 3). All were in the heterozygous state. The CYB5A T60A SNP, which was originally identified in an African American male [17], was not found in this population, although only 15 AA subjects were available.
Table 3.
Sequence variants found in human CYB5A and CYB5R3 cDNA in 111 subjects.
| Gene | Sequence change |
Heterozygosity | Amino acid change |
Ref SNP ID |
Number of subjects |
Minor allele frequency |
Genotype frequency1 |
||
|---|---|---|---|---|---|---|---|---|---|
| AA | CA | AA | CA | ||||||
| CYB5A | c.13T>G | Het | S5A | novel | 2 | 0.067 | 0.000 | 0.124 | 0.000 |
| c.36C>T | Het | None | rs1051236 | 28 | 0.100 | 0.156 | 0.180 | 0.263 | |
| Hom | 2 | N.O. | 0.022 | ||||||
| c.288G>A | Het | None | rs7238987 | 31 | 0.167 | 0.156 | 0.278 | 0.263 | |
| Hom | 2 | N.O. | 0.022 | ||||||
| c.369C>T | Het | None | rs2276275 | 12 | ND | ND | ND | ND | |
| CYB5R3 | c.132G>A | Het | rs5996200 | 22 | 0.067 | 0.097 | 0.124 | 0.192 | |
| c.176G>A | Het | R59H | novel | 1 | 0.000 | 0.005 | 0.000 | 0.011 | |
| c.350C>G | Het | T117S3 | rs1800457 | 7 | 0.200 | 0.005 | 0.320 | 0.011 | |
| c.890G>A | Het | R297H | novel | 1 | 0.000 | 0.005 | 0.000 | 0.011 | |
Het: heterozygous for SNP; Hom: homozygous for SNP
N.O.: not observed; ND: not determined
All polymorphisms were in Hardy-Weinberg equilibrium
Liver was of unknown ethnicity
Equivalent to published T116S [14]
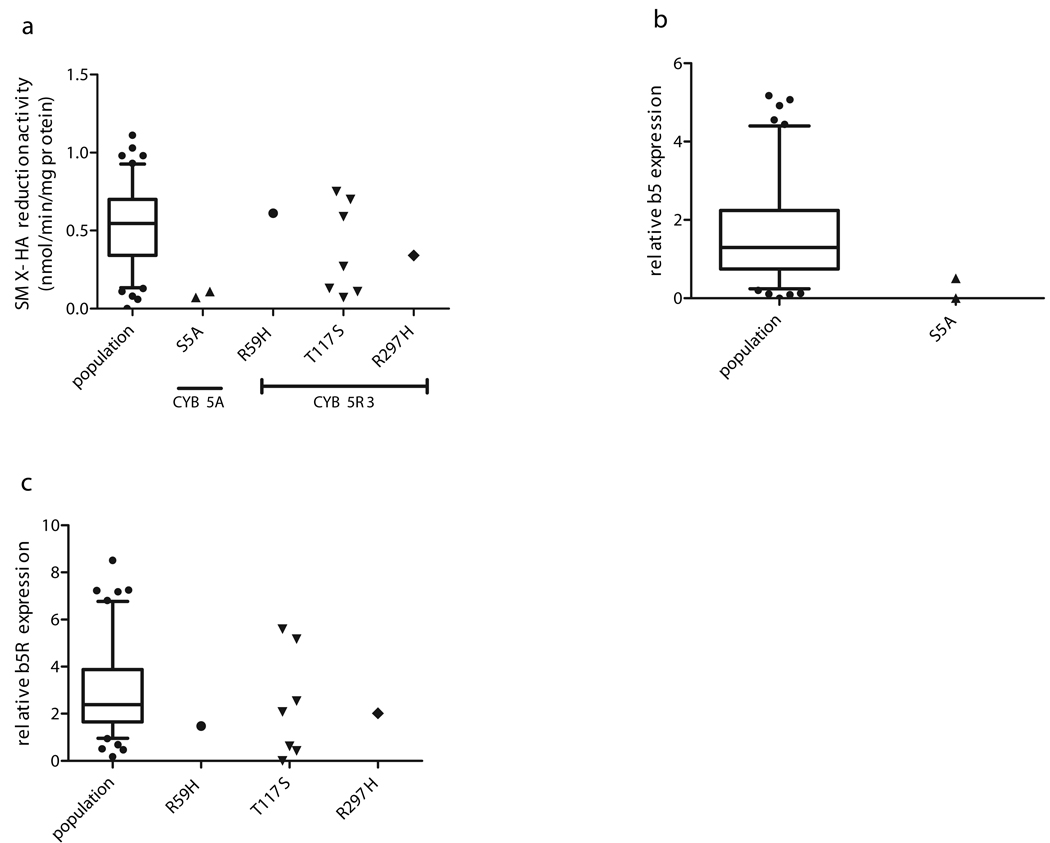
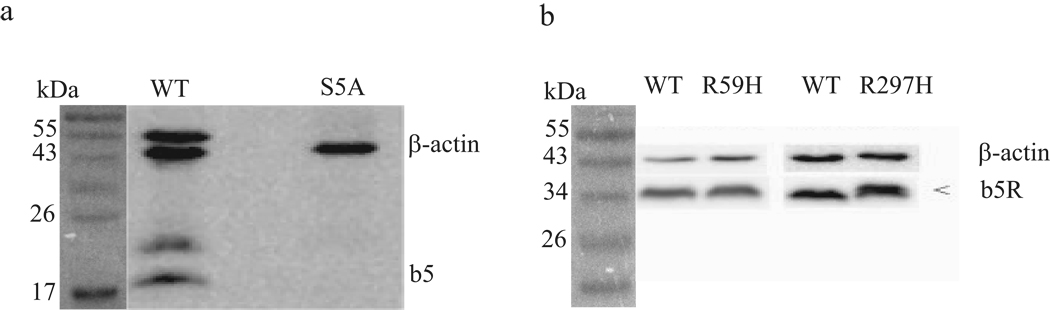
The CYB5A S5A SNP was found in two African-American individuals, one male and one female. Activities for these two livers were 0.07 and 0.11 nmole/min/mg protein, which was in the lower 5th percentile of the population (Fig. 3a) and immunoreactive b5 and b5R protein for the 2 livers was low or virtually undetectable (Fig. 3b). Interestingly, two bands at 22 and 54 kD that were also recognized by α-b5 antibody in all blots were absent or markedly decreased in the S5A patients (Fig. 4a). Despite low activities and expression in both livers carrying this SNP, the novel CYB5A S5A SNP was not predicted to be functionally significant by any of the computational methods used (Table 4), nor was it predicted to be a significant phosphorylation site (NetPhos score = 0.157).
Fig 3.
SMX-HA reduction activity (a), b5 protein (b), and b5R protein expression (c), in livers heterozygous for non-synonymous cSNPs in CYB5A and CYB5R3, compared to WT population data. Experimental data for the population is shown as a whisker-box plot with the box representing median and interquartile range, and the whiskers representing 95th and 5th percentiles. Outlying values for the WT population are shown as individual points outside the whiskers.
Fig 4.
Expression of b5 and b5R immunoreactive protein in three human livers with non-synonymous cSNPs in either CYB5A or CYB5R3. Representative immunoblots shown for assays done in duplicate; 30 µg microsomal protein loaded and probed with rabbit polyclonal anti-human b5 or anti-human b5R antibody. (a) wild-type (WT) CYB5A homozygote, and S5A heterozygote in CYB5A showing absence of b5 expression at 17 kDa. Additional bands (22 and 54 kD) represent as yet unidentified proteins that cross-react with anti-human b5 antibody. (b) wild-type CYB5R3 homozygote, and R59H heterozygote in CYB5R3, showing modest but significantly decreased b5R protein expression (37 kDa), when normalized to β-actin (42 kDa). Separate blot with wild-type wild-type CYB5R3 homozygote, and R297H heterozygote in CYB5R3, showing two bands for the R297H subject.
Table 4.
Computational prediction of impact on protein function for non-synonymous cSNPs in the genes encoding b5 and b5R.
| Gene SNP |
Scores for variant alleles1 | ||
|---|---|---|---|
| SIFT | PolyPhen | PMut | |
| CYB5A S5A |
0.61 | 0.63 | 0.10 (8) |
| CYB5R3 R59H |
0.04 | 0.44 | 0.53 (0) |
| CYB5R3 T117S |
0.18 | 0.24 | 0.03 (9) |
| CYB5R3 R297H |
0.12 | 1.67 | 0.40 (2) |
For SIFT, the tolerance index cutoff value was 0.05. For PolyPhen, a position-specific independent count or PSIC>1.5 was considered to be damaging. For PMut, a Neural Network Score, or NN output > 0.5 indicated a pathological mutation, while the value in parentheses refers to the prediction reliability index (0 (unreliable) to 9 (highly reliable)).
The two novel CYB5R3 SNPs (R59H and R297H) were found in one Caucasian-American subject each. R59H was predicted to be deleterious by SIFT and pathologically significant by PMut, while the R297H was predicted to be probably damaging by PolyPhen (Table 4); however, screening reduction activities for both cSNPs were comparable to the population mean (Fig. 3a). The relative expression of immunoreactive b5R protein for the R59H liver was modestly but significantly low (Fig 3c); interestingly, b5R protein from the R297H liver was resolved into two distinct bands (Fig. 4b).
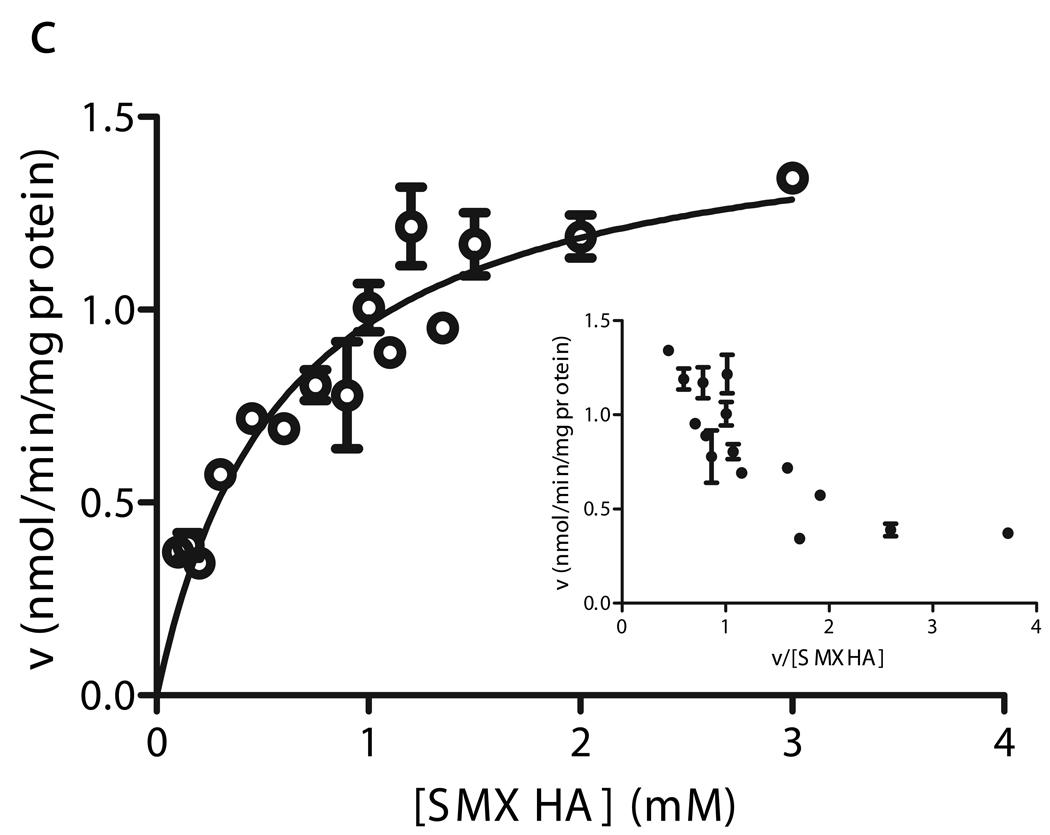
The CYB5R3 T117S genotype, which was previously reported in an African American population [27], was found with a much higher frequency among AA (40%) than CA (1.1%) subjects in our group of livers (p < 0.0001). Two of the five AA individuals with the b5R T117S genotype also had the b5 S5A SNP. Although the T117S SNP was predicted to be a potential phosphorylation site and a target for phosphokinase C (NetPhos and NetphosK scores = 0.58 and 0.78), activities and protein expression for the livers heterozygous for T117S, but without the S5A SNP, were not significantly different from the population mean and median, respectively (Figure 3).
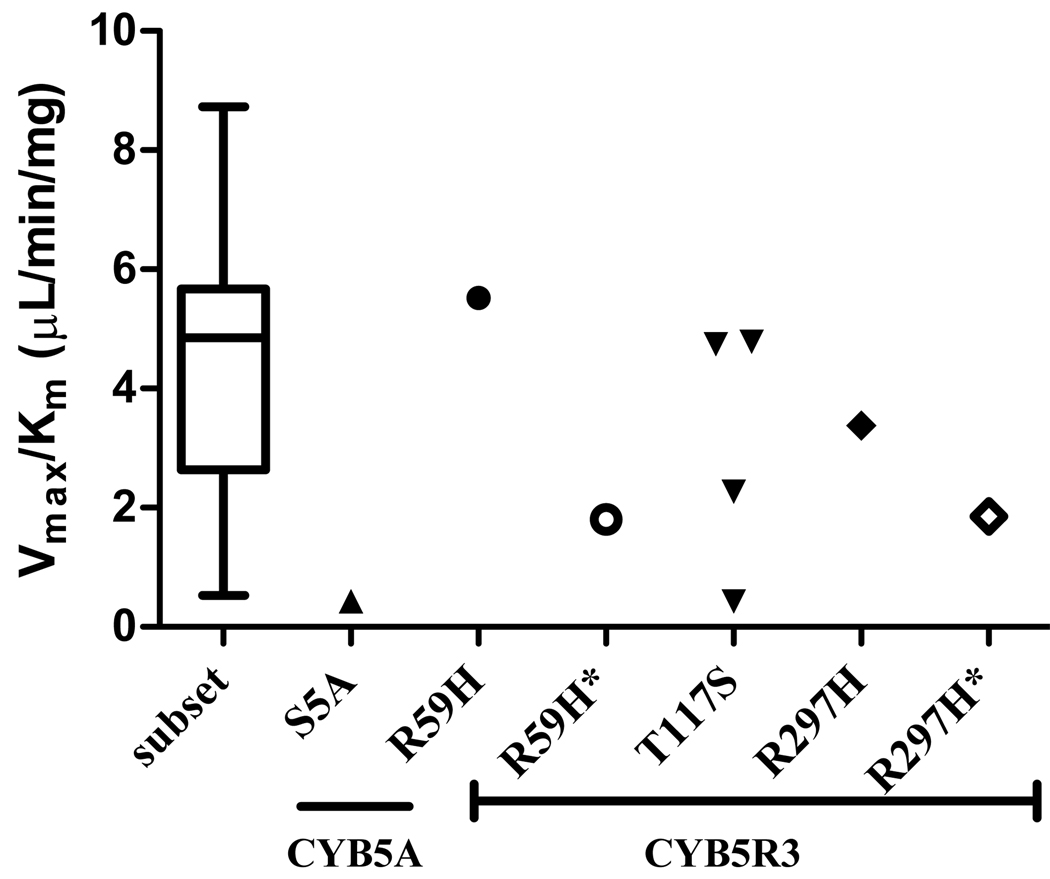
The possible effect of coding SNPs on SMX-HA reduction was further assessed by investigating the kinetics for this reaction in all livers with non-synonymous cSNPs, as well as in a subset of WT livers with outlying high ( >90th percentile for the population; n=7) or low activities (<10th percentile, n=5), as well as a control subset of WT livers with intermediate activities (45th-55th percentile, n=6). The reduction activities in this subset of 18 WT livers were normally distributed, as in the full population, and there were no differences in activities, b5 expression, or b5R expression between this subset and the full study population (p = 0.24, 0.15, 0.34 respectively). Activity data for this control subset of 18 WT livers followed Michaelis-Menten kinetics, with apparent Km values of 277 ± 107 µM (range: 88–472), and Vmax values of 1.21 ± 0.80 nmol/min/mg (range: 0.20–2.77).
One S5A liver had an apparent Km for SMX-HA reduction of 395 µM and an apparent Vmax (0.17 nmol/min/mg), resulting in an enzymatic efficiency (Vmax/Km) that fell in the lower 5th percentile of the calculated efficiencies for the WT subpopulation (Fig. 5). Activities for the second S5A heterozygote, as well as for the liver with undetectable activity on initial screening (which was WT for coding SNPs), were too low to calculate kinetic data.
Fig 5.
Enzyme efficiency (Vmax/Km) for SMX-HA reduction from kinetic determinations in a subset of 18 WT human livers, compared to data obtained from livers with CYB5A or CYB5R3 cSNPs. Reduction activities for human livers in the subset were in three defined ranges (>90th percentile, n=7; 45th–55th percentile, n=6; <10th percentile, n=5). *R59H and *R297H denote separate calculated efficiencies for the high affinity components observed in these heterozygous livers. Individual apparent Km and Vmax values are given in the text.
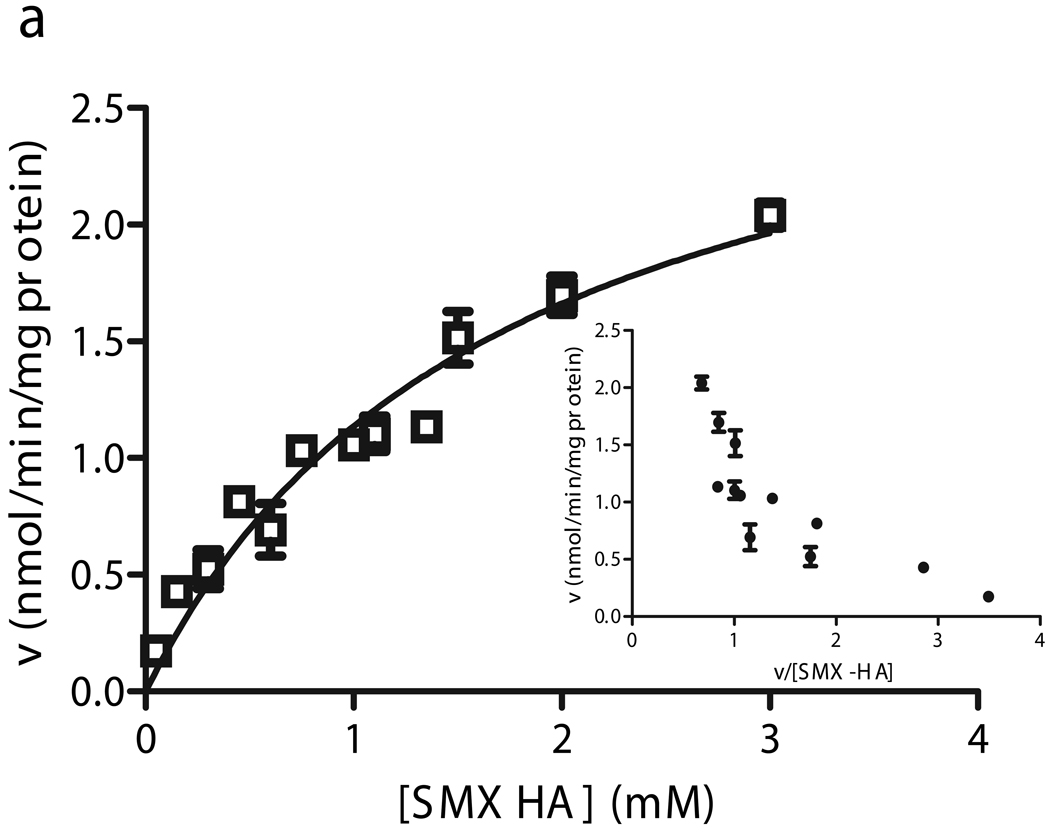
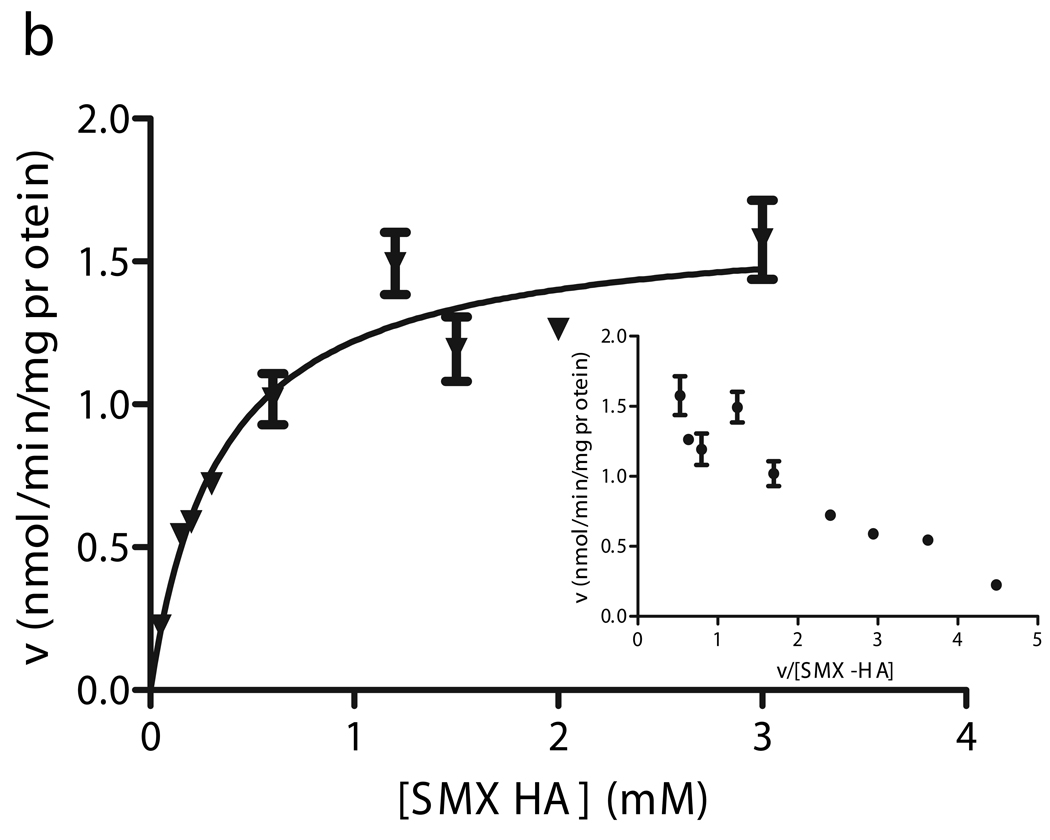
The two novel CYB5R3 cSNPs (R59H and R297H) exhibited biphasic kinetics (Fig. 6a, c). The apparent Km1 and Vmax1 for the R59H liver were 373 µM and 2.06 nmol/min/mg, while the estimated parameters for the R297H liver were 355 µM and 1.20 nmol/min/mg. These values were comparable to WT livers. The kinetic parameters for the low-affinity components (Km2, Vmax2) observed in these heterozygous livers were 1627 µM and 2.94 nmol/min/mg (R59H), and 955 µM and 1.77 nmol/min/mg (R297H). These second apparent Km’s were 4–6 times higher than the mean apparent Km of the subpopulation of 18 WT livers. This resulted in approximately four- and two-fold reductions in enzyme efficiency for the low-affinity components for R59H and R297H, respectively, compared to high affinity (presumably WT) components (Fig. 5).
Fig 6.
Fitted velocity versus substrate concentration curves and Eadie-Hofstee plots for SMX-HA reduction in human liver microsomes heterozygous for the R59H (a), T117S (b), and R297H (c) CYB5R3 cSNPs.. Kinetic experiments were performed for individual livers with (□) R59H, (▼) T117S, and (○) R297H CYB5R3 allozymes. Data for the T117S livers was fit to the Michaelis-Menten equation (data from one representative liver shown), while the kinetic constants for the high and low-Km components of the R59H and R297H livers were derived from Hanes plots.
For 4 livers with only the CYB5R3 T117S SNP, the mean apparent Km (295 ± 97 µM) and apparent Vmax (0.97 ± 0.64 nmol/min/mg) values were not significantly different from WT livers (p = 0.77, 0.57 respectively). Michaelis-Menten kinetics were observed for livers with this nonsynonymous CYB5R3 SNP (Fig. 6b).
Designation of haplotypes was based on the nucleotide sequence of the specific allozyme, with the wild-type sequence designated as *1A [28]. Haplotype names were designated with letters after the number, with decreasing allele frequency corresponding to increasing alphabetical order. For the coding region of CYB5A there were 5 haplotypes each for the AA and CA groups; however, only a small number of AA subjects were evaluated (Table 5). For CYB5R3, the coding region included 6 distinct haplotypes, one of which was only observed in AA. Synonymous CYB5A SNPs c.36C>T and c.288G>A were in linkage disequilibrium in the CA and AA groups (D' = 0.8, r2 = 0.70; D' = 1.0, r2 = 0.56, respectively). None of the other CYB5A or CYB5R3 SNPs appeared to be linked. No significant differences were found among the various CYB5A *1 allele haplotypes with respect to activity and b5 protein expression.
Table 5.
Haplotypes observed in coding regions of the genes for b5 (CYB5A) and b5R (CYB5R3) in 111 human liver samples.
| Allele | AA frequency | CA frequency | CYB5A cDNA nucleotide position | |||
|---|---|---|---|---|---|---|
| 13 | 36 | 288 | 369 | |||
| c.*1A | 0.60 | 0.67 | T | C | G | C |
| c.*1B | 0.20 | 0.25 | T | T | A | C |
| c.*1C | 0.07 | 0.04 | T | C | A | C |
| c.*1D | 0.04 | T | T | G | C | |
| c.*1E | 0.01 | T | C | G | T | |
| c.*2A | 0.07 | G | C | G | C | |
| c.*2B | 0.07 | G | C | A | C | |
| Allele | AA frequency | CA frequency | CYB5R3 cDNA nucleotide position | |||
| 132 | 176 | 350 | 890 | |||
| c.*1A | 0.53 | 0.77 | G | G | G | G |
| c.*1B | 0.07 | 0.20 | A | G | G | G |
| c.*2A | 0.33 | 0.01 | G | G | C | G |
| c.*2B | 0.07 | A | G | C | G | |
| c.*3A | 0.01 | G | A | G | G | |
| c.*4A | 0.01 | G | G | G | A | |
The “wild-type” for each position is indicated in bold type with shaded background. White letters on a dark background indicate the presence of a nonsynonymous SNP. Bold letters on a white background indicate the presence of a synonymous SNP. A blank cell indicates that the haplotype was not observed for that racial group. For allele numbering criteria, please refer to the text.
Discussion
The microsomal b5/b5R reduction pathway is important for the detoxification of the hydroxylamine metabolites of arylamine drugs such as SMX and dapsone [9], as well as the pro-carcinogenic hydroxylamine metabolites of 4-aminobiphenyl and PhIP [10]. Genetic sequence variation in either CYB5A or CYB5R3 could therefore influence the risk of toxicity from these bioactivated compounds. Although at least 39 exonic and 8 intronic mutations in CYB5R3, together with an intronic mutation in CYB5A [29], have been associated with disease (hereditary methemoglobinemia) to date (reviewed in [30]), little is known about the functional significance of less catastrophic or heterozygous SNPs in these genes, which may be more frequent in the population and devoid of any overt clinical symptoms. The present study adopted a genotype-phenotype approach to test the hypothesis that coding sequence variability in CYB5A and CYB5R3 contributes to variability in b5 and b5R protein expression and SMX-HA reduction activity in human liver. In the 111 human livers studied, SMX-HA reduction in human liver microsomes varied about 19-fold, with at least one liver having undetectable activity.
Enzyme activities correlated significantly with the expression of b5 and b5R protein, suggesting that variability in the expression of either enzyme could affect hydroxylamine reduction capacity. In an earlier study on the role of b5 and b5R in hydroxylamine reduction by our laboratory, b5R immunoreactive protein did not correlate with hepatic arylhydroxylamine reduction, a negative finding that may have been due to the narrow range of b5R content in the small number of human livers (n=11) studied [10]. The present study involved a greater number of subjects with a wider range in b5R expression.
Lower SMX-HA reduction activities were observed for liver samples from AA subjects; in fact, 27% of the liver activities from this group were in the lower 5% of the activity distribution, compared to 1% of CA. Erythrocyte ferrihemoglobin reductase activity (a measure of b5 and b5R activity) was previously shown to be increased in a group of clinically ill AA patients in a veterans hospital, relative to CA patients [31], a finding that is discordant with our results. This suggests that the effects of illness on b5 or b5R expression and function require further investigation.
Although the number of AA subjects in this population was quite small (n=15), the decreased SMX-HA reduction observed in this group, in addition to a trend towards decreased b5 protein expression in AA females, warrants further investigation. While racial differences in SMX hypersensitivity have not been documented, the b5/b5R pathway also detoxifies procarcinogenic N-hydroxylamine metabolites of aromatic and heterocyclic amine carcinogens [10]. It is possible that inefficient b5/b5R–mediated xenobiotic reduction may be a contributory factor to the higher rates of several cancer types seen in AA patients [32,33]; this hypothesis is presently under investigation in our laboratory.
The coding region of the microsomal b5 isoform is made up of five exons (exons 1–4, and 6) comprising a total of 402 bases. Including the results from this study, there are now nine known coding SNPs in CYB5A, four of which are non-synonymous: S5A in exon 1 (this study), R52K (rs1803366) and T60A [16] in exon 2, and the formation of a premature stop codon instead of Y130 in exon 6 (rs19711860). Although the S5A SNP was not predicted in silico to impact reduction activity, both individuals who were heterozygous for this polymorphism displayed markedly low SMX-HA reduction activities. Due to very low activities, it was difficult to obtain accurate kinetic data in more than one S5A liver, although the results in hand indicate a likely decrease in enzymatic efficiency associated with this SNP. Since biphasic kinetics were not observed in the heterozygous state, one can hypothesize that the S5A allozyme is essentially inactive or is rapidly degraded; the latter is supported by the low b5 protein expression observed.
The serine 5 residue is at the N-terminus of the b5 protein, and sequence alignment by ClustalW shows that it is conserved across mammals and birds. The crystal structure of human oxidized microsomal cytochrome b5 (PDB ID: 2i96) indicates that the Ser is located in the center of a ‘W’-shaped loop with its hydroxyl group within 2.5 Å of the amine group of Lys10, which is the final residue forming this loop. The substitution of Ser by Ala would prevent hydrogen-bond formation and potentially disrupt tertiary structure. However, this possibility was not considered significant by the PMut and PolyPhen prediction algorithms.
Interestingly, the levels of b5R expression were also significantly low or undetectable for both individuals with the S5A SNP in the b5 gene. In view of the strong correlation between b5 and b5R protein expression observed in this study, the low activity observed for both S5A heterozygotes could be due, in all or in part, to a deficiency of transcription factor(s) necessary for both genes, rather than the altered amino acid sequence S5A. The functional significance of S5A for b5 function is currently under investigation using site-directed mutagenesis.
An intriguing finding was that two additional proteins that were consistently recognized by α-b5 antibody, at 22 and 54 kD, were absent or markedly decreased in the S5A subjects (Fig. 4). We suspect that these bands represent endoplasmic reticular proteins with cytochrome b5 domains, such as mPR (membrane-associated progesterone component, 22 kD) and possibly NCB5OR (the b5/b5R fusion protein; 59.7 kD); mass spectrometry is underway to confirm this. However, it is as yet unclear why these two other proteins would also be undetectable in S5A subjects, unless these proteins are subject to similar transcriptional regulation, as for b5 and b5R, in these individuals.
The coding sequence of CYB5R3 is comprised of 9 exons and spans a total of 903 bases. Excluding those variants associated with hereditary methemoglobinemia, there are now 14 coding CYB5R3 SNPs known, which include 10 non-synonymous sequence variants: R59H (this study) and S66P (rs1130706) in exon 3; T117S (rs1800457) and Q135R (rs11541434) in exon 5; R160G (rs61732609), D162Y (ENSSNP11933047), R170S (rs61743717) in exon 6; E213Q or E213X (rs61745147) in exon 8; and R297H (this study) and V300L or V300F (rs61743746) in exon 9.
The nonsynonymous c.350G>C polymorphism in CYB5R3 (T117S) was found exclusively in the heterozygous state, at an allele frequency of 0.20 in AA in our study. This is similar to the allele frequency of 0.23 that had been previously found in 112 AA subjects [34]. The present study also documented the occurrence of CYB5R3 T117S in Caucasian-Americans, albeit at a very low incidence (one individual). This may account for the low allele frequency (0.08) for T117S reported by the National Institute of Environmental Health Sciences (NIEHS) SNPs program, which is derived from the genotyping of 90 individuals of unrecorded ethnicity but ‘representative of the US population’ (available at http://egp.gs.washington.edu/data/dia1/). The T117S sequence variation was not associated with significant changes in reduction kinetics or b5R protein expression. For the synonymous CYB5R3 c.132G>A SNP, the reported allele frequency from dbSNP and the NIEHS SNPs program is 0.075, which is in agreement with the values obtained in this study for both AA and CA subjects (0.067, 0.097; Table 3).
The wild type amino acids at the sites of the non-synonymous CYB5R3 cSNPs found in this study are well conserved across species, suggesting that these may be functionally important regions. R59H lies at the start of the second β-sheet in the FAD-binding domain of the reductase, and this SNP was predicted to have a pathological and deleterious effect by PMut and SIFT. A homozygous mutation in the preceding arginine residue (R58Q, FTID: VAR004619) is associated with Type I congenital methemoglobinemia [35,36], possibly due to instability and increased proteolytic susceptibility of the mutant protein. Recombinant enzymatic activity for this adjacent R58Q SNP was 62% of wild-type [37]; in our study, the low affinity microsomal component for heterozygous R59H showed a 67% reduction in efficiency relative to the higher affinity (presumably WT) component. However, the overall reduction activity for R59H was not significantly lower than the mean population activity, indicating that a biologically significant deleterious effect might only be observed with the SNP in a homozygous state.
The R297H SNP is found in the NADH-binding domain of b5R, close to the C-terminus. This region appears to be important for proper enzyme function since adjacent heterozygous G292D and homozygous F299del mutations result in hereditary methemoglobinemia [38,39], and both recombinant G292D and F299 del mutant b5R proteins exhibit decreased temperature stability [39,40]. In addition, the G292D mutation (FTID: VAR037316) in this region is associated with decreased NADH catalytic efficiency, due to misorientation of NADH as a result of disruption of hydrogen bonding between the C-terminus of b5R and the carboxamide group of NADH [40]. A similar defect may have been responsible for the high second Km for SMX-HA observed in the heterozygous R297H liver.
Our kinetic findings of distinct kinetic components for both R59H and R297H heterozygote livers is interesting in light of the immunoblot results for the R297H variant, which showed two b5R protein bands. This was unexpected, since any changes in the enzyme’s physical properties due to the single amino acid change would not typically result in visible electrophoretic separation. It is possible that a post-translational modification may have taken place that altered the migration of the mutant b5R allozyme. Additional studies are underway to generate purified expressed forms of both R59H and R297H variants, which may help to elucidate the biochemical reason for differential migration of these two apparent R297H allozymes, as well as confirm whether the low enzymatic efficiencies observed for these livers were due to the mutant b5R allozymes.
A number of livers with low activity and/or low b5 or b5R protein expression did not have any SNPs in the coding region (outlying points in Fig. 3a–c). The possibility of intronic SNPs leading to exon skipping, truncated protein, and low activity could be excluded since all amplicons of both b5 and b5R cDNAs were of the expected size. It is possible that these samples have sequence variations (rSNPs) in the promoter and/or 3´ untranslated region of the genes that affect transcriptional regulation. Experimental evidence exists for Sp binding sites in the CYB5R3 exon 1M [41] and CYB5A promoters, as well as for NF-1 and GATA-6 binding sites for CYB5A [42]. The high correlation of expression of b5 with b5R protein found in this study suggests shared transcriptional regulation. To this end, work is underway to characterize any common transcriptional regulatory regions for both CYB5A and CYB5R3.
In conclusion, this study demonstrated that SMX-HA reduction in human liver displays nearly 20-fold variation in activity, with lower reduction activities in African Americans compared to Caucasian subjects. Expression of hepatic b5 and b5R protein was also variable and in addition was positively correlated with SMX-HA reduction activities, and interestingly, expression of b5 and b5R were highly correlated with one another. A novel nonsynonymous SNP found in CYB5A, S5A, was found in two African American individuals and exhibited low reduction activity, and low expression of both b5 and b5R. The low substrate affinities and overall reduction efficiency observed in livers heterozygous for the CYB5R3 R59H and R297H SNPs may be due to the mutant b5R allozymes. Future work will focus on functional characterization of purified expressed forms of these newly discovered b5 and b5R sequence variants, along with analysis of allele frequencies in a larger number of African American subjects. Finally, since the novel cSNPs found in this study population were present at low allele frequencies, these cSNPs appear to contribute only a small amount to overall observed phenotypic variability. Work is underway to characterize additional polymorphisms in the promoter and 3” untranslated regions of both genes to further account for individual variability in hydroxylamine reduction.
Acknowledgements
Special thanks to Dr. Sharon Weber and Robert Rettammel, Division of General Surgery, University of Wisconsin-Madison, for providing human liver samples for this and a related study, Jennifer Simpson for microsome preparation, and to Nicholas Keuler, CALS computer lab, University of Wisconsin-Madison, for statistical assistance.
This work was supported by a grant from the National Institute of General Medical Sciences, National Institutes of Health, GM61753.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cribb AE, Lee BL, Trepanier LA, Spielberg SP. Adverse reactions to sulphonamide and sulphonamide-trimethoprim antimicrobials: clinical syndromes and pathogenesis. Adverse Drug Reactions and Toxicological Reviews. 1996;15:9–50. [PubMed] [Google Scholar]
- 2.Gordin FM, Simon GL, Wofsy CB, Mills J. Adverse reactions to trimethoprim-sulfamethoxazole in patients with AIDS. Annals of Internal Medicine. 1984;100:495–499. doi: 10.7326/0003-4819-100-4-495. [DOI] [PubMed] [Google Scholar]
- 3.Verhagen C, Stalpers L, dePauw B, Haanen C. Drug-induced skin reactions in patients with acute non-lymphocytic leukemia. Eur J Hematol. 1987;38:225–230. doi: 10.1111/j.1600-0609.1987.tb01168.x. [DOI] [PubMed] [Google Scholar]
- 4.Cribb AE, Spielberg SP, Griffin GP. N4-hydroxylation of sulfamethoxazole by cytochrome P450 of the cytochrome P4502C subfamily and reduction of sulfamethoxazole hydroxylamine in human and rat hepatic microsomes. Drug Metabolism and Disposition. 1995;23:406–414. [PubMed] [Google Scholar]
- 5.Cribb A, Miller M, Tesoro A, Spielberg S. Peroxidase-dependent oxidation of sulfonamides by monocytes and neutrophils from humans and dogs. Molecular Pharmacology. 1990;38:744–751. [PubMed] [Google Scholar]
- 6.Vyas PMRS, Woster PM, Svensson CK. Reactive oxygen species generation and its role in the differential cytotoxicity of the arylhydroxylamine metabolites of sulfamethoxazole and dapsone in normal human epidermal keratinocytes. Biochem Pharmacol. 2007;70:275–286. doi: 10.1016/j.bcp.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Cheng L, Stewart BJ, You Q, Petersen DR, Ware JA, Piccotti JR, et al. Covalent binding of the nitroso metabolite of sulfamethoxazole is important in induction of drug-specific T-cell responses in vivo. Mol Pharmacol. 2008;73:1769–1775. doi: 10.1124/mol.107.043273. [DOI] [PubMed] [Google Scholar]
- 8.Sanderson JP, Naisbitt DJ, Farrell J, Ashby CA, Tucker MJ, Rieder MJ, et al. Sulfamethoxazole and its metabolite nitroso sulfamethoxazole stimulate dendritic cell costimulatory signaling. J Immunol. 2007;178:5533–5542. doi: 10.4049/jimmunol.178.9.5533. [DOI] [PubMed] [Google Scholar]
- 9.Kurian J, Bajad S, Miller J, Chin N, Trepanier L. NADH cytochrome b5 reductase and cytochrome b5 catalyze the microsomal reduction of xenobiotic hydroxylamines and amidoximes in humans. J Pharmacol Exp Ther. 2004;311:1171–1178. doi: 10.1124/jpet.104.072389. [DOI] [PubMed] [Google Scholar]
- 10.Kurian JR, Chin NA, Longlais BJ, Hayes KL, Trepanier LA. Reductive Detoxification of Arylhydroxylamine Carcinogens by Human NADH Cytochrome b(5) Reductase and Cytochrome b(5) Chem Res Toxicol. 2006;19:1366–1373. doi: 10.1021/tx060106t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du M, Shirabe K, Takeshita M. Identification of alternative first exons of NADH-cytochrome b5 reductase gene expressed ubiquitously in human cells. Biochem Biophys Res Commun. 1997;235:779–783. doi: 10.1006/bbrc.1997.6873. [DOI] [PubMed] [Google Scholar]
- 12.Giordano S, Steggles A. The human liver and reticulocyte cytochrome b5 mRNA's are products from a single gene. Biochim Biophys Res Commun. 1991;178:38–44. doi: 10.1016/0006-291x(91)91776-9. [DOI] [PubMed] [Google Scholar]
- 13.Pietrini G, Aggujaro A, Carrera P, Malyszko J, Vitale A, Borgese N. A single mRNA, transcribed from an alternative, erythroid-specific, promoter, codes for two non-myristylated forms of NADH-cytochrome b5 reductase. Journal of Cell Biology. 1992;117:975–986. doi: 10.1083/jcb.117.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beretta m, Santachiara-Benerecetti A. Study of human red cell NADH diaphorase (Dia1) in the Italian population. Human Heredity. 1985;35:7–10. doi: 10.1159/000153506. [DOI] [PubMed] [Google Scholar]
- 15.Reghis A, Benabadji M, Tchen P, Kaplan J. Quantitative variations of red-cell cytochrome b5 reductase (NADA-methemoglobin reductase) in the Algerian population. Human Genetics. 1981;59:148–153. doi: 10.1007/BF00293065. [DOI] [PubMed] [Google Scholar]
- 16.Kurian JR, Longlais BJ, Trepanier LA. Discovery and characterization of a cytochrome b5 variant with impaired hydroxylamine reduction capacity. Pharmacogenetics and Genomics. 2006;19:1366–1373. doi: 10.1097/FPC.0b013e328011aaff. [DOI] [PubMed] [Google Scholar]
- 17.Kruglyak L, Nickerson D. Variation is the spice of life. Nature Genetics. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 18.Lavergne SN, Kurian JR, Bajad SU, Maki JE, Yoder AR, Guzinski MV, et al. Roles of endogenous ascorbate and glutathione in the cellular reduction and cytotoxicity of sulfamethoxazole-nitroso. Toxicology. 2006;222:25–36. doi: 10.1016/j.tox.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Spears G, Sneyd J, Loten E. A method for deriving kinetic constants for two enzymes acting on the same substrate. Biochem J. 1971;125:1149–1151. doi: 10.1042/bj1251149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Ferrer-Costa C, Orozco M, de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004;57:811–819. doi: 10.1002/prot.20252. [DOI] [PubMed] [Google Scholar]
- 23.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 26.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins MM, Prchal JT. A novel mutation found in the 3' domain of NADH-cytochrome B5 reductase in an African-American family with type I congenital methemoglobinemia. Blood. 1996;87:2993–2999. [PubMed] [Google Scholar]
- 28.Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, et al. Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. J Biol Chem. 2006;281:7364–7373. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- 29.Giordano S, Kaftory A, Steggles A. A splicing mutation in the cytochrome b5 gene from a patient with congenital methemoglobinemia and pseudohermaphrodism. Human Genetics. 1994;93:568–570. doi: 10.1007/BF00202825. [DOI] [PubMed] [Google Scholar]
- 30.Percy MJ, Lappin TR. Recessive congenital methaemoglobinaemia: cytochrome b(5) reductase deficiency. Br J Haematol. 2008;141:298–308. doi: 10.1111/j.1365-2141.2008.07017.x. [DOI] [PubMed] [Google Scholar]
- 31.Mansouri A, Nandy I. NADH-methemoglobin reductase (cytochrome b5 reductase) levels in two groups of American blacks and whites. Journal of Investigative Medicine. 1998;46:82–86. [PubMed] [Google Scholar]
- 32.Brinton LA, Sherman ME, Carreon JD, Anderson WF. Recent trends in breast cancer among younger women in the United States. J Natl Cancer Inst. 2008;100:1643–1648. doi: 10.1093/jnci/djn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams H, Powell IJ. Epidemiology, pathology, and genetics of prostate cancer among African Americans compared with other ethnicities. Methods Mol Biol. 2009;472:439–453. doi: 10.1007/978-1-60327-492-0_21. [DOI] [PubMed] [Google Scholar]
- 34.Jenkins M, Prchal J. A high-frequency polymorphism of NADH-cytochrome b5 reductase in African-Americans. Hum Genet. 1997;99:248–250. doi: 10.1007/s004390050347. [DOI] [PubMed] [Google Scholar]
- 35.Huang C, Xie Y, Wang Y, Wu Y, Ye Y, Zhu Z. [Arginine-glutamine replacement at residue 57 of NADH-cytochrome b5 reductase in Chinese hereditary methemoglobinemia] Zhonghua Xue Ye Xue Za Zhi. 1997;18:200–203. [PubMed] [Google Scholar]
- 36.Katsube T, Sakamoto N, Kobayashi Y, Seki R, Hirano M, Tanishima K, et al. Exonic point mutations in NADH-cytochrome B5 reductase genes of homozygotes for hereditary methemoglobinemia, types I and III: putative mechanisms of tissue-dependent enzyme deficiency. Am J Hum Genet. 1991;48:799–808. [PMC free article] [PubMed] [Google Scholar]
- 37.Shirabe K, Yubisui T, Borgese N, Tang CY, Hultquist DE, Takeshita M. Enzymatic instability of NADH-cytochrome b5 reductase as a cause of hereditary methemoglobinemia type I (red cell type) J Biol Chem. 1992;267:20416–20421. [PubMed] [Google Scholar]
- 38.Percy MJ, Gillespie MJ, Savage G, Hughes AE, McMullin MF, Lappin TR. Familial idiopathic methemoglobinemia revisited: original cases reveal 2 novel mutations in NADH-cytochrome b5 reductase. Blood. 2002;100:3447–3449. doi: 10.1182/blood-2002-05-1405. [DOI] [PubMed] [Google Scholar]
- 39.Shirabe K, Fujimoto Y, Yubisui T, Takeshita M. An in-frame deletion of codon 298 of the NADH-cytochrome b5 reductase gene results in hereditary methemoglobinemia type II (generalized type). A functional implication for the role of the COOH-terminal region of the enzyme. J Biol Chem. 1994;269:5952–5957. [PubMed] [Google Scholar]
- 40.Davis CA, Barber MJ. Cytochrome b5 oxidoreductase: expression and characterization of the original familial ideopathic methemoglobinemia mutations E255- and G291D. Arch Biochem Biophys. 2004;425:123–132. doi: 10.1016/j.abb.2003.12.041. [DOI] [PubMed] [Google Scholar]
- 41.Toyoda A, Fukumaki A, Hattori M, Sakaki Y. Mode of activation of the GC box/Sp1-dependent promoter of the human NADH-cytochrome b5 reductase-encoding gene. Gene. 1995;164:351–355. doi: 10.1016/0378-1119(95)00443-a. [DOI] [PubMed] [Google Scholar]
- 42.Huang N, Dardis A, Miller WL. Regulation of cytochrome b5 gene transcription by Sp3, GATA-6, and steroidogenic factor 1 in human adrenal NCI-H295A cells. Mol Endocrinol. 2005;19:2020–2034. doi: 10.1210/me.2004-0411. [DOI] [PubMed] [Google Scholar]