Summary
Delta gene expression in Drosophila is regulated by proneural bHLH transcription factors, such as acheate-scute. In vertebrates, multiple Delta-like and proneural bHLH genes are expressed during neurogenesis, especially in the retina. We recently uncovered a relationship between Acheate-scute like 1 (Ascl1), Delta-like genes, and Notch in chick retinal progenitors. Here, we report that mammalian retinal progenitors are also the primary source of Delta-like genes, likely signaling through Notch among themselves, while differentiating neurons expressed Jagged2. Ascl1 is co-expressed in Delta-like and Notch active progenitors, and required for normal Delta-like gene expression and Notch signaling. We also reveal a role for Ascl1 in the regulation of Hes6, a pro-neurogenic factor that inhibits Notch signaling to promote neural rather than glial differentiation. Thus, these results suggest a molecular mechanism whereby attenuated Notch levels coupled with reduced proneurogenic activity in progenitors leads to increased gliogenesis and decreased neurogenesis in the Ascl1 deficient retina.
Keywords: Notch activity, Delta-like, Dll1, Dll4, Dll3, Ascl1, Neurog2, Hes1, Hes5, Hes6, retinal development, mutual and lateral inhibition, molecular circuit
Introduction
Studies of neurogenesis in Drosophila have revealed that neuroblasts in the proneural cluster express Delta, a Notch ligand induced by a proneural bHLH transcription factor such as acheate-scute, which sends a lateral inhibitory signal to neighboring cells through the Notch receptor to prevent them from acquiring this fate (reviewed by Skeath and Carroll, 1994; Bertrand et al., 2002). The activation of Notch in neighboring cells induces a signal transduction cascade that upregulates expression of hairy and enhancer of split genes, which ultimately lead to the repression of proneural function. Since many components of the Notch signal transduction pathway are expressed in the vertebrate nervous system, and manipulations that activate or inhibit Notch signaling maintain progenitors in an undifferentiated state or promote their differentiation into neurons, respectively, the paradigm of Notch-Delta mediated lateral inhibition in Drosophila has been extended to vertebrate neurogenesis (reviewed by Lewis, 1996; Lowell, 2000; Louvi and Artavanis-Tsakonas, 2006).
The vertebrate retina has served as an excellent model system for studies of neurogenesis, and Notch signaling in particular. Notch signaling maintains the progenitor pool during the course of retinal development, and regulates the evolutionary conserved sequence of progenitor cell differentiation into the six types of neurons and one type of glia (Dorsky et al., 1995; Austin et al., 1995; Tomita et al., 1996a; Henrique et al., 1997; Dorsky et al., 1997; Furukawa et al., 2000; Hojo et al., 2000; Satow et al., 2001; Silva et al., 2003; Takatsuka et al., 2004; Nelson et al., 2006; Jadhev et al., 2006; Yaron et al., 2006; Nelson et al., 2007a). While most of these studies have demonstrated key functions of Notch and the downstream signaling components in progenitor cells, relatively little attention has been given to upstream components, such as the source of Delta ligands and their respective regulation. Over ten years ago, it was discovered that Delta-like 1 (Dll1) plays a key role in regulating Notch activity and maintaining a pool of progenitors during the period of retinogenesis (Dorsky et al., 1997; Henrique et al., 1997). Thus, a relatively simple lateral inhibitory model was proposed for vertebrates based on Drosophila, whereby differentiating neurons in the retina express Dll1 to laterally inhibit and maintain neighboring progenitors through activation of Notch signaling (Henrique et al., 1997).
Multiple Notch ligands are present during vertebrate retinal development, complicating this simple lateral inhibitory model (Lindsell et al., 1996; Bao and Cepko, 1997; Valsecchi et al., 1997; Henrique et al., 1997; Wang et al., 1998; Benedito and Duarte, 2005; Nimmagadda et al., 2007; Nelson et al., 2007a; Nelson and Reh, 2008). Vertebrates have two orthologous families of canonical Notch ligands, Delta-like and Jagged, but have different numbers of paralogs within each family depending on the species. For example, avians (chicken) have two Delta-like genes, Dll1 and Dll4, while mammals have three, Dll1, Dll3, and Dll4. Both chickens and mammals have two Jagged genes, Jagged1 and Jagged2 (Serrate1 and Serrate2 in chicken).
To understand how the functions of multiple Delta-like genes are coordinated during retinal development, we recently investigated how their expression patterns related to the pattern of neural differentiation in the chick. According to a previous model, Delta-like genes should be expressed in the differentiating neurons (Henrique et al., 1997). Surprisingly, we found that Dll1 was expressed in progenitor cells, and while Dll4 was also expressed in some progenitors, the cohort of differentiating newborn neurons primarily expressed Dll4 (Nelson and Reh, 2008). These results suggested that progenitor cells are themselves a primary source of Notch ligands and may mutually inhibit their own differentiation through activation of Notch receptors expressed in neighboring progenitors. Thus, together with lateral inhibition from newborn neurons, the mutual inhibition between progenitors may also be critical to maintain the progenitor pool and coordinate retinal histogenesis (Nelson and Reh, 2008).
From the analysis of developing chick retina, we also described a relationship between Dll1, Dll4, Acheate-scute like 1 (Ascl1) and Neurogenin2 (Neurog2), two vertebrate homologs of Drosophila proneural bHLH genes acheate-scute and atonal, respectively. Dll1, Dll4, Ascl1, and Neurog2 are all expressed in progenitors and exhibit similar expression kinetics during neuronal differentiation (Nelson and Reh, 2008). Moreover, we reported that over-expression of Ascl1 led to an increase in both Dll1 and Dll4 gene expression and Notch signaling activity (Nelson and Reh, 2008). These data suggested that Ascl1 and/or Neurog2 might normally regulate expression of Dll1 and Dll4 in the retina.
Here, we test the hypothesis that Ascl1 and/or Neurog2 are required for Delta-like gene expression and Notch signaling activity during mouse retinal development. We confirm and extend into mouse, our previous observations in the chick, that Delta-like genes are expressed in retinal progenitors, while newborn neurons express Jagged2. We report that Delta-like gene expression is significantly reduced in Ascl1 deficient mouse retinas, and that Ascl1 and Delta-like are co-expressed in progenitors. We also uncovered an additional role for Ascl1 in the regulation of Hes6, a pro-neurogenic factor that functions to promote neural differentiation by inhibiting Notch signaling and glial differentiation. These results establish that a conserved Ascl1/Delta-like/Notch/Hes molecular circuitry operates within the progenitor pool itself to coordinate retinal histogenesis. They also provide a molecular mechanism whereby attenuated Notch levels coupled with reduced proneurogenic activity may lead to increased gliogenesis and decreased neurogenesis in the Ascl1 knockout retina (Tomita et al., 1996b).
Methods
Animals
All mice were housed in the Department of Comparative Medicine and procedures were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of Washington, Seattle, WA. Wildtype C57BL and/or Swiss Webster mice were used for in situ hybridization, immunolabeling, and transfection experiments. Ascl1 knockout animals (B6.129-Ascl1tm1And/J, available from Jackson Labs; Guillemot et al., 1993) and Neurog2 knockout animals on a Swiss Webster background (gift from D. Anderson; Fode et al., 1998) were used for QPCR analysis (and Ascl1 was also used for in situ hybridization). Delta-like 3 pudgy (Dll3pu) mutant mice were obtained from Jackson Labs (stock no. 00306; Kusumi et al., 1998). Animals were genotyped according to their respective protocols (Guillemot et al., 1995; Fode et al., 1998; Kusumi et al., 1998; Hartman et al., 2007). Embryonic age was staged according the morning of vaginal plug date (E0.5) and Theiler Staging criteria. Bromodeoxyuridine (5-bromo-2-deoxyuridine, BrdU, Sigma, 10mg/ml, .15ml/animal) was injected intraperitoneally 2h prior to sacrifice into some animals.
In situ hybridization and immunolabeling
In situ hybridization and post-immunolabeling was performed as previously described (Nelson et al., 2007b). IMAGE clones (Open Biosystems) corresponding to Delta-like 1 (Dll1 (BC057400, IMAGE:6402691), Delta-like 3 (Dll3, BC052002, IMAGE:6404029), and Jagged2 (Jagged2, BC009082, IMAGE:3598850) were used to generate DIG-labeled riboprobes as previously described (Hartman et al., 2007). We also obtained the following IMAGE clones corresponding to Delta-like 4 (Dll4, BC042497, IMAGE:4017786), Hairy and enhancer of split 5 (Hes5, BC103539, IMAGE:40039948), Hairy and enhancer of split 6 (Hes6, BC012897, IMAGE:4011223), and Notch1 (BC010325, IMAGE: 2651506), all of which were confirmed by DNA sequencing. Antibodies used for immunolabeling after in situ hybridization were rat anti-BrdU (1:200 dilution, Accurate Chemical, including 100 Kunitz units/ml DNAse1, Sigma), neuronal specific mouse anti-acetylated beta-III tubulin (Tuj1, 1:750 dilution, Covance), mouse anti-proliferating cellular nuclear antigen (PCNA, 1:100 dilution, DAKO), rabbit anti-phosphorylated Histone H3 (PH3, 1:750, Chemicon), and secondary species specific ALEXA 488 or 568 antibodies (Invitrogen). Colorimetric immuno-detection of Hes1 antigen with rabbit anti-Hes1 (H140; 1:250 dilution, Santa Cruz, Inc.) was performed as described (Hartman et al., 2007). Antibodies used for immunolabeling of cryosections and whole-mount retinas include goat anti-Jagged1 (1:300, Santa Cruz Biotechnology, Jag1 C-20 Cat. No. SC-6011); goat anti-Sox2 (1:300, Santa Cruz, Inc.), rabbit anti-Sox9 (1:300, Chemicon), rabbit anti-GFP (1:1000, U of Alberta, CA), chick anti-GFP (1:300, Abcam), rat anti-beta-galactosidase (LacZ gene product, 1:500 dilution, Saul et al., 2008), mouse anti-Ascl1 (1:100, Chemicon), and were detected with species-specific ALEXA 488/568/594 conjugated secondary antibodies (1:500, Invitrogen). Images were acquired with a Zeiss Axioplan2 epifluorescent microscope equipped with Normarski/DIC optics and a Spot camera, a Zeiss LSM 5 Pascal laser scanning confocal microscope, or Olympus FV1000 multiphoton microscope (MPM). Images were assembled in Adobe Photoshop and Illustrator.
Reporter constructs
We used a mouse Notch1 14.3Kb promoter construct driving GFP to identify Notch1 expressing progenitor cells (Notch1GFP; Lewis et al., 1998), a mouse Hes5 0.76Kb promoter driving destabilized GFP to identify Hes5 and active Notch signaling in progenitor cells (Hes5d2GFP, Takebayashi et al., 1995; Ohtsuka et al., 2006; Nelson et al., 2006), and a mouse Dll1 4.3Kb promoter construct driving LacZ to identify Dll1 expressing progenitor cells (Dll1LacZ; Beckers et al., 2000; Castro et al., 2006). We also created a Dll3 reporter construct, since Ascl1 loss of function especially affected Dll3 expression. To make this construct, we analyzed multiple alignments of vertebrate genomes to identify evolutionarily conserved regions (ECR, ECR Browser, http://ecrbrowser.dcode.org, Ovcharenko et al., 2004), and found one ~400bp proximal ECR with 68.6% identity between mouse and human. Within this ECR lies the Ascl1-specific E-box/octamer motif identified by Castro and colleagues, similar to the Ascl1-specific enhancer in the Dll1 locus (Castro et al., 2006). We used PCR (LA Taq, TaKaRa) to amplify ~1.0Kb of sequence upstream of the mouse Dll3 locus just proximal to the Dll3 start codon containing this ECR. BglII and EcoR1 restriction sites were included in the forward and reverse primer, respectively: BglII forward primer CGCGCGAGATCTTGGGATTACAGGTCTGCCAT, EcoR1 reverse primer CCCGGGGAATTCCAGGATGGGGAAATAGTCTCA. PCR product was restriction enzyme digested, and cloned into the BglII/EcoR1 sites in a destabilized CFP expression plasmid (pd2CFP, Invitrogen) to create Dll3d2CFP, which was verified by DNA sequencing.
Retinal transfection and explant culture
Transfection and explant culture of retinal explants was performed as previously described (Nelson et al., 2006; Nelson et al., 2007b). Briefly, mouse retinas from different embryonic to postnatal ages (E13.5, E17.5-P0) were collected, extra-ocular tissue and retinal pigmented epithelium were removed and transfered to a Milli-cell culture insert (Millipore) in a custom-built electroporation chamber. Explants were electroporated (5 pulses, 35V, 50ms pulse length, BTX ECM830 electroporator) with DNA solutions (~1μl of 2-5μg/μl per explant), and cultured overnight at 37°C 5%CO2 with nutation as described (Nelson et al., 2007b).
QPCR
Quantitave polymerase chain reaction (QPCR) was performed as previously described (Nelson et al., 2006; Nelson et al., 2007a; Nelson et al., 2007b). All primer sequences for QPCR were obtained from PrimerBank (http://pga.mgh.harvard.edu/primerbank, Wang and Seed, 2003), and obtained from Invitrogen. Eye pairs from Ascl1 and Neurog2 P0 animals were individually collected in HBSS+. Extra-ocular tissue, retinal pigmented epithelium, and lens were dissected, and Neurog2 sister retinas were pooled, and lysed in Trizol (Invitrogen). For the Ascl1 eye pairs, one eye was fixed in modified Carnoy's solution and prepared for in situ hybridization as described above, while the sister retina was dissected and lysed in Trizol. Total RNA was extracted, genomic DNA contamination was removed by digesting with RQ1 Rnase-free DNase (Promega), isolated (RNeasy RNA isolation, Qiagen), and converted to cDNA with SuperScriptII reverse transcriptase (RT, Invitrogen), except for RT minus controls. All sample concentrations were normalized to wildtype sibling glyceraldehyde-3-phosphate dehydrogenase (Gapdh) gene expression levels by QPCR with SYBR green PCR master mix (Applied Biosystems) and a DNA Engine Opticon System (Bio-Rad). For statistical analysis, gene expression levels from Ascl1+/+ (n=1) and Ascl1+/− (n=2) littermates were combined (Ascl1+/+,−/−, n=3) and compared to Ascl1−/− (n=5) expression levels. Gene expression levels from Neurog2+/+ (n=4) were compared to Neurog2−/− (n=5) littermates; no statistically significant differences were observed between Neurog2+/+ and Neurog2+/− (data not shown). Student's T-test was used to determine significant differences between samples means, error bars represent standard deviation of the mean, and differences with p≤0.05 were considered significant.
Results
All Delta-like and Jagged genes are expressed in the developing mouse eye
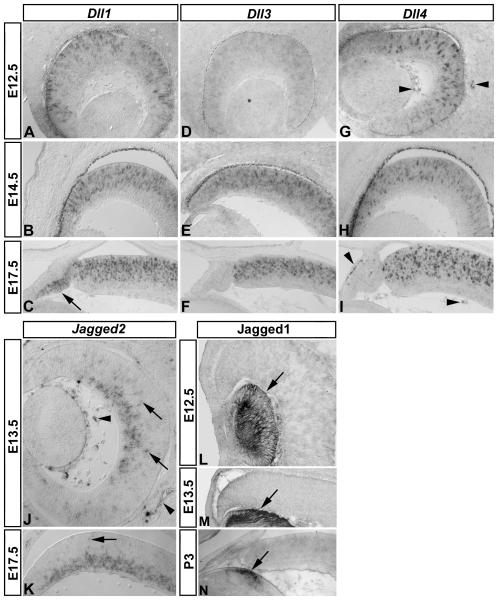
As a first step towards determining the nature of cells expressing Notch ligands in the developing mouse eye, we examined Dll1, Dll3, Dll4, Jagged1, and Jagged2 expression at early-, mid-, and late-stages of embryonic eye development. As a control for probe specificity, we first examined expression patterns for Dll1, Dll3, and Dll4 in the cortex, diencephalon, and rostral spinal cord (Supp Fig 1), which reveal probe specific signals. Within the eye, Dll1, Dll3, and Dll4 expression also exhibited unique expression patterns. At E12.5, both Dll1 and Dll4 were strongly expressed in single cells in the neurogenic region of the presumptive neural retina, compared to weaker Dll3 expression (Fig 1 A, G, D, respectively). Dll4 expression could also be detected in the vasculature (Fig 1 G, arrowheads). At E14.5 (Fig 1 B, E, H) and E17.5 (Fig 1 C, F, I), all Delta-like genes were strongly expressed in the neurogenic zone. Interestingly, at E17.5 Dll1 was also expressed in the peripheral non-neurogenic ciliary epithelium (Fig 1 C arrow). Jagged genes also exhibited unique expression patterns. At both E13.5 (Fig 1 J) and E17.5 (Fig 1 K), Jagged2 expression was strongest in the developing ganglion cell layer (gcl), but occasional cells were observed in the neural progenitor layer (Fig 1 J, K, arrows). Jagged2 was also expressed in the vasculature (Fig 1 J arrowheads), similar to Dll4. Jagged1 immunolabeling revealed expression in the embryonic lens at E12.5, E13.5, and postnatal day 3 (P3, Fig 1 L-N, respectively, arrows). These results confirm and extend earlier reports by showing that not only are all of the canonical Notch ligands expressed in the developing mouse eye, they have both overlapping and unique patterns that change over time.
Figure 1. Notch ligand expression defines discrete regions of the developing mouse eye.
In situ hybridization was used to detect Dll1 (A-C), Dll3 (D-F), Dll4 (G-I), Jagged2 (J, K), and immunolabeling was used to detect Jagged1 (L-N) expression at the indicated early stages of mouse eye development. Dll1 (A-C) and Dll4 (G-I) are strongly expressed in cells restricted to the central neurogenic region of the presumptive retina at embryonic day 12.5 (E12.5), E14.5, and E17.5: note that Dll1 expression is upregaluted in the peripheral non-neurogenic ciliary epithelium at E17.5 (C, arrow), and that Dll4 is expressed in the vasculature (G, I, arrowheads). Weak Dll3 expression at E12.5 (D) becomes upregulated by E14.5 (E), and within the developing retina, all Delta-like genes are restricted to the neuroblast layer (Dll1 B, C; Dll3 E, F; Dll4 H, I). Jagged2 (J, K) expression is restricted to the developing ganglion cell layer of the neurogenic zone, and in isolated cells in the neuroblast layer (arrows), as well as extra-ocular vasculature and cells in the vitreous (arrowheads), similar to Dll4. Jagged1 (L-N) immunolabeling was restricted to the lens at these ages (arrows).
Differentiating neurons express Jagged2
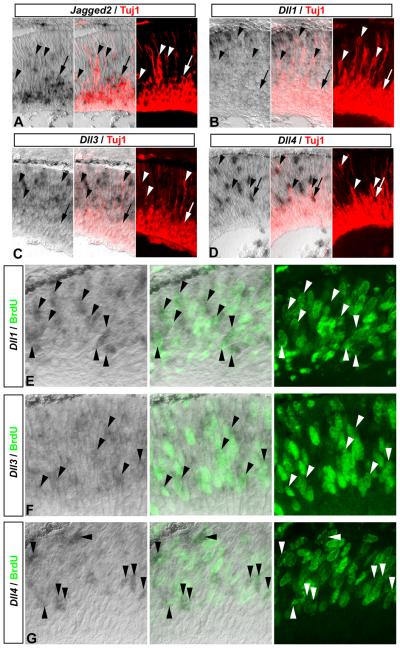
To determine the identity of Dll1, Dll3, Dll4, and Jagged2 cells in the neurogenic zone, we combined in situ hybridization with immunolabeling for neuronal-specific acetylated beta-III tubulin (Tuj1). When we analyzed the expression of Notch ligands with respect to differentiating neurons at E14.5, we found that strongest Jagged2 expression was localized to Tuj1+ neurons in the ganglion cell layer (gcl), and could even be detected in migrating Tuj1+ neurons (Fig 2 A, arrow and arrowheads, respectively). However, Dll1, Dll3, or Dll4 expression was not observed in Tuj1+ neurons in the ganglion cell layer, or in newborn Tuj1+ neurons in the progenitor zone (Fig 2 B-D, arrowheads and arrow, respectively). These data indicate that differentiating neurons express Jagged2 but not Delta-like genes.
Figure 2. Newborn neurons and proliferating progenitors express different Notch ligands.
(A-D) Jagged2 is expressed in postmitotic differentiating neurons. In situ hybridization for Jagged2 (A), Dll1 (B), Dll3 (C), and Dll4 (D) combined with acetylated beta-III tubulin immunolabeling (Tuj1, red), reveals that Jagged2, but not Delta-like genes, is expressed in Tuj1+ postmitotic differentiating neurons migrating to (arrowheads), and within the ganglion cell layer (arrow) at E14.5: note that the Dll4+ cell near the ganglion cell layer in D is not Tuj1+ (arrow). (E-G) Delta-like genes are expressed in retinal progenitor cells. E14.5 embryos received a 2h pulse of BrdU in utero prior to sacrifice, and in situ hybridization was used to detect Dll1 (E), Dll3 (F), and Dll4 (G) expression, followed by immunolabeling to detect BrdU incorporation (green). Individual and merged channels are shown, which reveal that Dll1 (E), Dll3 (F), and Dll4 (G) expressing cells at E14.5 can incorporate BrdU (arrowheads).
Progenitor cells express Delta-like genes
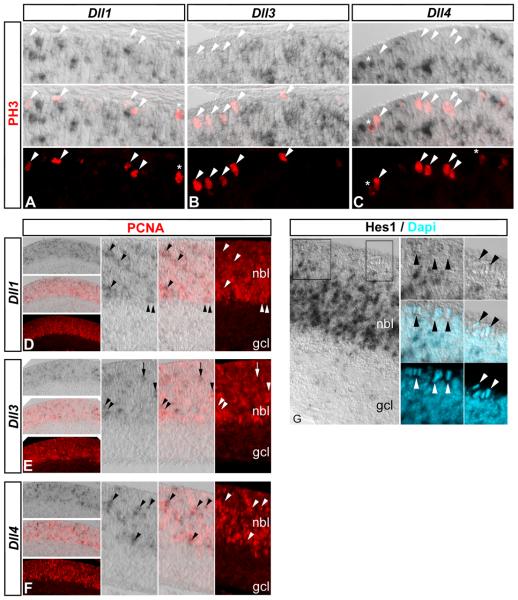
At the early stages of retinal development, most cells in the neurogenic region are either Tuj1+ neurons or Tuj1− progenitors, and the above analysis suggests the Delta-like expressing cells are actually progenitors. To confirm Delta-like genes are expressed in retinal progenitor cells, we first used in situ hybridization to detect Dll1, Dll3, or Dll4 expression coupled with immunolabeling for BrdU incorporation to detect S-phase progenitors. E14.5 and E17.5 embryos received a 2h pulse of BrdU in utero prior to sacrifice, and were prepared for in situ hybridization (Nelson et al., 2007b). At E14.5, analysis of Dll1, Dll3, and Dll4 expressing cells revealed that many had incorporated BrdU (Fig 2 E-G, arrowheads). Similarly, at E17.5, many Dll1, Dll3, and Dll4 expressing cells were found that had also incorporated BrdU (Supp Fig 2). We also noticed that Dll1, Dll3, and Dll4 cells were located at the apical surface, suggesting that they may be in M-phase; however, at E17.5, phospho-histone 3 (PH3) immunolabeling revealed few Dll1 and Dll4 cells that were PH3+ (Fig 3 A, C, asterisks, respectively). Thus, the majority of progenitors undergoing mitosis do not express Delta-like genes (Fig 3 A-C, arrowheads). Nevertheless, immunolabeling for proliferating cellular nuclear antigen (PCNA), a marker present throughout the progenitor cell cycle, revealed strong correlation between Delta-like gene expression and PCNA labeling at E18.5 (Fig 3 D-F, arrowheads), although we did observe occasional Dll3+ cells that were not PCNA+ (Fig 3 E, arrow). Hes1 immunolabeling at E17.5 confirmed that Notch signaling activity is normally lowest in mitotic progenitors at the apical surface, and highest in progenitors located in the neuroblast layer (Fig 3 G, arrowheads; Nelson et al., 2007a). These results indicate that progenitor cells in the retina are themselves the primary source of Delta-like genes, and likely regulate Notch signaling within their own pool; however, they may also receive lateral inhibitory signals via Jagged2 from differentiating neurons.
Figure 3. Delta-like gene expression and Notch signaling change during progenitor cell cycles.
In situ hybridization was used to detect Dll1 (A, D), Dll3 (B, E), and Dll4 (C, F) gene expression followed by phospho-histone H3 (PH3, E17.5, A-C, red) or proliferating cellular nuclear antigen (PCNA, E18.5, D-F, red) immunolabeling. (A-C) The majority of PH3+ mitotic progenitors at the apical surface do not express Delta-like genes (A-C, arrowheads): asterisks denote a few double-positive cells with weak Dll1, Dll4, and/or PH3 labeling. (D-F) Delta-like gene expression is restricted to the neuroblast layer (nbl) defined by PCNA immunolableing in contrast to the ganglion cell layer (gcl), and higher magnification views reveal that Delta-like genes are expressed in PCNA+ progenitors (arrowheads): note that one Dll3 cell is not labeled with PCNA (E, arrow). (G) Hes1 immunolabeling is also restricted to the region of active Notch signaling in progenitors in the nbl and not in the gcl: note that Hes1 protein is not detected in mitotic progenitors at the apical surface (arrowheads, Dapi counterstain, blue), similar to the pattern observed for Delta-like genes (A-C) and active Notch signaling (Nelson et al., 2006; Nelson et al., 2007a).
Ascl1 regulates Delta-like gene expression and Notch signaling activity
When we investigated the relationship between Delta-like and proneural bHLH genes during chick retinal development, we found that of all of the proneural bHLH genes, both Ascl1 and Neurog2 were expressed in progenitors that may also express Delta-like genes; however, only Ascl1 was found to upregulate Delta-like gene expression and Notch signaling activity in a gain-of-function assay (Nelson and Reh, 2008). To determine whether Ascl1 or Neurog2 are required for Delta-like gene expression, we used QPCR to measure the level of expression of Notch pathway components in the retinas from Ascl1 and Neurog2 mutant mice. Both Ascl1 and Neurog2 null (Ascl1−/− or Neurog2−/−) animals die at birth (Guillemot et al., 1995; Fode et al., 1998), so we collected animals at P0 and prepared one or both retinas from individual animals for QPCR (Ascl1 and Neurog2, respectively). We chose this age for our molecular analyses because the reported phenotype of the loss of Ascl1 in the retina arises after birth (Tomita et al., 1996b), and we did not want to introduce molecular changes that might arise due long-term explant cultures. QPCR analysis of Ascl1 and Neurog2 gene expression levels in retinas from newborn Ascl1 and Neurog2 +/+, +/−, and −/− animals, respectively, confirmed the changes predicted from genotyping (data not shown).
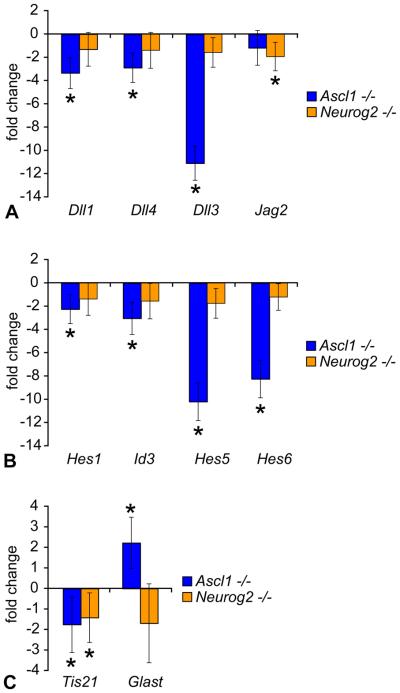
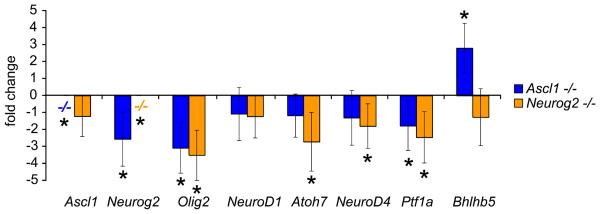
To test whether Ascl1 or Neurog2 regulated Notch ligands and Notch signaling, we first assayed for changes in expression of the Notch ligands Dll1, Dll3, Dll4, and Jagged2. QPCR analysis revealed that Dll1, Dll4, and particularly Dll3 gene expression levels were significantly decreased with loss of Ascl1, but not with loss of Neurog2 (Fig 4 A). By contrast, a small, but significant decrease in Jagged2 expression was detected in Neurog2 deficient, but not in Ascl1 deficient retinas (Fig 4 A). Decreased Notch ligand expression would predict a concomitant decrease in Notch signaling activity. To test whether Notch signaling activity was decreased as well, we measured expression levels of Notch target genes Hes1, Hes5, and Id3 (Nelson et al., 2007a). Hes1, Id3, and particularly Hes5 gene expression levels were significantly decreased with loss of Ascl1, but were unchanged in Neurog2 deficient reitnas (Fig 4 B).
Figure 4. Delta-like gene expression and Notch signaling are downregulated in Ascl1, but not Neurog2, deficient retina.
(A-C) QPCR analysis of Ascl1 and Neurog2 mutant retina. Individual retinas from postnatal day 0 (P0) Ascl1 and Neurog2 mice were prepared for QPCR analysis (see methods), and sample concentrations were normalized to Gapdh gene expression levels in sibling controls. Graphs depict fold change in expression levels of the indicated genes between Ascl1−/− (n=5) and littermate control Ascl1+/+,+/− (n=3) retinas (blue), and Neurog2−/− (n=5) and littermate control Neurog2+/+ (n=4) retinas (orange, error bars represent standard deviation of the mean, student's T-test was used to determine significant differences between sample means, and changes with p≤0.05 were considered significant (asterisks). Changes in gene expression levels of the Notch ligands Dll1, Dll4, Dll3, and Jagged2 (A); the Notch targets Hes1, Id3, Hes5, and the proneural target Hes6 (B); and neurogenic versus gliogenic progenitor markers Tis21 and Glast, respectively (C) due to loss of Ascl1 or Neurog2 function are depicted.
To determine whether Neurog2 contributed any role to this molecular circuitry, we analyzed Hes6 gene expression levels, a known target of Neurog2 in the spinal cord involved in a negative feedback inhibitory loop with Hes5 to regulate Notch signaling (Fior and Henrique, 2005). Surprisingly, Hes6 gene expression was strongly decreased with loss of Ascl1, rather than Neurog2 function (Fig 4 B). To determine whether loss of either Ascl1 or Neurog2 affected progenitor neural differentiation, we measured levels of Tis21, a gene that marks progenitors biased towards neurogenic divisions (Iacopetti et al., 1999; Attardo et al., 2008). Tis21 gene expression was decreased with loss of either Ascl1 or Neurog2 function (Fig 4 C). To determine whether the decreased bias towards neural differentiation resulted in an increased bias towards progenitor/glia differentiation, we measured Glast expression levels, a marker of progenitors at this age, and a Muller Glia marker later (Gotz and Huttner, 2005). Glast was significantly upregulated in the Ascl1 deficient mouse retina, but was unchanged in retinas lacking Neurog2 (Fig 4 C). These data indicate that Ascl1, rather than Neurog2, plays the primary role in regulating Notch ligands and Notch signaling in retinal progenitors, and biases them towards neural differentiation by regulating the proneurogenic factor Hes6.
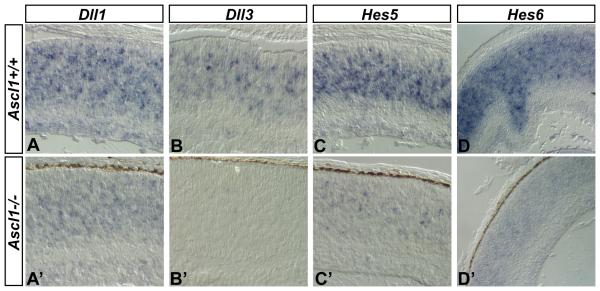
To confirm the changes in expression of Notch pathway components due to loss of Ascl1 function, we used in situ hybridization to visualize expression levels of Dll1, Dll3, Hes5, and Hes6 genes in the sister Ascl1+/+ and Ascl1−/− retinas. Neighboring sections from Ascl1+/+ (Fig 5 A-D) and Ascl1−/− retinas (Fig 5 A′-D′) were hybridized with Dll1, Dll3, Hes5, and Hes6 riboprobes, respectively, incubated in equal amounts of substrate, and developed for equivalent periods of time. Comparison of Dll1 (Fig 5 A, A′), Dll3 (Fig 5 B, B′), Hes5 (Fig 5 C, C′), and Hes6 (Fig 5 D, D′) gene expression levels demonstrates that these genes are downregulated with loss of Ascl1 function, confirming the gene expression changes quantified in the sister retinas by QPCR analysis. Thus, Ascl1 is required for normal levels of Delta-like gene expression and Notch signaling activity in the developing mouse retina.
Figure 5. Visualization of key downregulated genes in Ascl1 deficient retina.
(A-D) In situ hybridization was used to visualize changes in Dll1, Dll3, Hes5, and Hes6 gene expression levels between Ascl1+/+ (A-D) and Ascl1−/− (A′-D′) in the sister P0 eyes from the previous QPCR analysis. Dll1, Dll3, Hes5, and Hes6 gene expression levels are decreased in the Ascl1−/− retina compared to normal expression levels observed in the Ascl1+/+ retina.
Delta-like genes and Notch signaling components are expressed in Ascl1 retinal progenitors
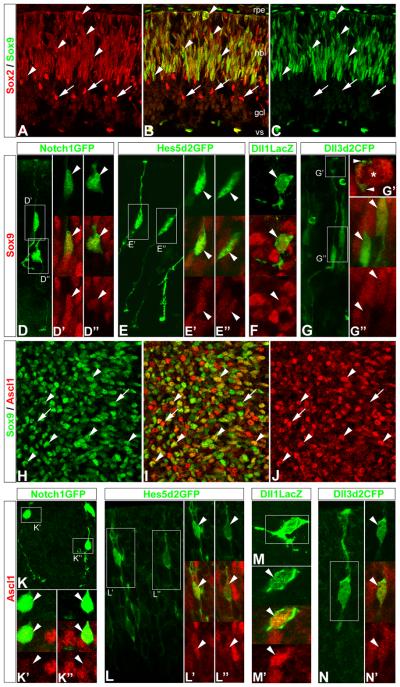
Since reliable reagents for immunolabeling most Notch pathway components are not available, we used reporter plasmids containing cis-regulatory elements from Notch1, Hes5, Dll1, and Dll3 to drive expression of fluorescent proteins (GFP/CFP) or beta-galactosidase (LacZ gene product) to further investigate the relationship between Delta-like genes, Notch signaling components, and Ascl1 expression in retinal progenitor cells Previous studies have demonstrated the specificity of these cis-regulatory elements, and we provide further documentation in Supp Fig 3 (Notch1GFP, Lewis et al., 1998), (Hes5d2GFP, Takebayashi et al., 1995; Ohtsuka et al., 2006; Nelson et al., 2006), (Dll1LacZ, Beckers et al., 2000; Castro et al., 2006), (Dll3d2CFP, this report, see methods). We used electroporation to transfect Notch1GFP, Hes5d2GFP, Dll1LacZ and Dll3d2CFP into embryonic retinal explants and cultured them overnight (≤24h) to allow reporter expression (Nelson et al., 2007b). High-resolution laser scanning confocal microscopy (LSCM) revealed that cells expressing Notch1GFP, Hes5d2GFP, Dll1LacZ, or Dll3d2CFP reporters in intact retinas typically have both an apical and basal process, indicative of progenitor cells (Supp Fig 3). We also noticed a surprising degree of morphological complexity in regards to the presence of multiple, short extensions and varicosities often observed along both apical and basal processes (Supp Fig 3). To confirm that Notch1GFP, Hes5d2GFP, Dll1LacZ, and Dll3d2CFP reporters are active in progenitor cells, we used antibodies to Sox2 and Sox9 to immunolabel retinal progenitors (Le Rouëdec et al., 2002; Taranova et al., 2006; Sakami et al., 2008; Moshiri et al., 2008; this report), along with BrdU incorporation. Immunolabeling of E17.5 retinal sections revealed that Sox2 and Sox9 identify the vast majority of the progenitor pool (Fig 6 A-C). Transfection of Notch1GFP, Hes5d2GFP, Dll1LacZ, and Dll3d2CFP reporters into E17.5 retinas revealed that transfected cells were also immunolabeled with the Sox9 antibody (Fig 6 D-G arrowheads) and could incorporate BrdU in a 2h pulse (data not shown). We next confirmed that Ascl1 is expressed in progenitor cells (Jasoni et al., 1994; Jasoni and Reh, 1996; Nelson and Reh, 2008) by labeling retinas with antibodies against Ascl1 and Sox9 (Fig 6 H-J), and then tested whether Dll1 and Dll3 are expressed in the Ascl1 progenitors by transfecting E17.5 retinas with the reporter constructs and immunolabeling with Ascl1 antibodies. We found that cells expressing either Dll1LacZ, and Dll3d2CFP were also labeled with Ascl1 (Fig 6 M,N); moreover, both Notch1GFP and Hes5d2GFP reporters were also expressed in Ascl1 labeled cells (Fig 6 K,L). These data altogether show that Delta-like genes and Notch signaling components are expressed in Ascl1 expressing retinal progenitors.
Figure 6. Delta-like and Notch pathway reporters are active in Ascl1 expressing retinal progenitor cells.
(A-C) Laser scanning confocal microscopy (LSCM) was used to image E17.5 retinal section co-immunolabeled with antibodies to Sox2 (A, B red) and Sox9 (B, C green). Sox2 and Sox9 co-expression identify the vast majority of the progenitor pool in the neuroblast layer (nbl, arrowheads), although some postmitotic amacrine cells retain high expression of Sox2 (arrows): note that Sox9 is also expressed in the retinal pigmented epithelium (rpe, top), both Sox2 and Sox9 are expressed in cells along the inner/vitreal surface (vs) outside of the ganglion cell layer (gcl), and that Sox9 expression levels in retinal progenitors are observed at low or high levels. (D-G) LSCM was used to image E17.5 wholemount mouse retinas transfected with Delta-like and Notch pathway reporters, and co-immunolabeled with antibodies to GFP or LacZ, and Sox9. LSCM was used to visualize transfected cells in their intact environment, either in transverse orientation with apical and basal processes located at the top and bottom, respectively (D, E, G), or enface (top-down, F). (D) Maximum intensity projection (MIP) of neighboring Notch1GFP transfected cells (green) that are Sox9+ (red): D′, high Sox9, D″ lower Sox9, arrowheads; single optical slices showing individual and merged channels, respectively. (E) MIP of neighboring Hes5d2GFP transfected cells (green) that are also Sox9+ (red): E′ high Sox9, E″, lower Sox9, arrowheads; single optical slices showing individual and merged channels, respectively. (F) Enface view of a Dll1LacZ transfected cell (green) that is Sox9+ (red, arrowhead): single optical slices showing individual and merged channels, respectively. (G) Single optical slice of neighboring Dll3d2CFP transfected cells (green) that are Sox9+ (red): G″ high Sox9, top arrowhead; low Sox9, bottom arrowhead; single optical slices showing individual and merged channels, respectively: note that some basal progenitors can make extensive contact with apical progenitors at the ventricular surface (G′ arrowheads and asterisk, respectively). (H-J) LSCM was used to image E17.5 whole-mount mouse retinas co-immunolabeled with antibodies to Sox9 (green) and Ascl1 (red). Enface view of single optical slice through the neuroblast layer identifies many progenitors that co-express Sox9 and Ascl1 (arrowheads), and subsets of progenitors that express higher-to-lower levels of either Sox9 or Ascl1, respectively (arrows). (K-N) LSCM was used to image E17.5 whole-mount mouse retinas transfected with Delta-like and Notch pathway reporters, and co-immunolabeled with antibodies to GFP or LacZ, and Ascl1. LSCM was used to visualize transfected cells in their intact environment, either in transverse orientation with apical and basal processes located at the top and bottom, respectively (K, L, N), or enface (M). (K) MIP of neighboring Notch1GFP transfected cells (green) that are Ascl1+ (red): K′, low Ascl1, K″ higher Ascl1, arrowheads; single optical slices showing individual and merged channels, respectively. (L) MIP of neighboring Hes5d2GFP transfected cells (green) that are also Ascl1+ (red): L′ higher Ascl1, L″, lower Ascl1, arrowheads; single optical slices showing individual and merged channels, respectively. (M) Enface MIP view of a Dll1LacZ transfected cell (green) that is Ascl1+ (red, arrowhead): M′ single optical slices showing individual and merged channels, respectively. (N) MIP of a Dll3d2CFP transfected cell (green) that is Ascl1+ (red, arrowhead): N″ single optical slices showing merged and individual (red) channels, respectively; note that N′ depicts a Dll3d2CFP+ progenitor contacting a Sox9+ mitotic progenitor at the ventricular surface.
Ascl1 and Neurog2 are upstream proneural bHLH transcription factors
Synchronizing progenitor cell differentiation by timing Notch signaling inactivation revealed that a transient and sequential cascade of proneural bHLH transcription factors underlies the progenitor neural differentiation program (Nelson et al., 2007a; Nelson and Reh, 2008). For example, an immediate transient wave of increased Ascl1 and Neurog2 expression is observed in progenitors due to a rapid loss of Notch activity that precedes the later transient increases observed in downstream proneural bHLH genes such as NeuroD4, NeuroD1, and Atoh7, which are normally expressed in differentiating, postmitotic neurons (Nelson et al., 2007a; Nelson and Reh, 2008).
To determine whether the loss of Ascl1 or Neurog2 affects other downstream proneural bHLH transcription factors, we quantified differences in expression levels of Ascl1, Neurog2, Olig2, NeuroD1, Atoh7, NeuroD4, Ptf1a, and Bhlhb5 between Ascl1 or Neurog2 deficient retinas using QPCR as described above. As expected Ascl1 and Neurog2 expression were not detected in Ascl1 or Neuorg2 deficient retinas, respectively (Fig 7). Ascl1 expression levels were unchanged in Neurog2 deficient retinas (Fig 7). By contrast, Neurog2 expression was decreased in Ascl1 deficient retinas (Fig 7). Olig2 expression levels were decreased in both Ascl1 and Neuorg2 deficient retinas (Fig 7). NeuroD1 expression levels were unchanged (Fig 7). Atoh7, NeuroD4, and Ptf1a expression was downregulated in Neurog2 deficient retinas, but only Ptf1a expression was decreased in Ascl1 deficient retinas (Fig 7). Bhlhb5 expression was increased with loss of Ascl1 function, but was unchanged in Neurog2 deficient retinas (Fig 7). Upregulation of other Ascl1 and Neuorg2 bHLH family paralogs normally not expressed in the retina was not observed (data not shown). Thus, loss of the upstream Ascl1 or Neurog2 bHLH transcription factors can affect expression of other downstream bHLH transcription factors (Akagi et al., 2004; Cho et al., 2007), and is consistent with their placement, particularly Ascl1, at the top of a molecular hierarchy regulating neural differentiation in the retina.
Figure 7. Ascl1 and Neuorg2 are upstream bHLH transcription factors.
QPCR was used to measure changes in other bHLH transcription factor family members expressed during retinal development due to loss of Ascl1 or Neurog2 (P0, see Figure 4, and Methods). Ascl1 was not expressed in the Ascl1−/− retina, and was not changed with loss of Neurog2 function. Likewise, Neurog2 was not expressed in the Neurog2−/− retina, however Neurog2 expression was decreased in the Ascl1−/− retina. Olig2 expression was decreased in both Ascl1 and Neurog2 knockout retina. NeuroD1 expression was not changed in either Ascl1 or Neurog2 knockout retina. Atoh7 and NeuroD4 expression were decreased in the Neurog2−/− retina, but not changed in the Ascl1−/− retina. Ptf1a expression was decreased in both Ascl1−/− and Neurog2−/− retina. Bhlhb5 expression was increased in the Ascl1−/− retina, but was not changed in the Neurog2−/− retina. These changes are consistent with the roles of Ascl1 in particular, as well as Neurog2, as upstream bHLH transcription factors expressed in progenitor cells that regulate a downstream proneural bHLH transcription factor cascade underlying the transition to differentiating neurons (Nelson et al., 2007a).
Discussion
We report here that progenitor cells in the mammalian retina are the primary source of Delta-like expression, and likely regulate Notch signaling among themselves. The progenitors may also receive lateral inhibitory signals via Jagged2 from differentiating neurons. We also found that loss of Ascl1 function down regulates Delta-like gene expression and Notch signaling in progenitors. These data together with our previous studies (Nelson et al., 2006; Nelson et al., 2007a; Nelson and Reh, 2008), demonstrate that a conserved Ascl1/Delta-like/Notch/Hes molecular circuitry operates within the progenitor pool itself to coordinate retinal histogenesis. This conserved Ascl1 function, combined with its proneural role, may be one molecular mechanism that underlies the phenotype observed in the Ascl1 mutant retina: a loss of late arising neurons with a concomitant increase in gliogenesis (Tomita et al., 1996b).
Mouse retinal progenitor cells express Delta-like genes
Within the neurogenic domain of the developing mouse retina, we found that Delta-like genes were expressed in progenitors throughout the embryonic period of histogenesis. Cells expressing Dll1, Dll3, or Dll4 genes were not labeled with Tuj1, which marks differentiating neurons, but rather incorporated BrdU (Fig 2, Supp Fig 2) and were immunoreactive for PCNA (Fig 3), identifying them as progenitors. Using a different approach, based on plasmids expressing reporter molecules under control of cis-regulatory elements from Dll1 and Dll3 (as well as Notch1 and Hes5), we found that Dll1+ and Dll3+ cells were progenitors based on morphology (Supp Fig 3) and immunoreactivity for Sox9 and Ascl1 (Fig 6) and BrdU incorporation (data not shown). By contrast, Jagged2 expression was restricted to the inner region of the Tuj1+ ganglion cell layer, and in scattered Tuj1+ cells in the outer neuroblast region (Fig 2), which are likely newly generated ganglion or amacrine cells migrating to their final position. Thus, mouse retinal progenitor cells are the source of Delta-like genes and likely mutually inhibit themselves, while newborn neurons express Jagged2 that may feedback to laterally inhibit neighboring progenitors, and function together to maintain the progenitor pool and regulate retinal histogenesis.
The notion that Delta-like genes are expressed in retinal progenitor cells in both chick and mouse (Nelson and Reh, 2008; this report) shows that some, and perhaps most, of the Notch signaling in the developing retina works through a “mutual inhibition” rather than a “lateral inhibition” mechanism, as is observed in the developing proneural clusters of Drosophila. Although this concept is novel to the vertebrate retina, Muskavitch and colleagues proposed a similar model for the Drosophila eye imaginal disc over a decade ago. In this system, the majority of undifferentiated cells in the morphogenetic furrow initially express Delta, suggesting that rather than a lateral inhibitory signal from the initial differentiated photoreceptor (R8), Delta-Notch functions in a mutual inhibitory manner to prevent them from responding to the initial neural inductive cue and maintain their uncommitted state (Parks et al., 1995). The reliance on mutual inhibition of differentiation may thus represent a conserved mechanism for patterning larger neurogenic epithelia in which not all of the progenitor cells can maintain contact with the differentiating neuroblast, as they do in the proneural clusters.
Although our studies in both chick and mouse show that vertebrate retinal progenitor cells express Notch ligands, there are differences between mouse and chick. First, the chick retina does not express Jagged (Serrate) genes (Myat et al., 1996; Hayashi et al., 1996). Second, the mammalian-specific ligand Dll3 is expressed in the mouse retina (this report), while the chicken genome does not have a Dll3 homolog (Pintar et al., 2007; Nelson and Reh, 2008). Third, newborn neurons in the chick express Dll4, whereas newborn neurons in the mouse express Jagged2 (Nelson and Reh, 2008; this report), which may be a consequence of the more limited repertoire of available Notch-ligands in the chick retina compared to the mouse retina. Nevertheless, one emerging theme from these studies is that within the neurogenic zone of the developing retina, different cell types express different Notch-ligands specifically during the transition from progenitor to differentiating neuron.
This theme may extend into the progenitor pool, particularly in mammals, since all three Delta-like genes are expressed in progenitors. These results raise an interesting possibility that different Delta-like genes may sort out different subpopulations, or mark different steps in the transition from multipotency to neurogenic dividing progenitors in the retina. In this regard, it is interesting that occasional Dll3 expressing cells were not labeled with progenitor markers (PCNA); Dll3 is not able to activate Notch signaling, and may even attenuate Notch signaling levels (Ladi et al., 2005; Geffers et al., 2007). These data suggest that Dll3 may be involved in the terminal neurogenic step of progenitor differentiation. However, we did not observe any major phenotype in retinas deficient for Dll3 (data not shown), similar to our observations in the cochlea (Hartman et al., 2007). Dll3 may mark progenitors that will undergo a neurogenic division like the neurogenic progenitor marker Tis21 (Iacopetti et al., 1999; Attardo et al., 2008), although live-cell imaging would be necessary to test this hypothesis. Nevertheless, our demonstration that retinal progenitors from both chick and mouse are a source of Notch-ligands (this report; Nelson and Reh, 2008), together with recent studies showing that mouse cortical progenitors themselves are also a source of Notch-ligands (Shimojo et al., 2008; Yoon et al., 2008; Kawaguchi et al., 2008a; Kawaguchi et al., 2008b), change the view of Notch signaling in the vertebrate nervous system.
Conserved Ascl1/Delta-like/Notch/Hes molecular circuitry in retinal progenitors
Another aspect of Notch signaling in the retina that has received little attention is the factor(s) that regulate Delta-like gene expression. Studies of neurogenesis in Drosophila demonstrate that Delta is regulated by proneural bHLH transcription factors such as acheate-scute or atonal (reviewed by Skeath and Carroll, 1994; Bertrand et al., 2002). Many bHLH homologs are expressed in the vertebrate retina, but only Ascl1 and Neurog2 are expressed in mitotically active progenitors (Jasoni et al., 1994; Jasoni and Reh, 1996; Perron et al., 1998; Yan et al., 2001; Marquardt, et al., 2001; Ma and Wang, 2006; Le et al., 2006; Nelson and Reh, 2008). While the functions of proneural bHLH genes have been extensively investigated with respect to their role in retinal cell fate specification (reviewed by Cepko, 1999; Vetter and Brown, 2001; Hatakeyama and Kageyama, 2004; Yan et al., 2005; Ohsawa and Kageyama, 2007; Harada et al., 2007), their function in the regulation Notch ligands have not been described in the developing retina. Recently, we found in the developing chick retina that the expression pattern and expression kinetics of Dll1 during synchronized progenitor differentiation most closely matched that of Ascl1, similar to observations in the frog retina (Perron et al., 1998). Although these patterns suggested a specific role for Ascl1 in Dll1 regulation, over-expression of Ascl1, but not Neurog2, upregulated both Dll1 and Dll4, as well as Hes5 gene expression levels, indicating that Ascl1 may play a more general role in regulating Delta-like genes and Notch signaling activity in the retina (Nelson and Reh, 2008).
In this report, we analyzed retinas from Ascl1 or Neurog2 mutant mice for changes in Delta-like gene expression and Notch signaling. Dll1, Dll4, and particularly Dll3 gene expression levels were significantly downregulated in Ascl1−/− mice, but not in Neurog2−/− mice. These results demonstrate that Ascl1 does have a conserved regulatory input into Delta-like gene expression and Notch signaling activity, although there must be additional positive regulators of at least Dll1 and Dll4 expression in the mouse retina. Nevertheless, decreased expression of Delta-like genes predicts decreased levels of Notch signaling activity. In support of this notion, the classic Notch target genes Hes1, Id3, and particularly Hes5 were all downregulated in Ascl1−/− mice, indicative of decreased Notch signaling activity. These results are complementary to our over-expression experiment in the chick retina (Nelson and Reh, 2008). These results are also consistent with the well-documented role and pattern of Ascl1 regulation of Delta-like gene expression and Notch signaling in other regions of the mammalian nervous system (Ma et al., 1997; Casarosa et al., 1999; Beckers et al., 2000; Cau et al., 2002; Yun et al., 2002; Mizuguchi et al., 2006; Wildner et al., 2006; Castro et al., 2006). Neurog2 also has well-documented roles in regulating Delta-like gene expression (Ma et al., 1996; Fode et al., 1998; Beckers et al., 2000; Bertrand et al,. 2002; Castro et al., 2006); however, it is not required for Delta-like gene expression in the retina. Interestingly, regions of high Jagged expression are correlated with Neurogenin expression in the CNS (Ma et al., 1997), and we did observe decreased Jagged2 expression in the Neurog2, but not Ascl1, mutant retina.
Levels of Notch signaling regulate maintenance of the progenitor pool
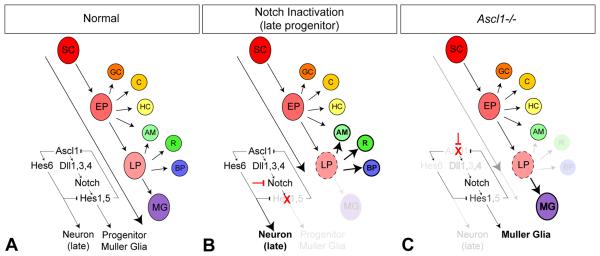
Our data show that the loss of Ascl1 leads to a decrease in Notch signaling. Previous analyses of loss of Notch signaling during retinal development have consistently shown increased neural differentiation and inhibition of progenitors/gliogenesis (Dorsky et al., 1995; Austin et al., 1995; Tomita et al., 1996a; Henrique et al., 1997; Dorsky et al., 1997; Furukawa et al., 2000; Hojo et al., 2000; Satow et al., 2001; Silva et al., 2003; Takatsuka et al., 2004; Nelson et al., 2006; Jadhev et al., 2006; Yaron et al., 2006; Nelson et al., 2007a). Notch inactivation in early embryonic progenitors increases early retinal neurons (Nelson et al., 2007a), whereas Notch inactivation in later postnatal progenitors increases differentiation of late retinal cell types and decreases glial differentiation (Fig 8 A normal, Fig 8 B Notch inactivation in LP; Nelson et al., 2007a). By contrast, loss of Ascl1 results in decreased neural differentiation of retinal cell types, such as rods and bipolar cells in particular, with a concomitant increase in gliogenesis (Fig 8 C; Tomita et al., 1996b; Tomita et al., 2000 Hatakeyama et al., 2001). How might these opposing Notch and Ascl1 phenotypes be resolved? One answer may lie in the fact that Hes6 gene expression is strongly downregulated with loss of Ascl1 function. Hes6 is regulated by upstream proneural bHLH genes, and is sufficient to promote neural and prevent glia differentiation (Bae et al et al., 2000; Koyano-Nakagawa et al., 2000; Gratton et al., 2003; Fior and Henrique, 2005; Jhas et al., 2006). Thus, decreased expression of pro-neurogenic Hes6 coupled with attenuated Notch signaling levels in the progenitor pool provides one molecular mechanism that may explain the observed increase in gliogenesis in the Ascl1 deficient retina (Fig 8 C). In support of this view, although the cellular phenotype of decreased neurogenesis and increased gliogenesis arises somewhat later in the postnatal Ascl1 mutant retina, we can already detect molecular changes indicative of this switch at birth, with reduced Tis21 and increased Glast expression levels in Ascl1 mutant retina.
Figure 8. Summary of Ascl1 function in the conserved Delta-like/Notch/Hes molecular circuitry, and comparison of retinal phenotypes due to loss of Notch versus Ascl1 activity.
(A) Model of vertebrate retinal development under normal conditions (adapted from Reh and Fischer, 2006). Ascl1 contributes to the regulation of Delta-like genes to activate Notch signaling. Ascl1 also drives proneurogenic Hes6, which functions to inhibit Notch signaling, creating a balance between neural differentiation and maintenance of progenitors. (B) Inactivating Notch signal transduction in late progenitors (LP, broken circle) blocks Hes1/5 gene function, and increases Ascl1/Hes6 activity, shifting the balance to force differentiation of later born neurons at the expense of Muller glia (Nelson et al., 2007a). (C) By contrast, loss of Ascl1 decreases Delta-like gene expression and attenuates Notch activity in progenitors. However, the balance of neurogenic versus gliogenic differentiation signals shift in the LP (broken circle) towards gliogenesis due to downregulation of proneurogenic Hes6 and other upstream proneural differentiation functions of Ascl1, while residual lower-levels of Notch activity (Hes1) suffice to generate Muller glia. Hence Muller glia are increased at the expense of later born neurons with loss of Ascl1 (Tomita et al., 1996). Abbreviations: SC, stem cell; EP, early progenitor; LP, late progenitor; GC, ganglion cell; C, cone; HC, horizontal cell; AM, amacrine; R, rod; BP, bipolar cell; MG, Muller glia.
Taken together with previous studies, our data show that an Ascl1/Delta-like/Notch/Hes molecular circuitry regulates neurogenesis. In vertebrates, this pathway regulates retinal histogenesis by coordinating progenitor differentiation, and in higher order vertebrates, this circuitry is used to maintain the progenitor pool during their protracted period of retinogenesis. The fact that this molecular circuit operates within progenitor cells themselves shines new light into how vertebrate visual systems normally develop, and provides a simple framework for understanding neurogenesis in the retina. Additionally, recent findings indicate that this normally quiescent developmental circuit is reactivated following damage in adult retinas, regulating the capacity for retinal regeneration in fish and chick (Fischer and Reh, 2001; Hayes et al., 2007; Fausett et al., 2008). Since retinal regeneration is more limited in mammals (see Lamba et al., 2008 for review), re-establishment of this Ascl1/Delta-like/Notch/Hes molecular circuit may serve as a focal point for developing strategies to stimulate endogenous mechanisms or guide cell-based approaches toward mammalian retinal repair therapies.
Supplementary Material
Expression of Dll1, Dll3, Dll4, and Jagged2 in the CNS. At embryonic day 12.5 (E12.5), Dll1 expression was restricted to the ventricular zone of the cortex, diencephalon, and rostral spinal cord (A, arrow, asterisk, inset, respectively). Dll3 expression was restricted to the ventricular zone of the cortex, more strongly expressed in the dorsal and ventral regions of the diencephalon, but in a pattern similar to Dll1 in the rostral spinal cord (B, arrow, asterisk, inset, respectively). Dll4 expression was not observed in either the cortex or diencephalon, but was detected in a small region in the middle of the rostral spinal cord (C, arrow, asterisk, inset, respectively), as recently reported (Del Barrio et al., 2007). In situ hybridization for Jagged2 revealed a largely complementary pattern with respect to Dll1 and Dll3 in the brain and cortex. For example, at E13.5, Jagged2 expression was restricted the mantle layer of the cortex and diencephalon, where postmitotic differentiating neurons reside, in contrast to the expression of Dll1 and Dll3 in ventricular zone, where mitotically active progenitors are located (D, arrows, asterisk).
Delta-like genes are expressed in E17.5 retinal progenitor cells. E17.5 embryos received a 2h pulse of BrdU in utero prior to sacrifice, and in situ hybridization was used to detect Dll1 (A), Dll3 (B), and Dll4 (C) expression, followed by immunolableling to detect BrdU incorporation (green). Individual and merged channels are shown, which reveal that Dll1 (A), Dll3 (B), and Dll4 (C) expressing cells at E17.5 can incorporate BrdU (arrowheads).
Reporter constructs used to identify Delta-like and Notch-active progenitor cells. (A, B) Schematic of Dll1LacZ, Hes5d2GFP, and Notch1GFP reporter constructs used to identify Dll1, Dll3, Notch1, and Hes5 active cells. Reporters were electroporated into mouse retinal explants (E17.5-E18.5), cultured overnight (≤24h) to allow expression, fixed, and then imaged directly in intact retinas with high-resolution multiphoton microscopy to detect endogenous fluorescent signal (Notch1GFP C, Hes5d2GFP D, Dll3d2CFP F), and/or after immunolabeling and imaging with laser scanning confocal microscopy (LSCM, Dll1LacZ E). Reporter constructs are drawn to scale. (B) A mouse Dll3 reporter plasmid with 1.0Kb upstream the sequence containing the evolutionarily conserved cis-regulatory element with the preferred octamer/E-box Ascl1 binding site (Castro et al., 2006) was constructed and used to drive destabilized CFP in Dll3 expressing progenitor cells. (C- F) Fluorescent panels demonstrate typical morphology of Notch1GFP (C), Hes5d2GFP (D), Dll1LacZ (E), and Dll3d2CFP (F) expressing cells with apical and basal processes indicative of progenitor cells: note that small extensions and varicosities are often observed in the apical and/or basal processes (F, inset, arrowheads, arrows, respectively; ae, apical endfoot; d2GFP/d2CFP refer to destabilized GFP or CFP proteins with ~2h half-lives).
Cover art. Retinascape: Multiphoton microscopy was used to look deep into the retina, at the dawn of neurogenesis in the embryonic mouse eye: mitotic progenitors (anti-phosphohistone H3, green) and newborn neurons (beta-III tubulin, red). View is of new neurons originating in the central neurogenic domain of the embryonic day 12 (E12) retina surrounded by mitotic progenitors, looking from within the retina itself. New data from Nelson and colleagues (page #) reveals that, in addition to Notch and Ascl1, mammalian retinal progenitors also express multiple Delta-like genes, and that a conserved Ascl1/Delta-like/Notch/Hes molecular circuitry operates between progenitors themselves to coordinate retinal histogenesis.
Acknowledgements
We thank Drs. A. Gosslar, R. Kageyama, W. Gao, and D. Anderson for use of reagents, and J. Brzezinski for critical reading of the manuscript. This work was supported by NRSA postdoctoral fellowship F32 EY15631 to B. R. N., NICHD predoctoral fellowship T32 HD007183 to B. H. H., and NIH grant EY13475 to T. A. R.
References
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279(27):28492–8. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Attardo A, Calegari F, Haubensak W, Wilsch-Bräuninger M, Huttner WB. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS ONE. 2008;3(6):e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121(11):3637–50. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127(13):2933–43. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17(4):1425–34. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers J, Caron A, Hrabé de Angelis M, Hans S, Campos-Ortega JA, Gossler A. Distinct regulatory elements direct delta1 expression in the nervous system and paraxial mesoderm of transgenic mice. Mech Dev. 2000;95(1-2):23–34. doi: 10.1016/s0925-4773(00)00322-1. [DOI] [PubMed] [Google Scholar]
- Benedito R, Duarte A. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr Patterns. 2005;5(6):750–5. doi: 10.1016/j.modgep.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3(7):517–30. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–34. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11(6):831–44. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129(8):1871–80. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9(1):37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Cho JH, Klein WH, Tsai MJ. Compensational regulation of bHLH transcription factors in the postnatal development of BETA2/NeuroD1-null retina. Mech Dev. 2007;124(7-8):543–50. doi: 10.1016/j.mod.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Rapaport DH, Harris WA. Xotch inhibits cell differentiation in the Xenopus retina. Neuron. 1995;14(3):487–96. doi: 10.1016/0896-6273(95)90305-4. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature. 1997;385(6611):67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28(5):1109–17. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev Biol. 2005;281(2):318–33. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Müller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4(3):247–52. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20(3):483–94. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000;26(2):383–94. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Geffers I, Serth K, Chapman G, Jaekel R, Schuster-Gossler K, Cordes R, Sparrow DB, Kremmer E, Dunwoodie SL, Klein T, Gossler A. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. 2007;178(3):465–76. doi: 10.1083/jcb.200702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6(10):777–88. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gratton MO, Torban E, Jasmin SB, Theriault FM, German MS, Stifani S. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol Cell Biol. 2003;23(19):6922–35. doi: 10.1128/MCB.23.19.6922-6935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75(3):463–76. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Harada T, Harada C, Parada LF. Molecular regulation of visual system development: more than meets the eye. Genes Dev. 2007;21(4):367–78. doi: 10.1101/gad.1504307. [DOI] [PubMed] [Google Scholar]
- Hartman BH, Hayashi T, Nelson BR, Bermingham-McDonogh O, Reh TA. Dll3 is expressed in developing hair cells in the mammalian cochlea. Dev Dyn. 2007;236(10):2875–83. doi: 10.1002/dvdy.21307. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15(1):83–9. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131(22):5539–50. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Mochii M, Kodama R, Hamada Y, Mizuno N, Eguchi G, Tachi C. Isolation of a novel chick homolog of Serrate and its coexpression with C-Notch-1 in chick development. Int J Dev Biol. 1996;40(6):1089–96. [PubMed] [Google Scholar]
- Hayes S, Nelson BR, Buckingham B, Reh TA. Notch signaling regulates regeneration in the avian retina. Dev Biol. 2007;312(1):300–11. doi: 10.1016/j.ydbio.2007.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signaling in the embryonic chick retina. Curr Biol. 1997;7(9):661–70. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127(12):2515–22. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, Huttner WB. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. PNAS. 1999;96(8):4639–44. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133(5):913–23. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- Jasoni CL, Reh TA. Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J Comp Neurol. 1996;369(2):319–27. doi: 10.1002/(SICI)1096-9861(19960527)369:2<319::AID-CNE11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Jasoni CL, Walker MB, Morris MD, Reh TA. A chicken achaete-scute homolog (CASH-1) is expressed in a temporally and spatially discrete manner in the developing nervous system. Development. 1994;120(4):769–83. doi: 10.1242/dev.120.4.769. [DOI] [PubMed] [Google Scholar]
- Jhas S, Ciura S, Belanger-Jasmin S, Dong Z, Llamosas E, Theriault FM, Joachim K, Tang Y, Liu L, Liu J, Stifani S. Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J Neurosci. 2006;26(43):11061–71. doi: 10.1523/JNEUROSCI.1358-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008a;135(18):3113–24. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kawaguchi D, Yoshimatsu T, Hozumi K, Gotoh Y. Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development. 2008b;135(23):3849–58. doi: 10.1242/dev.024570. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000;127(19):4203–16. doi: 10.1242/dev.127.19.4203. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet. 1998;19(3):274–8. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170(6):983–92. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba D, Karl M, Reh T. Neural regeneration and cell replacement: a view from the eye. Cell Stem Cell. 2008;2(6):538–49. doi: 10.1016/j.stem.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295(2):764–78. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Le Rouëdec D, Rayner K, Rex M, Wigmore PM, Scotting PJ. The transcription factor cSox2 and Neuropeptide Y define a novel subgroup of amacrine cells in the retina. J Anat. 2002;200(Pt 1):51–6. doi: 10.1046/j.0021-8782.2001.00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AK, Frantz GD, Carpenter DA, de Sauvage FJ, Gao WQ. Distinct expression patterns of notch family receptors and ligands during development of the mammalian inner ear. Mech Dev. 1998;78(1-2):159–63. doi: 10.1016/s0925-4773(98)00165-8. [DOI] [PubMed] [Google Scholar]
- Lewis J. Neurogenic genes and vertebrate neurogenesis. Curr Opin Neurobiol. 1996;6(1):3–10. doi: 10.1016/s0959-4388(96)80002-x. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8(1):14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signaling in vertebrate neural development. Nat Rev Neurosci. 2006;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lowell S. Stem cells: You make me feel so glial. Curr Biol. 2000;10(16):R595–7. doi: 10.1016/s0960-9822(00)00636-9. [DOI] [PubMed] [Google Scholar]
- Ma W, Wang SZ. The final fates of neurogenin2-expressing cells include all major neuron types in the mouse retina. Mol Cell Neurosci. 2006;31(3):463–9. doi: 10.1016/j.mcn.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87(1):43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17(10):3644–52. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105(1):43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9(6):770–8. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Moshiri A, Gonzalez E, Tagawa K, Maeda H, Wang M, Frishman LJ, Wang SW. Near complete loss of retinal ganglion cells in the math5/brn3b double knockout elicits severe reductions of other cell types during retinal development. Dev Biol. 2008 Apr 15;316(2):214–27. doi: 10.1016/j.ydbio.2008.01.015. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat A, Henrique D, Ish-Horowicz D, Lewis J. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev Biol. 1996;174(2):233–47. doi: 10.1006/dbio.1996.0069. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Reh TA. Relationship between Delta-like and proneural bHLH genes during chick retinal development. Dev Dyn. 2008;237(6):1565–80. doi: 10.1002/dvdy.21550. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Gumuscu B, Hartman BH, Reh TA. Notch Activity Is Downregulated Just prior to Retinal Ganglion Cell Differentiation. Dev Neurosci. 2006;28(1-2):128–41. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007a;304(2):479–98. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Hayes S, Hartman BH, Reh TA. Fliesler SJ, Kisselev O, editors. Signal transduction in retinal progenitor cells. Signal transduction in the retina. 2007b CRC Press. [Google Scholar]
- Nimmagadda S, Geetha-Loganathan P, Prols F, Scaal M, Christ B, Huang R. Expression pattern of Dll4 during chick embryogenesis. Histochem Cell Biol. 2007;128(2):147–52. doi: 10.1007/s00418-007-0306-6. [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2007;1192:90–8. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Imayoshi I, Shimojo H, Nishi E, Kageyama R, McConnell SK. Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol Cell Neurosci. 2006;31(1):109–22. doi: 10.1016/j.mcn.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;1:W280–6. doi: 10.1093/nar/gkh355. 32 (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Turner R, Muskavitch MAT. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev. 1995;50:201–216. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- Perron M, Kanekar S, Vetter ML, Harris WA. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol. 1998;199(2):185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- Pintar A, De Biasio A, Popovic M, Ivanova N, Pongor S. The intracellular region of Notch ligands: does the tail make the difference? Biol Direct. 2007;2:19. doi: 10.1186/1745-6150-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reh TA, Fischer AJ. Retinal stem cells. Methods Enzymol. 2006;419:52–73. doi: 10.1016/S0076-6879(06)19003-5. [DOI] [PubMed] [Google Scholar]
- Sakami S, Etter P, Reh TA. Activin signaling limits the competence for retinal regeneration from the pigmented epithelium. Mech Dev. 2008;125(1-2):106–16. doi: 10.1016/j.mod.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satow T, Bae SK, Inoue T, Inoue C, Miyoshi G, Tomita K, Bessho Y, Hashimoto N, Kageyama R. The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J Neurosci. 2001;21(4):1265–73. doi: 10.1523/JNEUROSCI.21-04-01265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul SM, Brzezinski JA, 4th, Altschuler RA, Shore SE, Rudolph DD, Kabara LL, Halsey KE, Hufnagel RB, Zhou J, Dolan DF, Glaser T. Math5 expression and function in the central auditory system. Mol Cell Neurosci. 2008;37(1):153–69. doi: 10.1016/j.mcn.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58(1):52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Silva AO, Ercole CE, McLoon SC. Regulation of ganglion cell production by Notch signaling during retinal development. J Neurobiol. 2003;54(3):511–24. doi: 10.1002/neu.10156. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB. The achaete-scute complex: generation of cellular pattern and fate within the Drosophila nervous system. FASEB J. 1994;8(10):714–21. doi: 10.1096/fasebj.8.10.8050670. [DOI] [PubMed] [Google Scholar]
- Takatsuka K, Hatakeyama J, Bessho Y, Kageyama R. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004;1004(1-2):148–55. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- Takebayashi K, Akazawa C, Nakanishi S, Kageyama R. Structure and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-5. Identification of the neural precursor cell-specific promoter element. J Biol Chem. 1995;270(3):1342–9. doi: 10.1074/jbc.270.3.1342. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20(9):1187–202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996a;16(4):723–34. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Tomita K, Nakanishi S, Guillemot F, Kageyama R. Mash1 promotes neuronal differentiation in the retina. Genes Cells. 1996b;1(8):765–74. doi: 10.1111/j.1365-2443.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19(20):5460–72. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi C, Ghezzi C, Ballabio A, Rugarli EI. JAGGED2: a putative Notch ligand expressed in the apical ectodermal ridge and in sites of epithelial-mesenchymal interactions. Mech Dev. 1997;69(1-2):203–7. doi: 10.1016/s0925-4773(97)00146-9. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12(6):491–8. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Research. 2003;31(24):1–8. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21(1):63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wildner H, Müller T, Cho SH, Bröhl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133(11):2105–13. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- Yan RT, Ma WX, Wang SZ. neurogenin2 elicits the genesis of retinal neurons from cultures of nonneural cells. PNAS. 2001;98(26):15014–9. doi: 10.1073/pnas.261455698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan RT, Ma W, Liang L, Wang SZ. bHLH genes and retinal cell fate specification. Mol Neurobiol. 2005;32(2):157–71. doi: 10.1385/MN:32:2:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to Suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133(7):1367–78. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58(4):519–31. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129(21):5029–40. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of Dll1, Dll3, Dll4, and Jagged2 in the CNS. At embryonic day 12.5 (E12.5), Dll1 expression was restricted to the ventricular zone of the cortex, diencephalon, and rostral spinal cord (A, arrow, asterisk, inset, respectively). Dll3 expression was restricted to the ventricular zone of the cortex, more strongly expressed in the dorsal and ventral regions of the diencephalon, but in a pattern similar to Dll1 in the rostral spinal cord (B, arrow, asterisk, inset, respectively). Dll4 expression was not observed in either the cortex or diencephalon, but was detected in a small region in the middle of the rostral spinal cord (C, arrow, asterisk, inset, respectively), as recently reported (Del Barrio et al., 2007). In situ hybridization for Jagged2 revealed a largely complementary pattern with respect to Dll1 and Dll3 in the brain and cortex. For example, at E13.5, Jagged2 expression was restricted the mantle layer of the cortex and diencephalon, where postmitotic differentiating neurons reside, in contrast to the expression of Dll1 and Dll3 in ventricular zone, where mitotically active progenitors are located (D, arrows, asterisk).
Delta-like genes are expressed in E17.5 retinal progenitor cells. E17.5 embryos received a 2h pulse of BrdU in utero prior to sacrifice, and in situ hybridization was used to detect Dll1 (A), Dll3 (B), and Dll4 (C) expression, followed by immunolableling to detect BrdU incorporation (green). Individual and merged channels are shown, which reveal that Dll1 (A), Dll3 (B), and Dll4 (C) expressing cells at E17.5 can incorporate BrdU (arrowheads).
Reporter constructs used to identify Delta-like and Notch-active progenitor cells. (A, B) Schematic of Dll1LacZ, Hes5d2GFP, and Notch1GFP reporter constructs used to identify Dll1, Dll3, Notch1, and Hes5 active cells. Reporters were electroporated into mouse retinal explants (E17.5-E18.5), cultured overnight (≤24h) to allow expression, fixed, and then imaged directly in intact retinas with high-resolution multiphoton microscopy to detect endogenous fluorescent signal (Notch1GFP C, Hes5d2GFP D, Dll3d2CFP F), and/or after immunolabeling and imaging with laser scanning confocal microscopy (LSCM, Dll1LacZ E). Reporter constructs are drawn to scale. (B) A mouse Dll3 reporter plasmid with 1.0Kb upstream the sequence containing the evolutionarily conserved cis-regulatory element with the preferred octamer/E-box Ascl1 binding site (Castro et al., 2006) was constructed and used to drive destabilized CFP in Dll3 expressing progenitor cells. (C- F) Fluorescent panels demonstrate typical morphology of Notch1GFP (C), Hes5d2GFP (D), Dll1LacZ (E), and Dll3d2CFP (F) expressing cells with apical and basal processes indicative of progenitor cells: note that small extensions and varicosities are often observed in the apical and/or basal processes (F, inset, arrowheads, arrows, respectively; ae, apical endfoot; d2GFP/d2CFP refer to destabilized GFP or CFP proteins with ~2h half-lives).
Cover art. Retinascape: Multiphoton microscopy was used to look deep into the retina, at the dawn of neurogenesis in the embryonic mouse eye: mitotic progenitors (anti-phosphohistone H3, green) and newborn neurons (beta-III tubulin, red). View is of new neurons originating in the central neurogenic domain of the embryonic day 12 (E12) retina surrounded by mitotic progenitors, looking from within the retina itself. New data from Nelson and colleagues (page #) reveals that, in addition to Notch and Ascl1, mammalian retinal progenitors also express multiple Delta-like genes, and that a conserved Ascl1/Delta-like/Notch/Hes molecular circuitry operates between progenitors themselves to coordinate retinal histogenesis.