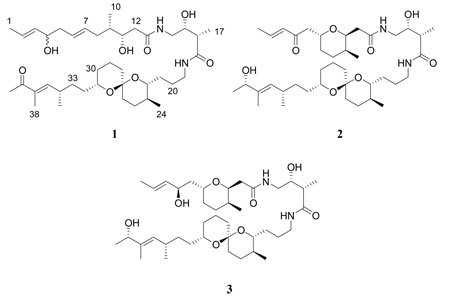
Abstract
Bioassay-guided fractionation of a marine extract from Trididemnum cyclops afforded the new lipopeptide 39-oxobistramide K (1) and the known bistramides A (2) and D (3). Structure elucidation of 1 was carried out by analysis of one and two-dimensional NMR spectroscopy and HRMS data. Bistramides have been reported to exhibit antiproliferative activity in the nanomolar range against a number of tumor cell lines in vitro and in vivo. The isolate 1 was tested for antiproliferative activity against the A2780 cell line, and exhibited an IC50 value of 0.34 µM.
In our continuing search for biologically active natural products from Madagascar as part of an International Cooperative Biodiversity Group (ICBG) program,1 an extract was obtained from the tunicate Trididemnum cyclops Michaelsen 1921 (Didemnidae). The marine invertebrate Trididemnum has been the source of a number of potently bioactive compounds, of which the most frequently reported in the literature and subsequently found to be the most bioactive are a class of cyclic depsipeptides known as didemnins. Rinehart et al. reported the first occurrence of these amino acid macrocycles from Trididemnum solidum.2 One of the isolates was didemnin B, the first marine-derived compound to enter Phase I and Phase II clinical trials by the National Cancer Institute as a potential antitumor agent.
In the current study, an extract of T. cyclops showed significant antiproliferative activity against the A2780 ovarian cancer cell line, and it was thus selected for study. Fractionation yielded one new and two known bistramides. Bistramides are a class of polyether amides with methylated tetrahydropyran and spiroketal subunits. The first of its kind, bistramide A (2), was discovered in 1988 by Gouiffes et al. from the ascidian Lissoclinum bistratum, also of the family Didemnidae.3 The bistramides have been shown to display diverse bioactivities, including immunomodulating and weak antimalarial activity,4,5 but their most notable trait is their antitumor properties. Statsuk et al. reported that actin, a protein essential for cellular motility and division, is the primary cellular receptor of bistramide A.6 In a series of cell-based and in vitro studies in conjunction with affinity-based protein isolation, it was determined that bistramide A (2) aggravates the actin cytoskeleton by depolymerizing filamentous F-actin and binding to monomeric G-actin. An actin-bistramide A complex was later crystallized, allowing for detailed analysis of the binding interactions of this marine metabolite with its monomeric G-actin counterpart.7 Recently, Rizvi et al. described the mechanism of action of bistramide A, namely, the “severing of actin filaments and covalent sequestration of monomeric actin in the cell.”8 They postulated the C-1-C-4-enone subunit to be crucial for covalent protein modification, an event that helps trigger actin depolymerization.
Since the identification of bistramide A there has been only one report of the isolation of additional bistramides as natural products (analogs B-D and K, also from L. bistratum),9 in comparison with nearly 40 journal articles focusing on their synthesis and bioactivity. To date, the only natural source of bistramides has been from L. bistratum, and the first report of these potential antitumor compounds from another source is described herein.
Bioassay-guided fractionation of an extract from a Trididemnum cyclops afforded 300 µg of the new compound 1, together with the known bistramides A (2) and D (3). Compound 1 was obtained as an amorphous solid (0.3 mg). HRFABMS (positive-ion mode) analysis indicated a molecular formula of C40H68N2O8. Its 1H NMR spectrum exhibited large solvent peaks and was noisy because of the limited sample size, and it was not possible to obtain a 13C NMR spectrum, but the 13C NMR shifts could be inferred from the HMBC and g-HSQC spectra of 1. Analysis of data from these spectra indicated the presence of two amide carbonyls (δC 173.2, C-13; 177.3, C-18), one ketone carbonyl (δC 202.5, C-39), and six sp2 olefinic carbons (δC 126.8, C-2; 134.7, C-3; 128.7, C-6; 132.1, C-7; 151.4, C-36; 137.1, C-37). There was also evidence of six sp3 oxygenated carbons (δC 73.4, C-4; 72.3, C-11; 73.1, C-15; 75.6, C-22; 96.7, C-27; 70.0, C-31), and two sp3 nitrogenated carbons (δC 44.1, C-14; 40.2, C-19) (Table 1). The 1H NMR resonances of 1 in CD3OD showed evidence for five unsaturated methines (δH 5.61, dq, J = 15.1, 6.4 Hz, H-2; 5.45, m, H-3; 5.45, m, H-6; 5.45, m, H-7; 6.53, d, J = 10.3 Hz, H-36) and seven methyl groups (δH 1.67, br d, J = 6.4 Hz, H3-1; 0.90, d, J = 6.8 Hz, H3-10; 1.13, d, J = 7.0 Hz, H3-17; 0.83, d, J = 6.5 Hz, H3-24; 1.06, d, J = 6.6 Hz, H3-35; 1.75, s, H3-38; 2.31, s, H3-40) (Table 1). There were eight degrees of unsaturation for the proposed molecular formula of 1, which corresponded to three double bonds, three carbonyl groups, and two rings.
Table 1.
NMR Data of Compound 1a
| Position | 1Hb | 13Cc |
|---|---|---|
| 1 | 1.67 br d (6.4) | 17.6 |
| 2 | 5.61 dq (15.1, 6.4) | 126.8 |
| 3 | 5.45 m | 134.7 |
| 4 | 3.98 dd (13.0, 6.0) | 73.4 |
| 5 | 2.18 m; 2.32 m | 41.7 |
| 6 | 5.45 m | 128.7 |
| 7 | 5.45 m | 132.1 |
| 8 | 1.85 m; 2.20 m | 37.4 |
| 9 | 1.55 m | 39.8 |
| 10 | 0.90 d (6.8) | 14.2 |
| 11 | 3.92 dd (10.9, 4.2) | 72.3 |
| 12 | 2.18 m | 37.4 |
| 13 | - | 173.2 |
| 14 | 3.22 m; 3.38 dd (13.5, 3.4) | 44.1 |
| 15 | 3.71 m | 73.1 |
| 16 | 2.38 m | 45.2 |
| 17 | 1.13 d (7.0) | 14.8 |
| 18 | - | 177.3 |
| 19 | 3.20 m | 40.2 |
| 20 | 1.50 m; 1.85 m | 26.6 |
| 21 | 1.32 m; 1.72 m | 31.3 |
| 22 | 3.18 m | 75.6 |
| 23 | 1.25 m | 35.9 |
| 24 | 0.83 d (6.5) | 18.1 |
| 25 | 1.48 m; 1.55 m | 28.9 |
| 26 | 1.45 m; 1.58 m | 36.8 |
| 27 | - | 96.7 |
| 28 | 1.32 m; 1.52 m | 36.2 |
| 29 | 1.52 m; 1.83 m | 19.8 |
| 30 | 1.18 m; 1.52 m | 32.2 |
| 31 | 3.54 m | 70.0 |
| 32 | 1.36 m | 35.0 |
| 33 | 1.52 m; 1.58 m | 34.7 |
| 34 | 2.62 m | 34.7 |
| 35 | 1.06 d (6.6) | 20.2 |
| 36 | 6.53 d (10.3) | 151.4 |
| 37 | - | 137.1 |
| 38 | 1.75 s | 11.4 |
| 39 | - | 202.5 |
| 40 | 2.31 s | 25.3 |
In CD3OD.
δ(ppm) 600 MHz; multiplicities; J values (Hz) in parentheses.
δ (ppm); shifts inferred from HMBC and g-HSQC spectra.
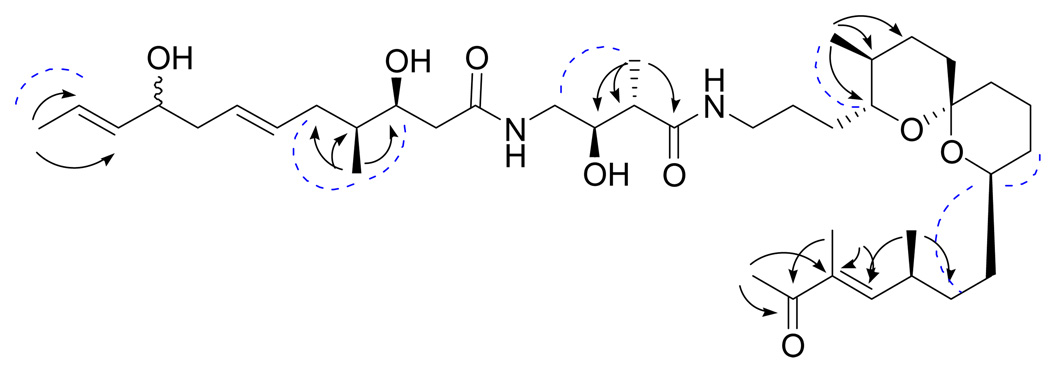
The aforementioned components of 1 were pieced together using both g-COSY and g-HMBC data, as well as comparing 1H and 13C NMR chemical shift values with those of bistramide K,9 although noticeable differences in chemical shifts suggested the occurrence of slightly different functionalization. Most importantly, an additional ketone carbonyl signal (C-39) was observed instead of a hydroxylated carbon signal at C-39. The molecular weight of 1 was found to be two mass units less than that of bistramide K. The absence of one sp3-oxygenated hydrogen resonance in the 1H NMR spectrum (H-39, bistramide K), in addition to the deshielded nature of H-40 (δH 2.31, s) and a downfield shift of the C-36 olefinic carbon (δC 151.4) signal as the result of becoming integrated into an α,β-unsaturated ketone system, signified that the sole structural difference between 1 and bistramide K is the oxidation of the C-39 hydroxyl moiety to a ketone functional group. Some key HMBC correlations are listed in Figure 1. Notably, HMBC correlations of H3-40 and H3-38 to the carbonyl at C-39 and the α-quaternary olefinic carbon (C-37), in addition to correlations of H3-40 and H3-38 to C-36, assisted in making the assignments at the tail end of the polyketide chain of 1.
Figure 1.
Key HMBC (arrows) and ROESY (dashed curves) correlations for compound 1
Because of the small amount of compound isolated, spectroscopic methods were relied on to establish the relative configuration of 1. The planar structures of 1 and 2 are identical from C-12 to C-37, and superimposing their 1H NMR spectra showed no significant differences between chemical shifts and coupling patterns, indicating that the two compounds share the same relative configuration at these positions. A correlation in the ROESY spectrum (axial H-22 to equatorial H3-24) also supported this hypothesis, and similarities were observed between the 1H NMR chemical shifts and coupling constants (JH-9–H-11) of 1 with those found in the literature for bistramide K.9 Despite overlap of the H-6 and H-7 olefinic resonances in the 1H NMR spectrum, the regiochemistry of the double bond was assigned as E due to similar chemical shifts of these two positions to those of bistramide K and the observance of ROESY correlations of H-6 with H4/H-8, and H-7 with H-5/H-9. Finally, although the small amount of sample available precluded the running of a quantitative CD spectrum of 1, the qualitative CD spectra of 1 and 2 were similar, with each showing a broad negative Cotton effect between 210 and 240 nm. These data suggested that bistramide K and compound 1 have the same absolute and relative configurations. The stereochemistry of the new chiral center at C-4 could not be determined from the available data.
Bistramides A (2) and D (3) were identified on the basis of comparison of MS and one and two-dimensional NMR spectroscopic data to those reported by Biard et al.9
The IC50 values for the antiproliferative activities of compounds 1 and 2 against the A2780 human ovarian cancer cell line were 0.34 and 0.26 µM, respectively. Compared with IC50 values against other cell lines reported in the literature (45, 20, and 22 nM against the KB, P388, and normal endothelial cells, respectively),10 the values against the A2780 cell line were about ten-fold larger.
In addition to bistramides 1–3, 1H NMR spectroscopic analysis of other minor peaks from the HPLC separation indicated the presence of several other bistramide analogs or bistramide-derived secondary metabolites; we estimate that this extract may contain as many as ten additional bistramides. Since only five known naturally occurring bistramides have been isolated over the past 20 years, this suggests that this structural class is more diverse than previously known. Given their nanomolar potency against selected cell lines and their well-defined mechanism of action against cancer, the isolation of additional bistramide derivatives will increase the chances of discovering analogs that exhibit similar activity with less toxicity.
The current work has proven a Trididemnum cyclops to be a source of small quantities of structurally unique bistramides. Since Trididemnum spp. are also sources of the didemnins, this genus is a versatile producer of bioactive metabolites. This raises the question of the true origin of the bistramides, which were originally isolated from the ascidian Lissoclinum bistratum.4,9 It has been reported that other species of Lissoclinum, as well as a few species of Trididemnum, contain obligate Prochloron spp. bacterial symbionts,10 and the collection used for this work also had a crust of a Prochloron sp. These cyanobacteria act as important nutrient sources for their host ascidians (particularly of the family Didemnidae) via photosynthesis and nitrogen fixation.11 They have been implicated in the production of cyclic peptides,12,13 a class of compounds that includes both bistratamides (Lissoclinum) and didemnins (Trididemnum). It is thus very possible that the bistramides are in fact products of microbial origin.
Experimental Section
General Experimental Procedures
Optical rotations were recorded on a JASCO P-2000 polarimeter. IR and UV spectra were measured on MIDAC M-series FTIR and Shimadzu UV-1201 spectrophotometers, respectively. CD analysis was performed on a JASCO J-720 spectropolarimeter. NMR spectra were obtained on JEOL Eclipse 500, Varion Inova 400, and Bruker 600 spectrometers (1.7 mm probe). Chemical shifts are given in δ (ppm), and coupling constants (J) are reported in Hz. Mass spectra were obtained on a JEOL JMS-HX-110 instrument and a Finnigan LTQ LC/MS. HPLC was performed using Shimadzu LC-10AT pumps coupled with analytical and semi-preparative Phenomenex Luna C18 columns (250 × 4.6 and 250 × 10 mm, respectively) and a semi-preparative Varian Lichrosorb diol phase column (250 × 10 mm). The HPLC system employed a Shimadzu SPD-M10A diode array detector and a Shimadzu FRC-10A fraction collector.
Material
Samples of an organism (wet weight 778 g) identified as a Trididemnum cyclops Michaelsen (Didemnidae) were collected by SCUBA from the waters around Nosy-Be, Madagascar, at −13.39 S, 48.05 E, on April 6, 2005. The organism formed colonies with a thin crust of bright green color due to the symbiotic cyanobacteria Prochloron; small zooïds 1.5 mm in length; branchial sac with tree stigmata rows. Its assigned collection number was PHIL_28. Identification was made by J. Maharavo, and a voucher specimen is preserved at the Centre National de Recherches Océanographiques, Nosy-Be, Madagascar. The sample was soaked for 24 h in 1:1 EtOH-seawater, the supernatant was decanted, and the damp organism was then extracted with EtOH, to yield 11.5 g of extract, 3.3 g of which were supplied to Virginia Polytechnic Institute and State University as extract NB 04-05-31.
Antiproliferative Bioassay
The A2780 ovarian cancer cell line assay was performed at Virginia Polytechnic Institute and State University as previously reported.14 The A2780 cell line is a drug-sensitive ovarian cancer cell line.15
Extraction and Isolation
Extract NB 04-05-31 (2.5 g; IC50 2.0 µg/mL) was suspended in aqueous MeOH (MeOH-H2O, 9:1, 1,000 mL) and extracted with hexanes (2 × 500 mL; IC50 14 µg/mL). The aqueous layer was then diluted to 40% water and extracted with dichloromethane (2 × 500 mL). The dichloromethane-soluble fraction (269 mg) displayed antiproliferative activity (IC50 = 0.1 µg/mL), thus it was further chromatographed over a reversed-phase C18 column to yield six fractions (E-J). Fraction G (30.3 mg, IC50 0.1 g/mL) was determined as being the most active. Fraction G was also found to be extremely complex, and a direct HPLC separation method required an abnormally long run time (approximately 155 min). Consequently, semi-preparative reversed-phase C18 HPLC on a Phenomenex Luna column with an isocratic flow of 73% aqueous methanol was used to separate fraction G. This technique afforded two pure compounds, 39-oxobistramide K (1, fraction 23, tR 112 min, 0.3 mg) and bistramide A (2, fraction 27, tR 135 min, 1.1 mg). Other pure compounds were collected, but the very small quantities obtained prevented their structure elucidation.
In an attempt to isolate greater quantities of these bioactive molecules, a re-extraction of NB 04-05-31 was undertaken. Extract NB 04-05-31 (0.76 g) was suspended in aqueous MeOH (MeOH-H2O, 9:1, 500 mL) and extracted with hexanes (2 × 250 mL). The aqueous layer was then diluted to 40% water and extracted with dichloromethane (2 × 250 mL). The dichloromethane fraction (92 mg) was chromatographed over a reversed-phase C18 column to yield four fractions (I-IV). Fraction II (50 mg, IC50: 0.2 g/mL) was the most active. Semi-preparative reversed phase C18 HPLC using 73% aqueous MeOH resulted in the collection of fifty fractions from fraction II. Based on sample weight and its 1H NMR spectrum, fraction 37 (tR 195 min, 0.8 mg) was purified on a semi-preparative diol phase HPLC column using an isocratic flow of 87% CH2Cl2/MeOH to afford bistramide D (3, tR 7.0 min, 0.7 mg).
39-Oxobistramide K (1)
amorphous powder; α25D -72 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 200 (4.1), 240 (3.9) nm; 1H NMR (600 MHz, CD3OD) and 13C NMR (150 MHz, CD3OD) see Table 1; HRFABMS m/z 727.4864 (calcd for C40H68N2O8Na, 727.4873).
Bistramide A (2)
amorphous powder; CD (MeOH, c 0.0077) [θ]215 -5.44, [θ]246 1.87, [θ]259 2.68; 1H data, see Supporting Information; LCEIMS m/z 705.5 [M+H]+ (calcd for C40H69N2O8, 705.5).
Bistramide D (3)
amorphous powder; 1H NMR data, see Supporting Information; LCEIMS m/z 707.6 [M+H]+, 729.5 [M+Na]+ (calcd for C40H70N2O8Na, 729.5).
Supplementary Material
Acknowledgment
This International Cooperative Biodiversity Group project was supported by the Fogarty International Center, the National Cancer Institute, the National Science Foundation, the National Heart, Lung and Blood Institute, the National Institute of Mental Health, the Office of Dietary Supplements, and the Office of the Director of NIH, under Cooperative Agreement U01 TW000313 from the National Institutes of Health, and this support is gratefully acknowledged. We also thank Mr. Bill Bebout and Dr. M. Ashraf-Khorassani for obtaining mass spectra, and Mr. Kim Harich for obtaining CD data. We gratefully acknowledge courtesies extended by the Government of Madagascar (Ministère des Eaux et Forêts).
Footnotes
Supporting Information Available: Spectroscopic data, consisting of 1H NMR spectroscopic data of bistramides A (2) and D (3), 1H NMR, ROESY, COSY, HSQC, HMBC, and CD spectra of 39-oxobistramide K (1), 1H NMR, 13C NMR, COSY, HMBC, and CD spectra of bistramide A (2), the 1H NMR spectrum of bistramide D (3), and a photograph of a sample of Trididemnum cyclops are available as Supporting Information. This material is available free of charge via the internet at http://pubs.acs.org.
References and Notes
- 1.Biodiversity Conservation and Drug Discovery in Madagascar, Part 36. For Part 35, see: Hou Y, Cao S, Brodie PJ, Miller JS, Callmander MC, Ratovoson F, Rakotobe E, Rasamison VE, Ratsimbason M, Alumasa J, Roepe PD, Kingston DGI. Bioorg. Med. Chem. 2009;17:2871–2876. doi: 10.1016/j.bmc.2009.02.022.
- 2.(a) Rinehart KL, Gloer JB, Cook JC. J. Am. Chem. Soc. 1981;103:1857–1859. [Google Scholar]; (b) Rinehart KL, Gloer JB, Hughes RG, Renis HE, McGovren PJ, Swynenberg EB, Stringfellow DA, Kuentzel SL, Li LH. Science. 1981;212:933–935. doi: 10.1126/science.7233187. [DOI] [PubMed] [Google Scholar]
- 3.Gouiffes D, Moreau S, Helbecque N, Bernier JL, Henichart JP, Barbin Y, Laurent D, Verbist JF. Tetrahedron. 1988;44:451–459. [Google Scholar]
- 4.Pusset J, Maillere B, Debitus C. J. Nat. Toxins. 1996;5:1–6. [Google Scholar]
- 5.Gautret P, Le Pape P, Biard JF, Menard D, Verbist JF, Marjolet M. Acta Parasitol. 1998;43:50–53. [Google Scholar]
- 6.Statsuk AV, Baj R, Baryza JL, Verma VA, Hamel E, Wender PA, Kozmin SA. Nature Chem. Biol. 2005;1:383–388. doi: 10.1038/nchembio748. [DOI] [PubMed] [Google Scholar]
- 7.Rizvi SA, Tereshko V, Kossiakoff AA, Kozmin SA. J. Am. Chem. Soc. 2006;128:3882–3883. doi: 10.1021/ja058319c. [DOI] [PubMed] [Google Scholar]
- 8.Rizvi SA, Courson DS, Keller VA, Rock RS, Kozmin SA. Proc. Natl. Acad. Sci., USA. 2008;105:4088–4092. doi: 10.1073/pnas.0710727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biard JF, Roussakis C, Kornprobst JM, Gouiffes-Barbin D, Verbist JF, Cotelle P, Foster MP, Ireland CM, Debitus C. J. Nat. Prod. 1994;57:1336–1345. doi: 10.1021/np50112a002. [DOI] [PubMed] [Google Scholar]
- 10.Münchoff J, Hirose E, Maruyama T, Sunairi M, Burns BP, Neilan BA. Envir. Microbiol. 2007;9:890–899. doi: 10.1111/j.1462-2920.2006.01209.x. [DOI] [PubMed] [Google Scholar]
- 11.Shimada A, Yano N, Kanai S, Lewin RA, Maruyama T. Phycologia. 2003;42:193–197. [Google Scholar]
- 12.Schmidt EW, Sudek S, Haygood MG. J. Nat. Prod. 2004;67:1341–1345. doi: 10.1021/np049948n. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc. Nat. Acad. Sci., USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao S, Brodie PJ, Randrianaivo R, Ratovoson F, Callmander M, Andriantsiferana R, Rasamison VE, Kingston DGI. J. Nat. Prod. 2007;70:679–681. doi: 10.1021/np060627g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Res. 1985;45:2110–2115. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.