Abstract
Several hundred thousand electronic pacemakers are implanted in the US each year to treat abnormally slow heart rates. Biological pacemaker research strives to replace this hardware, and the associated monitoring and maintenance, by using gene or cell therapy to create a permanent and autonomically responsive pacemaker. While there are numerous technological hurdles to overcome before this is a therapeutic reality, one critical issue is determining the optimal channel gene to employ in creating a biological pacemaker. This review discusses the pros and cons of various model systems for characterizing and evaluating the function of candidate channel genes. It is argued that a sequential approach that combines in silico, in vitro and in vivo models is required.
INTRODUCTION
Abnormally slow heart rates (bradyarrhythmias) are a serious and life threatening condition, requiring implantation of electronic pacemakers in over 300,000 people annually in the US. While highly successful, electronic pacing also is associated with complications related to the need for implanting and maintaining hardware, wires and batteries. For example, site placement is dictated by lead stability, but this can result in non-optimal ventricular activation and contraction, and eventually cardiac failure [1]. Lead fracture and the complications associated with lead replacement also can occur [2]. Further, although progress has been made in developing rate responsive electronic pacemakers, they still lack sensitivity to neurohumoral agonists. As a result, numerous laboratories worldwide are developing biological pacemakers as an alternative therapy (see [1,3]).
Proof of concept experiments have been conducted with both gene and cell therapies, and in the latter case with multiple cell types. The approaches taken thus far in developing a biological pacemaker can be broadly separated into two categories. One category relies on cells that are endogenously automatic, such as explanted sinoatrial node (SAN) cells or stem cells that have been driven down a cardiac lineage [3]. An inherent limitation of this approach is that one is restricted to the ion channels that are naturally expressed in the selected cell type. Thus, even if existing problems with cell source, cell differentiation and immune reaction are overcome, the final result may not be ideal because one cannot fine-tune function by altering the biophysical characteristics of the channels contributing to pacemaking. The other category employs genetic engineering to introduce, either directly into cardiac myocytes or into cells that are then implanted into the heart, genes that can initiate or enhance automaticity [1,3]. In this case one can employ all the tools of bioinformatics and molecular biology to engineer custom channels with distinct biophysical properties that may be best suited for a specific patient population or implant site. Thus, there is no inherent limit to the range of pacemaker function that can be created. However, thoroughly evaluating the functional characteristics of each channel mutation is an expensive and time consuming problem. Thus, model systems are needed to permit triage of candidate genes so that only the most promising ones are pursued at the level of intact animal studies and eventually in clinical trials.
This review focuses on methods of evaluating candidate biological pacemaker channels. It addresses only variations on the hyperpolarization-activated cyclic nucleotide gated (HCN) channel, since this is the channel backbone that has most often been employed in recent biological pacemaker studies. However the general approach outlined here would be equally applicable to exploration of other candidate channels. As summarized in Table I, this review compares in vivo, in vitro and in silico models for their utility in evaluating biological pacemakers.
Table I.
Model systems for evaluating biological pacemaker channels
| In Vivo | In Vitro | In Silico | |
|---|---|---|---|
| Pro | Most predictive of therapeutic outcome, particularly when large animal models are employed | Suitable for detailed biophysical characterization; can obtain both rate and rhythm information when employing a myocyte culture | Fastest and most economical means to compare different biophysical properties; can provide insight into which channel parameters are most useful to modify |
| Con | Expensive and time consuming if comparing many channel constructs | Removed from intact animal situation; rate and rhythm information still fairly time consuming to acquire; ideally should employ localized expression, which is more difficult | Highly dependent on accuracy of the model, particularly regarding non-equilibrium behavior of expressed channel; also dependent on accurate biophysical data from in vitro studies |
HCN channel structure-function and use as a biological pacemaker
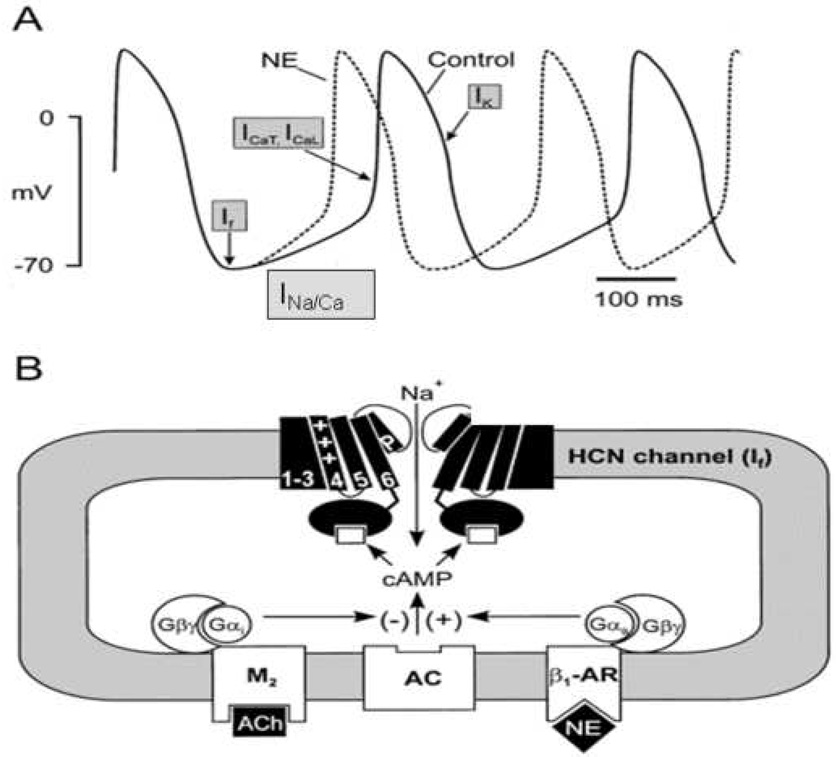
The HCN gene family encodes a series of 4 channel isoforms (HCN1-HCN4), 3 of which occur naturally in the heart but with regional differences in expression. Four subunits (of the same or a mix of isoforms) come together to form a functional channel, resulting in a current that has been called If or the pacemaker current [4]. While debate continues concerning the relative contribution of If to normal cardiac pacemaking [5], it is clear that the HCN channel has a number of characteristics that make it an ideal candidate for use as a biological pacemaker (Figure 1). The channels have mixed Na/K permeability and activate on hyperpolarization. Thus, they generate an inward depolarizing current at diastolic membrane potentials (i.e. the period between contractions) but rapidly deactivate upon action potential firing so that they do not contribute to potentially proarrhythmic action potential prolongation. In addition, they contain a cyclic nucleotide binding domain so that they respond directly to changes in sympathetic and parasympathetic nerve activity and corresponding changes in cAMP level, providing autonomically responsive pacemaking. Figure 2 schematically compares the normal cardiac pacemaker with HCN based biological pacemakers arising from gene and cell therapy. For gene therapy, a viral vector is typically employed to deliver the channel gene to the cardiac cells. For delivery as part of cell therapy, the channel gene is most commonly delivered within an adult mesenchymal stem cell. However, any cell type that is capable of electrically coupling to cardiac myocytes (I.e. expresses compatible connexin proteins) [6] can in theory serve as an appropriate delivery vehicle.
Figure 1.
Role of pacemaker channels in regulating normal sinoatrial node automaticity. A) Representative sinoatrial node action potential (control: solid lines) and some of the contributing ion channels and exchangers. Ifis activated on hyperpolarization and provides current to initiate diastolic depolarization. Na/Ca exchange current and T-/L-type Ca currents contribute to late diastole and threshold. The potassium current Ikrepolarizes the membrane. The effect of norepinephrine (NE) to increase diastolic slope and speed impulse initiation is represented by the dashed line. B) Depiction of an HCN pacemaker channel (2 of the 4 subunits composing a functional channel are shown), illustrating the 6 transmembrane spanning domains of each subunit. When the channel is open, Na influx depolarizes the cell. β1-adrenergic and M2-muscarinic receptors respond to NE and acetylcholine (ACh), respectively, to regulate adenylyl cyclase (AC) activity via G-protein coupling. AC, in turn, regulates intracellular cAMP level, which is sensed by a cAMP binding site in the carboxy terminus of the HCN subunit (adapted with permission from [16]).
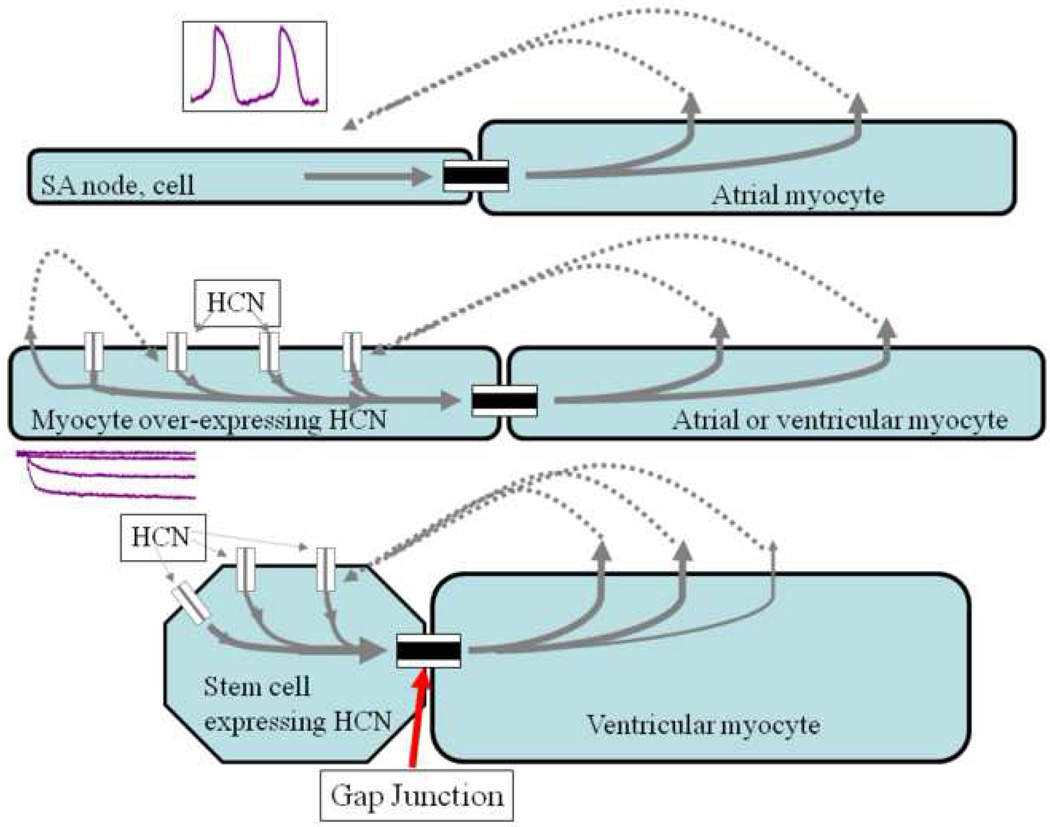
Figure 2.
Rationale for biological pacing. Top) Initiation of spontaneous rhythms by sinoatrial node cells. Action potentials (inset) are initiated via inward current flowing through transmembrane channels (see Figure 1). Current flowing via gap junctions to adjacent myocytes results in excitation and impulse propagation through the conducting system. Middle) Gene-based biological pacemaker implanted in atrium or ventricle. HCN channel genes are virally introduced into a group of myocytes (one shown), resulting in channel proteins incorporated in the myocyte membrane. When the membrane is hyperpolarized at the end of an action potential, the HCN channels open to induce inward current, which excites the myocyte to initiate an action potential that then propagates via gap junctions to neighboring cells. Bottom) Cell-based biological pacemaker shown implanted in ventricle. HCN channels are expressed in stem cells that are then implanted in myocardium, where they electrically couple to neighboring myocytes via gap junctions. When the stem cell membrane is hyperpolarized via current flow through the gap junctions from coupled myocytes, the HCN channels open to induce inward current, which travels through the gap junction to excite the coupled myocyte and initiate an action potential in the myocyte. In each depicted example, current spread requires the presence of gap junctions to electrically couple the cells (adapted with permission from [34]).
In vivo models
Any biological pacemaker (consisting of the candidate channel gene and the virus or cell in which it is packaged) will have to undergo in vivo testing in an animal model prior to clinical trials. There are economic advantages to small animal models, and indeed the first in vivo study of a channel based biological pacemaker was performed in such a system [7]. However, therapeutic applicability is limited both by the relatively rapid heart rates of small animals and their unsuitability for employing delivery methods relevant to human therapy. In this regard, the FDA guidance on cell therapies for cardiac disease states “large animal models (pig sheep, dog) provide information on safety and activity of cellular products and delivery systems, leading to the selection of a potentially safe starting dose for a Phase 1 clinical trial” (http://www.fda.gov/OHRMS/DOCKETS/98fr/FDA-2009-D-0132-gdl.pdf). Further, one typically wants to create a relevant disease model such as atrioventricular (AV) block for evaluating functionality of the biological pacemaker and this is more readily accomplished in large animals. Large animal models also are amenable to simultaneous implantation of an electronic pacemaker operating in back-up mode [8]. Not only is this the likely paradigm in initial clinical trials, but the electronic pacemaker memory provides valuable information on the history of biological and electronic pacing during an experiment.
We have found the canine to be a suitable large animal model for biological pacemaker studies [8–13], although others have used the pig [14,15]. In particular, we have used this model to compare biological pacemaker functionality with different HCN constructs. Our initial efforts focused on the native HCN2 isoform. In deciding which isoform to employ we considered several factors. First, we sought robust autonomic responsiveness, which favored selection of HCN2 or HCN4 over the other isoforms [4,16]. Second, we hypothesized that function would depend on 3 basic channel parameters: current magnitude (larger = faster rate), activation voltage (less negative voltage = faster rate) and activation kinetics (faster activation = faster rate). We assumed expression would not differ among isoforms, and also noted that the isoforms exhibited relatively modest differences in voltage dependence. However HCN2 activation kinetics are much faster than those of HCN4, leading us to favor HCN2 as our initial candidate channel. When delivered either as a gene or cell therapy in conjunction with AV block and a back-up electronic pacemaker, we obtained heart rates of at least 50 bpm as well as evidence of adrenergic responsiveness of rate [8,12].
While such a result could be considered therapeutically adequate, it would be preferable to be able to attain both higher basal rates and higher maximal rates (in response to autonomic stimulation), at least for some patient populations. For this reason, we and others have employed mutagenesis in efforts to engineer improved channel constructs [8,13,17]. We first introduced a mutation known to shift voltage dependence positive [18], reasoning that this would result in more current flowing during diastole. While this proved to be the case, it also resulted in pronounced reduction in channel expression and current magnitude, such that it inferred no advantage over wildtype HCN2 [8]. We next introduced the transmembrane portion of HCN1 into the HCN2 backbone. We reasoned this would provide the fast activation kinetics typical of HCN1 while preserving the strong autonomic responsiveness of HCN2 (since the cyclic nucleotide binding domain resides in the cytosolic C-terminal). The result was a biological pacemaker that “overshot” our target heart range, resulting in ventricular tachycardia interspersed with pauses [13].
These studies demonstrated several things: 1) localized HCN expression within the canine heart results in a functional biological pacemaker; 2) altering HCN channel biophysics (expression, voltage dependence, kinetics) results in different biological pacemaker outcome, both in terms of heart rate and heart rhythm (i.e. regularity of rate); 3) alternative models were needed to screen channel constructs more quickly and more economically than could be accomplished in large animals.
In vitro models
The first demonstration that HCN over-expression could serve as a biological pacemaker was performed in vitro, by expressing HCN2 in a monolayer culture of newborn rat ventricle (NBRV) myocytes [19]. These cells beat spontaneously in culture and HCN2 over-expression significantly increased the rate. A subsequent study demonstrated a similar effect for HCN4 [20]. Later studies have shown that this model system is predictive of how different HCN constructs will function in vivo. In our studies of the HCN2 point mutation with shifted voltage dependence [8], initial NBRV studies demonstrated reduced expression efficiency, which was born out in vivo when we failed to achieve higher rates despite the favorable voltage dependence. A more dramatic demonstration was seen in studies of the HCN2/HCN1 chimeric channel. The periods of rapid pacing interspersed with pauses observed in vivo [13] were reproduced in the cell culture system [21]. Thus, the in vitro model system can be employed to compare both mean rate and regularity of rate with different HCN constructs.
Further, the culture system is amenable to detailed biophysical analysis of different channel constructs, which is not possible in the intact animal. While this could in theory be done in any cell system, rather than in primary myocyte cultures, the known context dependence of HCN channel function (i.e. biophysical parameters are affected by the cellular environment) [22] argues for working with cardiac myocytes whenever possible, at least in the case of gene therapy. Cell therapy studies require additional data collection in the intended host cell type. The importance of being able to perform detailed biophysical analysis was again demonstrated in the studies of chimeric HCN channels, where comparison of the non-equilibrium (i.e. frequency-dependent) properties of the chimeric channel with those of HCN2 suggested an explanation for the difference in regularity of rhythm, and thus also suggested what further channel mutations might prove productive [23]. Finally, it also is possible, particularly in the case of cell therapy, to locally express the channel of interest within a monolayer NBRV culture, thereby permitting study of impulse initiation and propagation from the pacemaker site to surrounding myocardium [11,14].
Despite the practical advantages of the NBRV culture system for evaluating biological pacemaker function of different HCN constructs, one must remember that it is far removed from the intact animal or human situation. Thus, while a valuable screening tool, it must always be supplemented with in vivo experiments of the most promising constructs.
In silico models
While in vitro models are far removed from the in vivo situation, in silico models are even further removed. Yet they can be invaluable when trying to decide which biophysical alterations might favorably impact pacemaking function, and for understanding the results of in vivo and in vitro experiments.
The simplest computer simulation is to take one of the existing computer models of SAN automaticity, and alter the magnitude, voltage dependence and kinetics of the pacemaker current If to match the experimentally measured values of the expressed HCN current in whatever cell system is used to initially characterize a particular construct. We previously employed this approach to demonstrate that speeding activation kinetics should indeed lead to a faster biological pacemaker [21]. While providing confirmation that a particular channel biophysical parameter may be a useful target of mutagenesis, there are several limitations to such an approach. Most obviously, the implantation site of a biological pacemaker is not likely to be the SAN. Rather, gene or cell delivery will be to atrial muscle in the case of sinus node dysfunction with a healthy AV node, or – in the case of AV node disease, to the ventricular myocardium or conduction system. Thus, the computer model should be based on one or more of these cell types [24,25]. Alternatively, it might be based on the NBRV myocyte culture, since this is the common in vitro experimental system. Ideally, the investigator would have both models available. Comparison of an NBRV simulation to experimental results in vitro would validate the ability of the simulation to predict functionality of the tested HCN construct, and then extrapolation to models of different adult cardiac cell types would predict in vivo outcome of delivering the construct to different heart regions.
A challenge of developing such simulations is that most existing cardiac action potential models that incorporate pacemaker current use a Hodgkin-Huxley formulism [26]. However, such a model does not accurately predict the non-equilibrium characteristics of HCN channels such as voltage hysteresis [27,28], and such non-equilibrium behavior has been demonstrated to be important to biological pacemaker outcome [21]. A better solution is to employ an allosteric model or other HCN channel model that accounts for the observed non-equilibrium behavior. While several laboratories have developed allosteric models [29–31], only recently has an HCN channel model incorporating non-equilibrium characteristics been integrated into a simulation of a cardiac action potential, in this case for SAN myocytes [32]. This study explored the effect of non-equilibrium HCN behavior on regularity of rhythm. This approach has significant potential for assessing the effectiveness of novel HCN constructs as biological pacemakers. However, to fully realize this potential, an accurate model of HCN channel behavior must be incorporated into action potential models of the NBRV myocyte culture and into action potential models of networks of adult cardiac cells representing distinct cardiac regions. Further, these models must be able to simulate localized over-expression of the HCN construct or local coupling to HCN-expressing stem cells, so as to allow analysis of how the pacemaker site drives coupled non-automatic tissue. While there has been one study to date addressing the issue of localized expression [33], this approach has not yet been synthesized with the models of non-equilibrium HCN channel function.
CONCLUSIONS
Biological pacemakers represent a disruptive technology with the potential of replacing electronic pacemakers for many patients. They hold the promise of fewer complications and more natural physiologic function, ultimately providing a lifetime cure rather than the palliation and periodic monitoring/maintenance of existing hardware based treatments.
To realize the potential of biological pacemakers, many technical issues must be overcome. These include developing the appropriate virus or cell delivery package and demonstrating the safety and persistence of that package – issues that are beyond the scope of the current review. However, it also is necessary to identify the optimal channel construct to be expressed within the delivery package. As detailed here, the most likely candidate channel will be derived from the HCN gene family, which is ideally suited to function as a pacemaker when over-expressed. However, achieving optimum functionality will probably involve mutagenesis of the native isoform to fine tune channel biophysics. In addition, the desired biophysical traits will not necessarily be the same for all implantation sites within the heart or for all patient populations, so multiple candidate channel constructs will need to be designed and tested. Thus, model systems are required to efficiently characterize and evaluate the function of candidate channel constructs. No single model system is ideal, but by combining all three approaches described here, one can create an integrated triage system that winnows out the less successful candidate genes at each step. One would progressively move from the in silico system, through the more complicated in vitro system, to the most complex and physiologically relevant in vivo system.
Finally, one must recognize the potential for an implanted biological pacemaker to be proarrhythmic. While the most common problem to date has been a somewhat slower than desired target heart rate, the HCN2/HCN1 chimeric channel did lead to periods of tachycardia interspersed with pauses, both in vivo and in vitro [13,21]. While this reinforces the predictive value of the in vitro system to identify problematic constructs early in the development cycle, it also highlights a potential risk of this therapeutic modality. However, the fact that current biological pacemakers are based on the HCN gene family also suggests a suitable response to a proarrhythmic outcome, since an HCN blocking drug is available that was able to stop the HCN2/HCN1 tachycardia in vivo [13]. Whether such a drug could be employed to slow an abnormally fast biological pacemaker down to the desired rate remains to be determined. If not, or if such pharmacological intervention proved inadequate, radiofrequency ablation could be employed to silence the biological pacemaker. In that case the electronic pacemaker that is likely to be implanted in tandem as a back-up in any initial clinical trials [8] would continue to maintain heart rate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rosen MR, et al. Cardiac pacing: from biological to electronic … to biological? Circulation Arrhythmia Electrophysiology. 2008;1:54–61. doi: 10.1161/CIRCEP.108.764621. [DOI] [PubMed] [Google Scholar]
- 2.Hauser RG, Hayes DL. Increasing hazard of Sprint Fidelis implantable cardioverter-defibrillator lead failure. Heart Rhythm. 2009;6(5):605–610. doi: 10.1016/j.hrthm.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Robinson RB, et al. I(f) and the biological pacemaker. Pharmacol.Res. 2006;53(5):407–415. doi: 10.1016/j.phrs.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Robinson RB, Siegelbaum SA. Hyperpolarization-activated cation currents: from molecules to physiological function. Ann.Rev.Physiol. 2003;65:453–480. doi: 10.1146/annurev.physiol.65.092101.142734. [DOI] [PubMed] [Google Scholar]
- 5.Lakatta EG, DiFrancesco D. What keeps us ticking: A funny current, a calcium clock, or both? J Mol.Cell Cardiol. doi: 10.1016/j.yjmcc.2009.03.022. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valiunas V, et al. Human mesenchymal stem cells make cardiac connexins and form functional gap junctions. J.Physiol.(London) 2004;555(3):617–626. doi: 10.1113/jphysiol.2003.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miake J, Marban E, Nuss HB. Gene therapy: Biological pacemaker created by gene transfer. Nature. 2002;419(6903):132–133. doi: 10.1038/419132b. [DOI] [PubMed] [Google Scholar]
- 8.Bucchi A, et al. Wild-type and mutant HCN channels in a tandem biological-electronic cardiac pacemaker. Circulation. 2006;114:992–999. doi: 10.1161/CIRCULATIONAHA.106.617613. [DOI] [PubMed] [Google Scholar]
- 9.Qu J, et al. Expression and function of a biological pacemaker in canine heart. Circulation. 2003;107(8):1106–1109. doi: 10.1161/01.cir.0000059939.97249.2c. [DOI] [PubMed] [Google Scholar]
- 10.Plotnikov AN, et al. Biological pacemaker implanted in canine left bundle branch provides ventricular escape rhythms that have physiologically acceptable rates. Circulation. 2004;109:506–512. doi: 10.1161/01.CIR.0000114527.10764.CC. [DOI] [PubMed] [Google Scholar]
- 11.Potapova I, et al. Human Mesenchymal Stem Cells as a Gene Delivery System to Create Cardiac Pacemakers. Circ.Res. 2004;94:952–959. doi: 10.1161/01.RES.0000123827.60210.72. [DOI] [PubMed] [Google Scholar]
- 12.Plotnikov AN, et al. Xenografted adult human mesenchymal stem cells provide a platform for sustained biological pacemaker function in canine heart. Circulation. 2007;116:706–713. doi: 10.1161/CIRCULATIONAHA.107.703231. [DOI] [PubMed] [Google Scholar]
- 13.Plotnikov AN, et al. HCN212-channel biological pacemakers manifesting ventricular tachyarrhythmias are responsive to treatment with If blockade. Heart Rhythm. 2008;5:282–288. doi: 10.1016/j.hrthm.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehat I, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat.Biotechnol. 2004;22(10):1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 15.Cai J, et al. Adenoviral gene transfer of HCN4 creates a genetic pacemaker in pigs with complete atrioventricular block. Life Sci. 2007;80(19):1746–1753. doi: 10.1016/j.lfs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Biel M, Schneider A, Wahl C. Cardiac HCN channels. structure, function, and modulation. Trends Cardiovasc.Med. 2002;12(5):206–212. doi: 10.1016/s1050-1738(02)00162-7. [DOI] [PubMed] [Google Scholar]
- 17.Tse HF, et al. Bioartificial sinus node constructed via in vivo gene transfer of an engineered pacemaker HCN Channel reduces the dependence on electronic pacemaker in a sick-sinus syndrome model. Circulation. 2006;114(10):1000–1011. doi: 10.1161/CIRCULATIONAHA.106.615385. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, et al. The S4-S5 linker couples voltage sensing and activation of pacemaker channels. Proc.Natl.Acad.Sci.U.S.A. 2001;98(20):11277–11282. doi: 10.1073/pnas.201250598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu J, et al. HCN2 over-expression in newborn and adult ventricular myocytes: distinct effects on gating and excitability. Circ.Res. 2001;89:e8–e14. doi: 10.1161/hh1301.094395. [DOI] [PubMed] [Google Scholar]
- 20.Er F, et al. Dominant-negative suppression of HCN channels markedly reduces the native pacemaker current I(f) and undermines spontaneous beating of neonatal cardiomyocytes. Circulation. 2003;107(3):485–489. doi: 10.1161/01.cir.0000045672.32920.cb. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X, et al. In vitro characterization of HCN channel kinetics and frequency-dependence in myocytes predicts biological pacemaker functionality. J.Physiol.(London) 2009;587:1513–1525. doi: 10.1113/jphysiol.2008.163444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu J, et al. Functional comparison of HCN isoforms expressed in ventricular and HEK 293 cells. Pflugers Arch. 2002;444:597–601. doi: 10.1007/s00424-002-0860-7. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Kryukova Y, Yang X, Robinson RB. Designing a biological pacemaker by altering HCN2 activation/deactivation kinetics. Heart Rhythm. 2009;6(5S):S292. (Abstract) [Google Scholar]
- 24.Ramirez RJ, Nattel S, Courtemanche M. Mathematical analysis of canine atrial action potentials: rate, regional factors, and electrical remodeling. Am.J Physiol Heart Circ.Physiol. 2000;279(4):H1767–H1785. doi: 10.1152/ajpheart.2000.279.4.H1767. [DOI] [PubMed] [Google Scholar]
- 25.Hund TJ, Rudy Y. Rate dependence and regulation of action potential and calcium transient in a canine cardiac ventricular cell model. Circulation. 2004;110(20):3168–3174. doi: 10.1161/01.CIR.0000147231.69595.D3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noble D, Noble SJ. A model of sino-atrial node electrical activity based on a modification of the DiFrancesco-Noble (1984)equations. Proc.R.Soc.Lond.B. 1984;222:295–304. doi: 10.1098/rspb.1984.0065. [DOI] [PubMed] [Google Scholar]
- 27.Azene EM, et al. Non-equilibrium behavior of HCN channels: insights into the role of HCN channels in native and engineered pacemakers. Cardiovasc.Res. 2005;67(2):263–273. doi: 10.1016/j.cardiores.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Elinder F, et al. Mode shifts in the voltage gating of the mouse and human HCN2 and HCN4 channels. J.Physiol.(London) 2006;575(Pt 2):417–431. doi: 10.1113/jphysiol.2006.110437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altomare C, et al. Integrated allosteric model of voltage gating of hcn channels. J.Gen.Physiol. 2001;117:519–532. doi: 10.1085/jgp.117.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiFrancesco D. Dual allosteric modulation of pacemaker (f) channels by cAMP and voltage in rabbit SA node. J.Physiol.(London) 1999;515(Pt 2):367–376. doi: 10.1111/j.1469-7793.1999.367ac.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, et al. Activity-dependent regulation of HCN pacemaker channels by cyclic AMP: signaling through dynamic allosteric coupling. Neuron. 2002;36(3):451–461. doi: 10.1016/s0896-6273(02)00968-6. [DOI] [PubMed] [Google Scholar]
- 32.Mannikko R, et al. Hysteresis in the voltage dependence of HCN channels: conversion between two modes affects pacemaker properties. J Gen.Physiol. 2005;125(3):305–326. doi: 10.1085/jgp.200409130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanani S, Pumir A, Krinsky V. Genetically engineered cardiac pacemaker: stem cells transfected with HCN2 gene and myocytes - a model. Physics Letters A. 2008;372:141–147. [Google Scholar]
- 34.Rosen MR, et al. Genes, stem cells and biological pacemakers. Cardiovasc.Res. 2004;64:12–23. doi: 10.1016/j.cardiores.2004.05.012. [DOI] [PubMed] [Google Scholar]