Abstract
This unit outlines the steps required to prepare a sample for MS analysis following protein separation or enrichment by gel electrophoresis, liquid chromatography, and affinity capture within the context of a bottom-up proteomics workflow in which the protein is first broken up into peptides, either by chemical or enzymatic digestion, prior to MS analysis. Also included are protocols for enrichment at the peptide level, including phosphopeptide enrichment and reversed-phase chromatography for sample purification immediately prior to MS analysis. Finally, there is a discussion regarding the types of MS technologies commonly used to analyze proteomics samples, as well as important parameters that should be considered when analyzing the MS data to ensure stringent and robust protein identifications and characterization.
Keywords: in-solution digestion, in-gel digestion, peptide desalting proteomics, mass spectrometry
INTRODUCTION
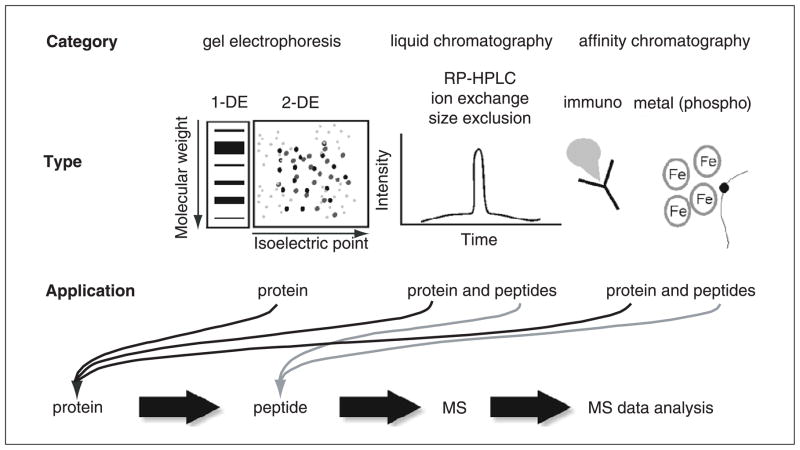
The use of mass spectrometry (MS) to identify and characterize biological molecules is a fundamental technology in protein biochemistry and proteomic analysis. The strategies used to prepare proteins or more complex proteomic samples for MS analysis involve many steps, and in bottom-up proteomics, the protein is first broken up into peptides, either by chemical or enzymatic digestion, prior to MS analysis (see Fig. 10.25.1 for workflow). The MS analysis is performed on the individual peptides, and the information is then stitched together to reveal the protein identity and/or characteristics (e.g., co-and post-translational modifications or isoforms). Key steps in this strategy include the preparation of the protein sample for digestion, enrichment for any particular peptides of interest, and cleanup or desalting of the final peptide mixture prior to MS analysis by either MALDI-TOF-MS/MS (matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry) or LC-ESI-MS/MS (liquid chromatography-electrospray ionization tandem mass spectrometry). Careful sample handling at the protein and peptide level is the key for successful analysis at the MS level. As such, it is necessary to take great care, and in some instances optimize each step involved, to obtain the maximum amino acid sequence coverage of the protein(s) of interest.
Figure 10.25.1.
Bottom-up proteomics workflow. Proteins are enzymatically or chemically cleaved into peptides which are then analyzed in the mass spectrometer. The proteins and peptides for analysis are often derived from separation/purification methods such as gel electrophoresis (UNIT 10.2A), liquid chromatography (UNITS 10.9–10.11A), and affinity chromatography (UNIT 10.11B).
This unit outlines the steps required to prepare a sample for MS analysis following protein separation or enrichment by gel electrophoresis (Basic Protocol 1), liquid chromatography (Basic Protocol 2), and affinity capture (Basic Protocol 3) within the context of a bottom-up proteomics workflow, whereby the proteins are enzymatically or chemically digested into peptides prior to MS analysis. Also included are protocols for the enrichment at the peptide level, including phosphopeptide enrichment (Basic Protocol 4) and reversed-phase chromatography (Basic Protocol 5) for sample purification immediately prior to MS analysis. Finally, there is a discussion regarding the types of MS technologies commonly used to analyze proteomics samples, as well as important parameters that should be considered when analyzing the MS data to ensure stringent and robust protein identifications and characterization.
BASIC PROTOCOL 1. PREPARATION OF PROTEIN FROM A POLYACRYLAMIDE GEL FOR MASS SPECTROMETRIC ANALYSIS
One standard method by which complex protein mixtures may be separated is polyacrylamide gel electrophoresis (PAGE). Proteins can be separated in a single dimension based on their molecular weight (1-DE or SDS-PAGE; UNIT 10.2A; Laemmli, 1970) or separated as a native complex (Blue Native PAGE; UNIT 10.2B; Schagger and von Jagow, 1991). Alternatively, proteins can be separated by two-dimensional electrophoresis (2-DE) (UNIT 10.4), where the first dimension separates proteins based on their isoelectric point (although other methods such as Blue Native PAGE may be used), and the second dimension separates by SDS-PAGE (Gorg et al., 2004). When preparing samples for downstream MS analysis (see Commentary), proteins can be visualized with an MS-compatible staining procedure such as glutaraldehyde-free silver staining protocols (Shevchenko et al., 1996), traditional Coomassie blue staining (TCS; UNIT 10.6, Chrambach et al., 1967), and colloidal Coomassie staining (CCS; Candiano et al., 2004). Traditional methods for silver staining that use glutaraldehyde to cross-link proteins to the acrylamide gel are not compatible with MS. Regardless of the type of gel, the preparation of proteins from acrylamide gels for eventual MS analysis requires a number of steps. This protocol describes the steps involved in destaining, digesting, and extracting peptides from a polyacrylamide gel to analyze the peptides by either MALDI-TOF-MS/MS or LC-ESI-MS/MS.
NOTE: Use only HPLC- and/or mass spectrometry–grade reagents throughout the protocol.
Materials
Polyacrylamide gel containing proteins of interest stained with MS-compatible stain: glutaraldehyde-free silver, traditional Coomassie stain (TCS; UNIT 10.6), or colloidal Coomassie stain (CCS)
Gel destain solution (see recipe specific for each type of stained gel: colloidal Coomassie destain, glutaraldehyde-free silver destain, or traditional Coomassie destain)
100% acetonitrile
10 mM dithiothreitol (DTT)
55 mM iodoacetamide (prepare fresh immediately prior to use; protect from light)
Gel wash solution (see recipe)
Gel enzyme solution (see recipe)
25 mM ammonium bicarbonate (NH4HCO3)
Gel extraction solution (see recipe)
Formic acid (optional)
Gloves
Glass plate
Light box
Sharp cutting tool (e.g., razor, scissors, scalpel)
0.6-ml low-binding, siliconized microcentrifuge tubes (Fisher Scientific)
Vacuum centrifuge
55 °C water bath
Additional reagents and equipment for desalting/cleanup prior to MS analysis (Basic Protocol 5)
NOTE: Wear gloves at all times to prevent contamination from proteins such as keratins.
Excise and destain gel bands/spots
-
1. Wearing gloves, and with the gel placed on a glass plate illuminated by a light box, use a razor blade or other sharp cutting tool to excise an individual band/spot from either a fresh or dried polyacrylamide gel and place it into a low-binding, siliconized microcentrifuge tube.
The size of microcentrifuge tube used depends upon the size/number of gel pieces being analyzed. The volumes listed in this protocol work well with 0.6-ml tubes. Volumes should be increased to reflect larger tubes/gel pieces.Protein spots or bands can be processed immediately after running the gel or can be taken from dried gels stored from previous experiments. The authors have had success drying the entire gel between sheets of cellophane such that gels stored for years at room temperature yield successful MS analyses that result in protein identifications.An automated robotic gel-cutting device can be used—e.g., EXQuest Spot Cutter (BioRad) or Ettan Spot Picker (GE Healthcare)—to reduce time and effort. These instruments can also be used to cut protein bands/spots from gels which are stained using only a fluorescent dye (e.g., CyDye, SYPRO Ruby) and are thus not visible to the eye. The disadvantage of gel spot pickers is that they only excise a small section of a spot or band. Thus, for low-abundance spots or bands, the resulting gel piece may not contain sufficient protein for successful MS analysis. Therefore, hand cutting may be advantageous, as one can ensure the maximum sample for extraction.For very low-abundance proteins, one may need to combine spots/bands from multiple gels or multiple lanes within the same gel. -
2. Destain in the microcentrifuge tube for 30 min with 100 μl of the appropriate gel destain solution (see Reagents and Solutions) with vigorous shaking (vortexing).
Regardless of whether the gel is stained with glutaraldehyde-free silver, TCS, or CCS, it is necessary to destain the gel pieces, as the stain will interfere with MS analysis. The choice of destaining protocol depends on the stain used, and there is a specific destain recipe for each type of gel stain (see Reagents and Solutions).Due to the difficulty in destaining CCS, it may be necessary to alternate between CCS and TCS destain solutions in order to fully remove color.The choice of protein stain depends on the protein concentration and the dynamic range of the sample. Silver and CCS stains offer a low limit of detection (1 to 10 ng; Shevchenko, et al., 1996; Candiano et al., 2004) compared to TCS (0.3 to 1 μg; UNIT 10.6), but quantitation by silver stain can be hampered by a smaller linear range. Of note, not all proteins stain equally by each dye, due to the intrinsic nature of the proteins. 3a. For glutaraldehyde-free silver stained gels: Remove destain solution, and wash gel piece with 400 μl water, then shake for 15 min at room temperature. Repeat wash at least twice or until gel pieces are colorless.
3b. For Coomassie stained gels (TCS or CCS): Remove destain solution and add 400 μl water. If the gel pieces are still blue, remove the water and repeat steps 2 and 3 until the gel pieces are colorless.
4. Dehydrate gel pieces with 400 μl of 100% acetonitrile for 10 min, then remove supernatant and dry gel pieces in vacuum centrifuge.
Reduce disulfide bonds and alkylate free cysteines
-
5. Add 100 μl of 10 mM DTT to the gel piece, then incubate 45 min at 55°C.
Dithiothreitol (DTT) is a reducing agent that converts cystine’s disulfide bond into cysteine’s free sulfhydryl groups. -
6. Remove solution, add 100 μl 55 mM iodoacetamide, and incubate 30 min at room temperature in the dark.
Iodoacetamide (IAA) is an alkylating agent that reacts with free sulfhydryl groups of cysteine residues to form S-carboxyamidomethyl-cysteine, which cannot be reoxidized to form disulfide bonds. This is important for allowing trypsin maximum access to cleavage sites within the protein. 7. Remove IAA solution and add 400 μl gel wash solution, incubate at room temperature, and shake for 15 min. Repeat this step twice.
8. Dehydrate gel pieces with 400 μl of 100% acetonitrile for 10 min, then remove supernatant and dry gel pieces using a vacuum centrifuge.
Perform enzymatic digestion
-
9. Dilute the gel enzyme stock solution ~1:1000 with 25 mM ammonium bicarbonate to obtain a 10 to 20 μg/ml working solution. Add a sufficient amount of the gel enzyme working solution to cover gel pieces and incubate on ice for 1 hr.
For a typical gel band/spot, this is ~10 to 20 μl.The enzyme solution diffuses into the gel pores containing the destained, reduced, and alkylated protein, and cleaves the protein into peptides, which must be extracted from the gel. -
10. Remove excess gel enzyme solution, then add sufficient 25 mM NH4HCO3 to cover the gel pieces.
The alkaline bicarbonate solution increases the pH and thereby inhibits the enzymatic digestion for those enzymes which have low pH optima. Furthermore, this step reduces the enzyme autolysis peaks that may complicate the MS spectra. 11. Incubate at 37°C overnight.
Extract peptides
12. Remove supernatant, which now contains peptides, to a new clean microcentrifuge tube.
-
13. Add 100 μl gel extraction solution to the gel pieces and incubate while shaking for 20 min at room temperature.
As an alterative to this extraction step, 50 μl of 5% (v/v) formic acid can be added for 15 min followed by the addition of 50 μl of 100% acetonitrile, for an additional 15 min. This alternative may be useful when samples will be analyzed by electrospray/nanospray-type MS, where TFA is not ideal.The volumes listed may be adjusted to ensure that the entire gel piece is covered with liquid. 14. Remove solution and combine with solution obtained in step 12.
15. Repeat steps 13 to 14.
-
16. Dry combined supernatants using a vacuum centrifuge.
This step can take up to several hours depending upon the volume used in step 13. 17. Desalt/clean peptides prior to MALDI-TOF-MS/MS or LC-ESI-MS/MS (see Basic Protocol 5).
BASIC PROTOCOL 2. PREPARATION OF PROTEIN FROM A LIQUID CHROMATOGRAPHY FRACTION FOR MASS SPECTROMETRIC ANALYSIS
Proteins can be separated or fractionated based on any of their physical properties including size/mass, charge, or hydrophobicity. In size-exclusion chromatography (SEC; UNITS 10.9 & 10.13), salt is present in the mobile phase to repress any specific charge interactions between the column matrix and the proteins. In ion-exchange chromatography (UNIT 10.10), proteins are eluted using an increasing salt gradient, and in chromatographic focusing, proteins are eluted based on a decreasing pH gradient in the presence of urea, which is used as a denaturant. Since high concentrations of salt and urea are incompatible with MS analysis as well as with many enzymatic digestions, the proteins must be desalted prior to further analysis. Reversed-phase high performance liquid chromatography (RP-HPLC; UNIT 10.14) is one method that is commonly used to concentrate and desalt intact proteins, either on its own or downstream from other types of chromatographic separations. RP-HPLC is a popular choice for proteomic workflows, as the mobile phases, which are volatile, are compatible with MS and may also be easily removed via vacuum centrifugation, if necessary. This protocol outlines the steps required to prepare a sample from an RP-HPLC separation for enzymatic digestion in preparation for MS analysis.
NOTE: Use only HPLC and/or mass spectrometry grade reagents throughout the protocol.
Materials
RP-HPLC fraction containing protein of interest (see Basic Protocols 1 and 5)
100 mM and 1 M ammonium bicarbonate (NH4HCO3), pH 8.5
Narrow-range pH paper
100 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) stock, or 100 mM dithiothreitol (DTT) stock (prepare fresh immediately prior to use)
100 mM iodoacetamide stock (prepare fresh immediately prior to use)
Proteolytic enzyme (e.g., chymotrypsin, trypsin, LysC, or AspN) or chemical (e.g., CNBr/formic acid)
1.0% (v/v) trifluoroacetic (TFA) acid in H2O
Low-binding, siliconized microcentrifuge tubes (Fisher Scientific)
End-over-end rotator
Additional reagents and equipment for desalting/cleanup prior to MS analysis (Basic Protocol 5)
Neutralize sample
The pH of the sample may be neutralized either prior to or after drying via vacuum centrifugation to remove acetonitrile and trifluoroacetic acid.
1a. To neutralize prior to drying: Add sufficient 1 M NH4HCO3, pH 8.5, to adjust final pH to ~8.0, then dry via vacuum centrifugation.
-
1b. To neutralize after drying: Resuspend dried protein in 100 mM NH4HCO3, pH 8.5. Spot an aliquot of the neutralized solution on narrow-range pH paper to validate that neutralization (to pH 8.0) has occurred.
RP-HPLC samples will likely contain aqueous trifluoroacetic acid and acetonitrile, since these are the most commonly used mobile phases (see Basic Protocol 5).The NH4HCO3 solution should be prepared fresh, as the pH of the solution will increase with age at room temperature. It is possible to store for several days at 4°C, but the pH should be checked prior to use.
Reduce disulfide bonds and alkylate free cysteine residues
Reduction of disulfide bonds can be achieved with either DTT or TCEP.
-
2. Add DTT to a final concentration of 10 mM or TCEP to a final concentration of 5 mM. Incubate sample at room temperature with agitation (e.g., end-over-end rotation, vortexing, sonication, or shaking), for 30 min with DTT or 10 min with TCEP. Add iodoacetamide to a concentration of 10 mM final and place at room temperature in the dark for 30 min.
Reduction and alkylation are not required prior to enzymatic or chemical cleavage, but typically will provide enhanced sequence coverage of the proteins if performed, as it may help to unfold the protein, thereby facilitating the cleavage. If analyzing proteins without cysteine residues, this step is unnecessary.Compared to DTT, which works optimally at pH 7 to 9, TCEP is typically more efficient, works at a wider pH range (pH 2 to 11), and has no offensive odor.The concentrations and incubation times for DTT, TCEP, and IAA may vary depending on the amount of protein to be treated. Typical concentrations range from 5 to 10 mM for DTT and TCEP, and 10 to 50 mM for iodoacetamide. Incubation times may range from 5 min to 1 hr for the reduction step and 10 min to 1 hr for the alkylation step. Similarly, the reduction can be performed at room temperature, 37°C or 56°C. Each parameter can be optimized depending on the nature of the protein sample. -
3. Optional: Following alkylation, adjust sample to 40 mM DTT.
This step will not affect covalently modified thiols, but will consume any remaining alkylating agent, thus preventing the alkylation of the enzyme used in the next step (e.g., trypsin). -
4. Digest the protein sample into peptides using an appropriate enzyme (e.g., chymotrypsin, trypsin, LysC, or AspN) or chemical (CNBr/70% formic acid). If using trypsin, add sufficient enzyme for a final trypsin:protein ratio of 1:20 to 1:100 (w/w); optimizing the amount of trypsin is discussed in the Commentary. Vortex briefly, seal tube with Parafilm, and incubate with end-over-end rotation at 37°C for 4 to 18 hr.
The amount of trypsin needed will vary depending on the amount of protein sample present and the desired speed of digestion. Many commercial vendors provide proteomics-grade trypsin in convenient aliquots (e.g., 1- to 25-μg vials), which eliminate the need to weigh out the enzyme. To minimize trypsin autolysis, which will add extra peaks to the MS spectra, use the minimum amount of trypsin. Typically, a ratio of 1:20 (w/w) is sufficient for complete digestion of 10 μg protein within 4 hr. If protein concentration is especially high or the accessibility of trypsin to the cleavage sites is hampered (e.g., due to insolubility), it may be helpful to add another aliquot of trypsin or perform a short digestion (4 hr) with LysC prior to the addition of trypsin. Finally, the activity of trypsin is enhanced in the presence of acetonitrile (10% to 50% v/v), and, thus, acetonitrile may be added to aid in the solubilization of the protein during tryptic digestion.Trypsin activity is greatest at pH 8.0. Thus, it is recommended that the pH of the solution be checked prior to the addition of the enzyme. In cases where sample volume is small, a 1- to 2-μl sample may be tested using pH paper.CNBr cleaves at the C-terminal side of Met residues, leading to a mixture of peptides with C-terminal homoserine and homoserine lactone, which are interconvertible. Met-Ser/Thr bonds are cleaved slowly. -
5. Stop reaction by adding 5 μl of 1.0% TFA (pH after TFA addition should be ~2 to 3).
This may be checked by applying a small drop of the solution to narrow-range pH paper. 6. Perform sample desalting, if necessary, prior to MALDI-TOF-MS/MS or LC-ESI-MS/MS (see Basic Protocol 5).
BASIC PROTOCOL 3. PREPARATION OF PROTEIN FROM AFFINITY CAPTURE FOR MASS SPECTROMETRIC ANALYSIS
Affinity capture is a traditional, widely used method to enrich/isolate intact proteins, to identify binding partners and protein complexes, or to investigate post-translational modifications. In cases where antibodies are used for the capture, the protein or protein complexes are often separated by gel electrophoresis prior to MS (see Basic Protocol 1). However, in some cases, it may be preferable to elute a sample from an affinity capture matrix and perform an in-solution digestion prior to MS analysis. In-solution digestion is often more efficient than in-gel digestion (Basic Protocol 1), since peptide extraction from a gel matrix is limited by diffusion kinetics. Hence, in-solution digestion produces greater peptide yields and can enhance peptide sequence coverage. Since virtually all detergents are incompatible with MS, great care should be taken to thoroughly wash the sample using nondetergent wash solutions prior to elution. Typically, low-pH solutions are effective for sample elution and do not interfere with MS analysis, although the elution conditions may vary depending on the method of affinity capture. This protocol outlines the steps required to efficiently elute a protein from an affinity matrix and perform an enzymatic digestion in preparation for MS analysis.
NOTE: Use only HPLC- and/or mass spectrometry–grade reagents throughout the protocol.
Materials
Affinity matrix with protein(s) of interest bound (see UNITS 10.11A & 10.11B)
Phosphate buffered saline (PBS) pH 7.4 (see recipe)
0.1 M glycine, pH 2.5
200 mM ammonium bicarbonate (NH4HCO3)
100 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) stock, or 100 mM dithiothreitol (DTT) stock (prepare fresh immediately prior to use)
100 mM iodoacetamide stock (prepare fresh immediately prior to use)
Proteolytic enzyme (e.g., trypsin, chymotrypsin, AspN, or LysC) or chemical (CNBr/70% formic acid)
1.0% (v/v) trifluoroacetic acid (TFA)
Low-binding, siliconized microcentrifuge tubes (Fisher Scientific)
End-over-end rotator
Additional reagents and equipment for protein assay (UNIT 10.1A) and desalting/cleanup prior to MS analysis (Basic Protocol 5)
-
Wash affinity matrix containing the adsorbed protein sample with the wash buffer (e.g., PBS) to elute the unbound or nonspecifically bound proteins from the matrix.
Beads may be washed by resuspending them in the wash buffer with gentle vortexing, then microcentrifuging at low speed (e.g., 800 rpm in a standard benchtop microcentrifuge) at room temperature to pellet the beads, and removing the supernatant by pipetting.If detergent is present in the starting material, it may be necessary to increase the number of washes to remove residual detergent. Often this can be difficult and may require up to ten additional 20× volume washes (i.e., 20 times the packed volume of the matrix). The choice of wash buffer will depend on the affinity matrix, but in general each solution should be free of detergent and contain as little salt as possible. For most immunocapture methods, PBS is an effective wash buffer, but other neutral-pH saline buffers such as HEPES or Tris·Cl can be used. If greater stringency is required, for example, a high-salt wash or inclusion of a chaotropic agent, perform more stringent washes first followed by additional PBS washes. It is often necessary to optimize wash conditions for individual affinity-capture methods. Elute protein from the matrix using a 5× volume of 0.1 M glycine, pH 2.5. Apply the elution buffer and allow the buffer to thoroughly wet the matrix for 5 to 10 min, and then collect the eluate. To enhance recovery, wash the matrix with an additional 5× volume of 0.1 M glycine, pH 2.5.
-
Adjust eluate to neutral pH by addition of an equal volume of 200 mM NH4HCO3.
The pH may be checked by applying a small drop of the solution to pH paper.This neutralization step is required to prevent denaturation of enzymes used for digestion in a subsequent step. However, if using CNBr for digestion, one can proceed directly to step 5, as reduction/alkylation is not required. In this case, disulfide-linked peptides may complicate the MS analysis, so depending on the peptide coverage sought from the MS analysis, a prior reduction and alkylation may be needed. -
To reduce disulfide bonds, add DTT to a final concentration of 10 mM or TCEP to a final concentration of 5 mM. Incubate sample at room temperature with agitation (e.g., end-over-end rotation, vortexing, sonication, or shaking) for 10 min with TCEP or 30 min with DTT. Next, to alkylate free cysteines, add iodoacetamide to a final concentration of 10 mM, and place at room temperature in the dark for 30 min.
For alternative approaches, see Basic Protocol 2. -
Digest the protein sample into peptides using an appropriate enzyme or chemical (e.g., see Basic Protocol 2). If using trypsin, add sufficient enzyme for a final trypsin:protein ratio of 1:20 to 1:100 (w/w). Vortex briefly, seal tube with Parafilm, and incubate with end-over-end rotation at 37°C for 4 to 18 hr.
It may be necessary to quantify the amount of protein present using, e.g., a BCA assay (UNIT 10.1A). Optimizing the amount of trypsin is described in the Commentary.For alternative approaches, see Basic Protocol 2. -
Stop reaction by adding 5 μl of 1.0% TFA (pH after TFA addition should be ~2 to 3).
The pH may be determined by applying a small drop of the solution to pH paper. Desalt sample, if necessary, prior to MS (see Basic Protocol 5).
BASIC PROTOCOL 4. PREPARATION OF PEPTIDE SAMPLES FOR MASS SPECTROMETRIC ANALYSIS USING PHOSPHOPEPTIDE ENRICHMENT METHODS
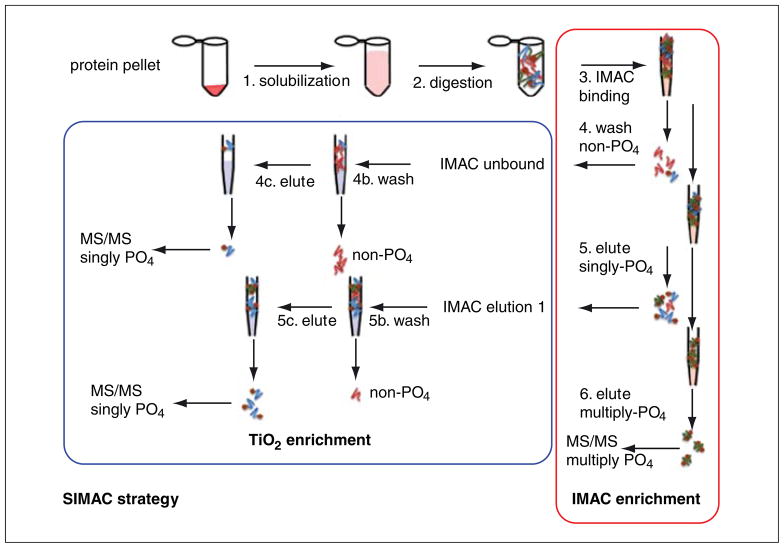
The ability to identify proteins post-translationally modified by phosphorylation is the goal of many proteomic studies. Immobilized metalion affinity chromatography (IMAC; UNIT 10.11B) has been shown to have a strong binding affinity for phosphorylated amino acid residues (see reviews by Porath, 1992; Smith and Figeys, 2008; Macek et al., 2009). However, it has been shown that IMAC has an equally strong affinity for peptides with low overall pI (peptides with a high content of Asp and Glu), and its relatively low binding capacity can be a limitation for complex samples (Arrell et al., 2006). An alternative to the usual metals used in IMAC (e.g., Fe3+, Ga3+, and Al3+) is the use of titanium dioxide (TiO2) beads, which have a very high affinity for phosphopeptides and are particularly efficient at enriching phosphopeptides from complex samples. By binding samples to the TiO2 beads in highly acidic buffers, the selectivity of TiO2 beads for negatively charged phosphopeptides is further improved (Larsen et al., 2005; Thingholm et al., 2006; Thingholm and Larsen, 2009). Complementarity between IMAC and TiO2 stems from the fact that, while both will bind both mono- and multiply phosphorylated peptides, TiO2 binds multiply phosphorylated peptides so tightly that their elution from TiO2 is nearly impossible. Therefore, the phosphoproteome sequence coverage can be increased when the two methods are combined, in an approach termed SIMAC (Sequential elution from IMAC; Thingholm et al., 2008, 2009; see Fig. 10.25.2). This protocol outlines the steps involved in performing SIMAC for the enrichment of mono- and multiply phosphorylated peptides, although either IMAC or TiO2 can be used independently following the same protocol.
Figure 10.25.2.
Strategies for the enrichment of phosphopeptides prior to MS analysis. It is possible to perform either IMAC enrichment or TiO2 enrichment alone, or in combination (SIMAC strategy). Following digestion of proteins, enrichment of phosphopeptides using the SIMAC strategy is achieved first by IMAC enrichment. Some phosphorylated peptides may not be captured by IMAC (IMAC unbound) and should therefore be enriched further by TiO2. This sample can be analyzed directly by MS/MS. Singly phosphorylated (singly PO4) peptides are eluted from IMAC (IMAC elution 1), then further enriched by TiO2 and analyzed by MS/MS. Finally, multiply phosphorylated peptides are eluted from IMAC and analyzed directly by MS/MS.
NOTE: Use only HPLC- and/or mass spectrometry–grade reagents throughout the protocol.
Materials
IMAC beads (e.g., PHOS-Select Iron Affinity Gel from Sigma)
IMAC wash solution (see recipe)
Sample containing phosphorylated peptides
Low-pH elution solution (see recipe)
High-pH elution solution (see recipe)
TiO2 beads (GL Science Inc., http://www.glsciences.com/)
StageTips packed with C8 disks (Proxeon Biosystems, http://www.proxeon.com/)
1% (w/v) sodium dodecyl sulfate (SDS)
TiO2 loading solution (see recipe)
TiO2 wash solution (see recipe)
Reversed phase resin for processing phosphopeptides (e.g., Oligo R3, Applied Biosystems)
0.1% (v/v) trifluoroacetic acid (TFA)
Phosphopeptide RP elution buffer for MALDI-TOF-MS/MS (see recipe)
Phosphopeptide RP elution buffer for LC-ESI-MS/MS (see recipe)
100% and 0.1% (v/v) formic acid in H2O
100% and 0.1% (v/v) trifluoroacetic acid (TFA)
Low-binding, siliconized microcentrifuge tubes (Fisher Scientific)
Orbital shaker
Combi-Syringe tips (e.g., Fisher Scientific)
Narrow-bore P-20 pipet tips
Vacuum centrifuge
Sample plate for MALDI
IMAC phosphopeptide enrichment
These steps can performed alone or as the first part of the SIMAC enrichment.
-
1. Wash IMAC beads (5 to 7 μl for simple mixtures; 30 to 50 μl for complex mixtures containing ≥120 μg protein) by applying 50 μl IMAC wash solution, vortexing, centrifuging at low speed (e.g., 800 rpm in a benchtop microcentrifuge) at room temperature to pellet the beads, and removing the supernatant by pipetting.
Too many IMAC beads may result in nonspecific binding of peptides because of reduced washing efficiency. A small amount of wash solution should be left in the tube with the beads, to avoid aspirating the beads. 2. Add protein sample to the IMAC beads in a volume of IMAC wash solution that is 10 times the volume of the packed beads. Incubate with constant, gentle agitation, at room temperature for 30 to 60 min (e.g., on an orbital shaker).
-
3. Using a Combi-Syringe and gentle pressure, pack the IMAC beads in a narrow bore P-20 tip by gradually adding the bead slurry in 20-μl aliquots.
Squeeze the tip of the narrow-bore P-20 pipet tip nearly shut, as this will prevent bead loss. The Combi-Syringe is only to be used for the generation of pressure in the column, not for measuring and loading the aliquots. The Combi-Syringe is reusable and should therefore not come into contact with any solution. -
4. Collect the column flow through in microcentrifuge tubes.
This will be referred to in subsequent steps as the IMAC-FT fraction. IMAC-FT can contain some phosphopeptides and should therefore be subsequently used for the SIMAC enrichment step (Fig. 10.25.2). -
5. Wash the IMAC column with 50 μl IMAC wash solution and pool the eluate with the IMAC-FT fraction from step 4.
This step should be performed slowly using a Combi-Syringe to apply pressure. -
6. Elute and collect the monophosphopeptides using 50 μl low-pH elution solution.
This is the IMAC–1% TFA fraction.This step should be performed quickly using a Combi-Syringe to apply pressure. -
7. Elute and collect the multiply phosphorylated peptides using 70 μl high-pH elution solution
This is the IMAC–pH 11 fraction.This step should be performed slowly using a Combi-Syringe to apply pressure. 8. Dry each eluate containing the multiply phosphorylated peptides (IMAC–pH 11 fraction), monophosphopeptides (IMAC–1% TFA fraction), and the IMAC flow through (IMAC-FT) separately using vacuum centrifugation.
Perform TiO2 enrichment
This method can be done alone or as the second step in SIMAC for further enrichment of monophosphopeptides (IMAC–1% TFA fraction), and those that are present in the IMAC-FT fraction from SIMAC (step 4; i.e., the “IMAC unbound” in Fig. 10.25.2).
-
9. Prepare a TiO2 bead slurry in 100% acetonitrile. Pack the TiO2 in StageTips that have been pre-packed with C8 disks.
The resulting TiO2 columns should be ~4 to 5 mm long. StageTips can be purchased with prepacked C8 disks to retain the TiO2 beads in the column.TiO2 is light sensitive, so keep powder in an amber or dark glass container or in a microcentrifuge tube covered with aluminum foil. When not in use, TiO2 should be in a container stored in a dark place (e.g., a drawer). It should not be exposed to light for longer than necessary. -
10. If samples have been dried, then resuspend in 3 μl 1 of % SDS. If peptide samples are in solution, then proceed to step 11.
For SIMAC, this includes monophosphopeptides (IMAC–1% TFA eluate) and the flow through (IMAC-FT eluate).CAUTION: SDS as a powder is toxic and should be handled with care, with additional airway protection. 11. Dilute IMAC–1% TFA and IMAC-FT samples each in 5 volumes (i.e., 15 μl) of TiO2 loading solution.
-
12. Load each of the samples onto one TiO2 column (for simple samples) or three to four TiO2 columns (for complex mixtures containing ≥120 μg protein).
The number of columns used will depend on the amount and complexity of sample. -
13. Collect TiO2 flow through in microcentrifuge tubes.
This is the TiO2FT fraction.This step should be performed slowly using a Combi-Syringe to apply pressure. -
14. Wash the TiO2 columns using 5 μl TiO2 loading solution and pool the eluates with the TIO2FT fraction collected in step 13.
This step should be performed slowly using a Combi-Syringe to apply pressure. -
15. Wash the TiO2 columns using 30 μl TiO2 wash solution.
This step should be performed slowly using a Combi-Syringe to apply pressure. It is not essential to keep the eluate from this step. -
16. Elute the phosphopeptides using 30 μl high-pH elution solution.
This is the TiO2E fraction.Use 30 μl per column and pool the eluates (TiO2E) from the same sample. This step should be performed slowly using a Combi-Syringe to apply pressure. 17. Acidify the eluates (TiO2E) using 100% TFA to a pH of ~2 to 3.
-
18. Prepare a slurry of Oligo R3 reversed-phase resin in 50% acetonitrile. Pack the Oligo R3 into a P-20 narrow-bore pipet tip.
Squeeze the tip of the narrow-bore P20 pipet tip to prevent the beads from leaking out. Sufficient Oligo R3 slurry should be added to make the columns ~5 mm long. It is important to have a good seal on these columns, as column breakthrough will block on-line liquid chromatography systems. -
19. Load each of the phosphopeptide-containing eluates that have been enriched with the TiO2 strategy (but only the IMAC–1% TFA and IMAC-FT fractions from the SIMAC protocol) onto one (for simple samples) or two to three (for complex mixtures) Oligo R3 columns depending on the amount of material.
This step should be performed slowly using a Combi-Syringe to apply pressure. -
20. Wash the Oligo R3 columns using 30 μl 0.1% TFA.
This step should be performed slowly using a Combi-Syringe to apply pressure. -
21. Prepare samples for MS:
-
For MALDI-TOF-MS/MS:
-
Elute the peptides from Oligo R3 column using 5 μl phosphopeptide RP elution buffer for MALDI-TOF-MS/MS for subsequent MALDI-TOF-MS/MS analysis.
The elution buffer contains the MALDI matrix. Spot directly onto MALDI sample plate, using a pipet.
As spot is drying, if matrix crystals are not forming well, spot an additional 0.5 μl phosphopeptide RP elution buffer for MALDI-TOF-MS/MS onto the phosphopeptide spot.
-
-
For LC-ESI-MS/MS:
Elute the peptides from the Oligo R3 column using 30 μl phosphopeptide RP elution buffer for LC-ESI-MS/MS for each column, and pool the eluates.
Dry pooled eluates by vacuum centrifugation.
-
Resuspend the monophosphopeptides in 0.5 μl 100% formic acid and add 10 μl of 0.1% formic acid immediately.
TFA may interfere with the ionization in LC-ESI-MS/MS; therefore, it is critical that formic acid be used for this preparation.The monophosphopeptides (IMAC–TFA 1% and IMAC-FT fractions) are now ready for LC-ESI-MS/MS analysis.
-
Elution of multiply phosphorylated peptides (IMAC–pH 11 fraction)
-
22. Resuspend the dried multiply phosphorylated peptides from the IMAC–pH 11 eluate in 0.5 μl of 100% formic acid and dilute this immediately using ultrapure water to obtain a final pH of 2 to 3.
The multiply phosphorylated peptides are now ready for LC-ESI-MS/MS analysis.
BASIC PROTOCOL 5. PEPTIDE SAMPLE CLEANUP PRIOR TO MS ANALYSIS, MS INSTRUMENT SELECTION, AND DATABASE SEARCHING
It is often necessary to desalt and/or concentrate peptide mixtures prior to analysis by MS (either MALDI-TOF-MS/MS or LC-ESI-MS/MS), as excess salts or trace amounts of detergents interfere with peptide ionization and also add to the chemical noise or background in the mass spectra. Sample cleanup can be performed either on-line, using a trap column or filter prior to the analytical column in a liquid chromatography setting (e.g., LC-ESI-MS/MS), or off-line using any number of commercially available devices. This protocol details the use of an off-line sample cleanup format. The most commonly used methods for off-line cleanup employ reversed-phase chromatography in a pipet tip, spin column, or syringe column format. Examples of commercially available devices used for peptide cleanup include Millipore ZipTips, Nest Group UltraMicrospin columns (http://www.nestgrp.com), and Waters Sep-Pak cartridges, among others. The choice of format often depends on the required binding capacity, although the protocol is generally applicable to all reversed phase–based sample cleanup devices.
NOTE: Use only HPLC and/or mass spectrometry grade reagents throughout the protocol.
Materials
100% acetonitrile
0.1% (v/v) and 10% (v/v) trifluoroacetic acid (TFA)
Sample containing peptides for MS analysis (see previous protocols)
RP LC-ESI-MS/MS elution solution (see recipe)
RP MALDI-TOF-MS/MS elution solution (see recipe)
Low-binding, siliconized microcentrifuge tubes (Fisher Scientific)
C18 spin column, cartridge, or pipet tip with sufficient binding capacity for peptides in sample
Vacuum manifold (optional)
-
Prepare the column by washing three times with 200 μl 100% acetonitrile. Discard flow through.
The processing of samples can be performed using a pipet or centrifuge, or with the aid of a vacuum manifold. If a centrifuge is used, a microcentrifuge tube can be used as the flow through receptacle. In cases where a C18 spin column is used, the solution is added to the top of the column and then forced through the matrix by applying a vacuum, centrifugal force, or manual force. All volumes listed in this protocol are appropriate for UltraMicrospin columns (Nest Group), but may be adjusted for other devices. Rinse the column three times with 200 μl 0.1% TFA. Discard flow through.
To the peptide sample, add 10% TFA to bring the final concentration to 0.1% TFA.
-
Apply the sample to the column and use a pipet to push sample through slowly.
The flow through may be collected and reapplied to the column if desired. -
Rinse column three to ten times with 200 μl 0.1% TFA.
The volume required for sufficient rinsing depends on the concentration of salts and other reagents in the sample.If electrospray/nanospray is to be performed, it is advisable to subsequently rinse the column three times with 200 μl water to remove the TFA, which may interfere with ionization in electrospray/nanospray. -
Elute peptides from the column using 30 μl of RP LC-ESI-MS/MS elution solution for samples that will be analyzed by LC-ESI-MS/MS, or RP MALDI-TOF-MS/MS elution solution for samples that will be analyzed by MALDI-TOF-MS/MS. Collect the eluate in a clean tube. The samples are now ready for MS analysis.
Sometimes it is necessary to further concentrate the eluate, using a vacuum centrifuge to reduce the volume, prior to analysis by MS.
SELECTING THE APPROPRIATE MS INSTRUMENTATION
The field of MS instrumentation has exploded in the past 10 years, and the development of technology platforms for handling biological samples was a driving force behind this expansion. While there are many different configurations or types of MS instruments available and each is suited to the analysis of particular types of samples, there are three components common to all of these instruments. The first step in MS involves generating the gas-phase charged species, i.e., the ion, and this is done at the first component of the mass spectrometer, the ionization source. The second step is the mass filter or mass analyzer, which functions to separate the ions by their mass-to-charge ratio (m/z). The third step is the detector, which converts the ion impact, for example, into an electronic signal.
The two most common types of ionization used in the analysis of proteins and peptides are matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI). MALDI is a technique that involves mixing the analyte (e.g., peptides or proteins) with a matrix, generally a small organic acid that absorbs in the UV region, and allowing them to co-crystallize on a stainless steel sample plate. The ionization is achieved by firing a pulsed UV-emitting laser at the sample to generate ionized peptides or proteins. In contrast to MALDI, ESI generates a continuous supply of ions from a liquid solution, where a liquid sample is emitted from a nozzle, needle, or capillary in the presence of a strong electric field. ESI is often coupled with on-line liquid chromatography to separate the peptides in complex mixtures. Furthermore, it is possible to separate peptides via liquid chromatography off-line, in order to generate less complex mixtures before the plating used for MALDI.
The most common types of mass analyzer include time-of-flight (TOF), the quadrupole, and the ion trap. As the TOF is capable of detecting very high masses (e.g., 300,000 Da), it is well suited to MALDI ionization, which typically generates singly charged species. Ion traps are typically limited to smaller mass ranges (e.g., 2000 to 4000 Da), and are well suited to ESI, which generates multiply charged species, allowing high-mass ions to be detected at their lower m/z values. Though traditional ion traps have a lower resolving power than TOF, the recently invented Orbitrap (Thermo) breaks this rule and offers mass resolution up to 100,000 Da.
In general, most commercially available mass spectrometry platforms can be used to analyze most types of peptide and protein samples. However, selecting the appropriate instrumentation often depends on availability, cost, ease of use, and performance characteristics required for the particular sample being analyzed. Important performance characteristics include whether the instrument is capable of tandem mass spectrometry (MS/MS), which is becoming a routine requirement for reporting peptide identifications, and is often used instead of or in addition to peptide mass fingerprinting (PMF), where patterns of peptide masses are used to obtain the identification. Other characteristics include the resolving power, mass accuracy, and linear dynamic range. Finally, depending on the type of sample analysis required, it may be important to consider the types of fragmentation, such as electron transfer dissociation (ETD; Mikesh et al., 2006), electron capture dissociation (ECD; Zubarev, 2004), or collision-induced dissociation (CID; Sleno and Volmer, 2004), as these can have an impact on the types of post-translational modifications that can be identified.
CONCEPTS TO CONSIDER WHEN PERFORMING THE MS DATABASE SEARCH
After the MS data are collected, the next step in a proteomics pipeline (Fig. 10.25.1) is to perform a database search to identify the proteins and/or peptides analyzed by MS. A number of parameters can affect the outcome of a database search, and, ultimately, search parameters should consider the history of the sample, such as organism, enzymatic or chemical digestion, and reduction/alkylation steps. Importantly, the summary below highlights a number of critical considerations for database searching for MS/MS data and does not discuss the often extensive amount of downstream data analysis required after the search results are returned, such as manual interpretation, filtering out low-scoring matches, and redundancy removal, all of which can have significant effects on the final output.
Protein database scope
The protein database is typically a collection of protein amino acid sequences (e.g., in FASTA format) which will be used to generate the in silico peptide database against which the MS data will be searched. If the database does not contain the protein that is in the sample, it will not find a suitable match. It is essential that the protein database contain sufficient coverage of the taxonomy (e.g., species) or related taxonomy represented in the sample. Not all databases are curated rigorously, and some are combinations of multiple databases. Table 10.25.1 highlights the main features of protein databases commonly used for searching proteomic MS data.
Table 10.25.1.
Features of Commonly Used Protein Sequence Databases
| Database | Source | Download site | Number of species/taxonomiesa | Number of entriesa | Description |
|---|---|---|---|---|---|
| Swiss-Prot | Swiss Institute of Bioinformatics, European Bioinformatics Institute | ftp://ftp.ebi.ac.uk/pub/databases/ | 11,661 | 412,525 | A curated protein sequence database which strives to provide a high level of annotation (such as the description of the function of a protein, its domain structure, post-translational modifications, variants, etc.), a minimal level of redundancy, and high level of integration with other databases |
| TrEMBL | Swiss Institute of Bioinformatics, European Bioinformatics Institute | ftp://ftp.ebi.ac.uk/pub/databases | 191,318 | 7,341,751 | A computer-annotated supplement of Swiss-Prot that contains all the translations of EMBL nucleotide sequence entries not yet integrated in Swiss-Prot |
| NCBInr | National Center for Biotechnology Information | ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz | 64 | 7,031,513 | Nonredundant protein sequence database with entries from GenPept, SwissProt, PIR, PDF, PDB, and RefSeq |
| IPI | European Bioinformatics Institute | ftp://ftp.ebi.ac.uk/pub/databases/IPI/current/ | 7 | 309,242 | Protein sets are made for a limited number of higher eukaryotic species whose genomic sequence has been completely determined but where there are a large number of predicted protein sequences that are not yet in UniProt. Nonredundant. Annotated splice variants are listed as separate entries. Assembled from Swiss-Prot, TrEMBL, RefSeq, Ensemble, TAIR, H-InvDB, Vega. |
| RefSeq | National Center for Biotechnology Information | ftp://ftp.ncbi.nih.gov/refseq/release/ | 7,773 | 6,413,124 | A nonredundant collection of richly annotated DNA, RNA, and protein sequences from diverse taxa. The collection includes sequences from plasmids, organelles, viruses, archaea, bacteria, and eukaryotes. Each RefSeq represents a single, naturally occurring molecule from one organism. |
| dbEST | National Center for Biotechnology Information | ftp://ftp.ncbi.nih.gov/blast/db/FASTA/estothers.gz | 1704 | 60,497,687 | Contains “single-pass” cDNA sequences, or Expressed Sequence Tags, from the EST divisions of GenBank |
Statistics valid as of 3/10/09.
Peptide database size
The user interface for the database search will typically allow for a range of parameters to be manually set by the user. Many of these parameters will be used to generate the in silico peptide database from the protein database and can greatly affect the sensitivity and search speed of the search algorithm. It is important to allow for those features or modifications that are realistic while conserving database size, as larger databases may require longer search times and lead to a loss in sensitivity.
Taxonomy
If sufficient representation of the appropriate taxonomy is included in the selected protein database, the peptide database can often be restricted at the species level (e.g., Homo sapiens) or class level (e.g., mammals).
Enzyme
Was an enzyme used to generate the peptides prior to MS analysis? If so, the peptide database can be generated using the cleavage specificity of the enzyme (e.g., the presence of a C-terminal Lys for trypsin- or LysC-cleaved peptides) or combination of enzymes when appropriate. If the peptides were not generated using an enzyme, a “no-enzyme” search can be performed. A no-enzyme search should not be performed on data generated using an enzymatic digest, as it will significantly increase the size of the database and can lead to random matches. “Semi-enzyme” searches, where the peptides generated will contain enzyme specificity at only one terminus, can be used to increase the peptide database size.
Missed cleavages
If an enzyme was used to generate the peptide sample, how many missed cleavages (i.e., sites the enzyme missed) are acceptable? For tryptic digests, this number is often set between 0 and 2.
Post-translational modifications
Modifications can increase the peptide database size greatly and are directly related to the sample treatment. Modifications can be either fixed (always on; e.g., carboxyamidomethylation on cysteine, which occurs after treatment with iodoacetamide), or variable (not always on, e.g., phosphorylation). Modifications should be chosen carefully because, if too many unrealistic options are allowed, the search times can be significantly increased, and there can be an increase in the number of false positive matches, which will require a lot of manual data interpretation to validate.
Instrumentation-specific parameters
Additional parameters often included in the search engine user interface include instrumentation-specific parameters. These are directly related to the expected or known performance of the mass spectrometer used to generate the data. Examples include whether the instrument is capable of recording either monoisotopic or average masses for the precursor and fragment ions. Also, what is the mass accuracy for the precursor and fragment ions? These affect the mass type (e.g., monoisotopic versus average) and mass tolerances (e.g., parent and fragment mass tolerances) allowed during the search. Other parameters that may be included are the charge states and fragmentation expected. For example, in MALDI, singly charged species are expected, whereas in ESI, multiply charged species are expected. Finally, the type of fragmentation used (CID, ETD, or ECD) and/or the type of mass spectrometer (e.g., TOF versus trap) will affect the types of fragment ions expected.
Search algorithm
There are currently more than a dozen commercial or open-source search algorithms available. The choice of search algorithm can be based on cost, accessibility, vendor compatibility, ease of use, features, performance with certain data types, or a combination of all of the above. However, all search algorithms are not created equal, and the outputs from each can vary widely depending on the type of data searched. Differences among search engines can include the scoring algorithm, whether it considers data quality, and whether it considers all fragment ion types or only a subset. Often, the best way to select an algorithm is to examine the details regarding data-format compatibility and necessary features, test it on a data set, and compare the results to what is expected. Finally, since the same MS data will often result in a different output depending on the search algorithm used, it is not uncommon to search the same data using multiple algorithms to obtain high-confidence matches. Table 10.25.2 outlines a number of commonly used search algorithms, but is not meant to be an exhaustive list.
Table 10.25.2.
Commonly Used Search Algorithms for MS/MS Data Searching
| Search algorithm | Availability | Compatible file formats | Vendor/source |
|---|---|---|---|
| Mascot | Commercial | pkm, mzData, mzXML, asc, pkl, dta, pks | http://www.matrixscience.com |
| OMSSA | Open source | dta, mzXML, mgf, pkl | http://pubchem.ncbi.nlm.nih.gov/omssa/ |
| Peaks | Commercial | pkl, dta, mgf, anz, mzXML, mzData, RAW, wiff, baf, yep, d, dat | http://www.bioinformaticssolutions.com |
| Phenyx | Commercial | dta, pkl, mgf, bdtx, mzdata, mzXML | http://www.genebio.com |
| ProteinLynx | Commercial | mzXML, mzData, pkl, txt | http://www.waters.com |
| Paragon (in ProteinPilot) | Commercial | wiff, mgf, ABI | http://www.appliedbiosystems.com |
| Sequest (in BioWorks) | Commercial | RAW | http://www.thermo.com |
| SorcererSequest | Commercial | RAW, mzXML, dta, out, srf, pkl, mgf | http://www.sagenresearch.com |
| Spectrum Mill | Commercial | Agilent, RAW, wiff, pkl | http://www.chem.agilent.com |
| X!Tandem | Open source | dta, pkl, mgf | http://www.thegpm.org/TANDEM/ |
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2; for suppliers, see APPENDIX 4. Acetonitrile is a flammable, organic solvent and as such should be disposed of according to individual institutional disposal procedures. Similar disposal considerations must be applied to solutions containing K3[Fe(CN)6] or methanol.
Colloidal Coomassie destain
50% (v/v) methanol
50% (v/v) H2O
Store indefinitely at room temperature
Gel enzyme stock solution
12.5 mg/ml trypsin
25 mM NH4HCO3
Stock enzyme solutions can be kept lyophilized at −20°C or −80°C for months
Depending on the complexity of the peptide mixture desired, different proteases may be chosen individually or sequentially. Chymotrypsin cleaves on the C-terminal side of Phe, Tyr, Leu, Trp, and Met residues; trypsin cleaves on the C-terminal side of Lys and Arg residues; LysC cleaves on the C-terminal side of Lys; and AspN cleaves on the N-terminal side of Asp. Each of these enzymes cleaves most efficiently at a particular pH. Chymotrypsin, trypsin, LysC, and AspN are most efficient at alkaline pH (e.g., pH 8).
Chemicals (e.g., cyanogen bromide/formic acid) are an alternative to enzymes. Cyanogen bromide may be useful when working with insoluble proteins, such as transmembrane proteins, or when the introduction of an enzyme is not desired. Common concentrations of cyanogen bromide range from 30 to 75 mg/ml in 70% aqueous formic acid.
Gel extraction solution
50% (v/v) acetonitrile
1% (v/v) trifluoroacetic acid
49% H2O
Store up to 1 month at room temperature in dark glassware
Gel wash solution
50% (v/v) acetonitrile
25 mM NH4HCO3
Store at room temperature for approximately 1 to 2 days
Glutaraldehyde-free silver destain
15 mM K3[Fe(CN)6]
50 mM Na2S2O3
100 mM Na2S2O3 and 30 mM K3[Fe(CN)6] should be prepared fresh, mixed in a 1:1 ratio, and used immediately.
High-pH elution solution
2% (v/v) NH4OH
98% H2O
Store indefinitely at room temperature
CAUTION: NH4OH is toxic and should be handled with care.
IMAC wash solution
50% (v/v) acetonitrile
0.1% (v/v) trifluoroacetic acid
49.9% H2O
Store up to 1 month at room temperature in amber glassware
Low-pH elution solution
20% (v/v) acetonitrile
1.0% (v/v) trifluoroacetic acid
79% H2O
Store up to 1 month at room temperature in dark glassware
Phosphate-buffered saline (PBS)
0.1 M sodium phosphate, pH 7.4 (APPENDIX 2)
0.15 M NaCl
Store up to 6 months at room temperature
Phosphopeptide RP elution buffer for LC-ESI-MS/MS
50% (v/v) acetonitrile
0.1% (v/v) formic acid
49.9% H2O
Store up to 1 month at room temperature in dark glassware
Phosphopeptide RP elution buffer for MALDI-TOF-MS/MS
20 mg/ml 2,5-dihydroxybenzoic acid (DHB)
50% (v/v) acetonitrile
1% (v/v) phosphoric acid
Phosphoric acid is toxic and should be handled with care.
RP LC-ESI-MS/MS elution solution
50% (v/v) acetonitrile
0.1% (v/v) formic acid
49.9% H2O
Store up to 1 month at room temperature in dark glassware
RP MALDI-TOF-MS/MS elution solution
50% (v/v) acetonitrile
0.1% (v/v) trifluoroacetic acid
49.9% H2O
Store up to 1 month at room temperature in dark glassware
TiO2 loading solution
1 M glycolic acid
80% (v/v) acetonitrile
5% (v/v) trifluoroacetic acid
Store up to 1 month at room temperature in amber glassware
TiO2 wash solution
80% (v/v) acetonitrile
1% (v/v) trifluoroacetic acid
19% H2O
Store up to 1 month at room temperature in amber glassware
Traditional Coomassie destain solution
50% (v/v) acetonitrile
50% 25 mM NH4HCO3
Solution can be stored at room temperature for ~1 to 2 days.
50 mM NH4HCO3 should be made fresh and diluted 1:1 with acetonitrile.
COMMENTARY
Background Information
MS is an essential analytical technique of modern protein biochemistry where it plays a principal role in protein identification and characterization. The preparation of proteins for eventual analysis by MS often involves first isolating or separating the proteins(s) in order to reduce the complexity and dynamic range (both concentration and mass) of the proteome. The primary advantage of separating the intact proteins prior to enzymatic or chemical digestion is that the intrinsic protein information is maintained. This increases the probability of obtaining a higher degree of amino acid sequence coverage, and therefore increases the probability of being able to identify any protein isoforms, polymorphisms, and post-translational modifications.
The most popular methods for protein separation in a proteomics workflow are electrophoresis and chromatography. Separation can be based on one or more of the four physical properties of proteins: size/mass, charge (pI), hydrophobicity, and biospecificity. One can carry out fractionation or enrichment under denaturing or nondenaturing conditions; nondenaturing conditions preserve protein–protein interactions. Combining different types of separation (i.e., number of modalities), either on-line or off-line, increases the overall resolving power. When gels are used, the challenges involve completely digesting the protein when it is embedded within the gel matrix, and subsequently extracting the maximum amount of the peptides following digestion. Liquid chromatography is also a popular method for separating proteins and peptides. RP-HPLC is the most commonly used chromatography method, and is often the final step in a multi-dimensional separation. This is due to the ability of RP-HPLC to both concentrate and desalt the sample with MS-compatible solvents. While protein and peptide separation in the context of proteomics continues to evolve, this unit outlines the most useful methods that have been extensively used to date.
Critical Parameters and Troubleshooting
One of the key, yet often overlooked, principles in preparation of a protein sample for MS analysis is that for a protein sample to be enzymatically digested, the enzyme must have access to the cleavage sites (i.e., the protein must be solubilized), and the solution must be at an appropriate pH for enzyme activity. Disulfide bonds tether different regions in a protein and thereby compact its three-dimensional structure. Such compacting inhibits the ability of proteases to access all possible cleavage sites, leading to incomplete digestion. As such, proteins with disulfide bonds may be reduced and alkylated prior to enzymatic digestion in order to enhance the efficiency of cleavage. One must remember, however, that when reduction and alkylation are performed, the alkylation results in carbamidomethyl-modified cysteine residues, which increase the mass of cysteine residues by 57.021 atomic mass units (amu), and this modification must be taken into account when performing the MS data searches. Similarly, a chemical cleavage with CNBr at Met results in C-terminal homoserine and homoserine lactone in equilibrium.
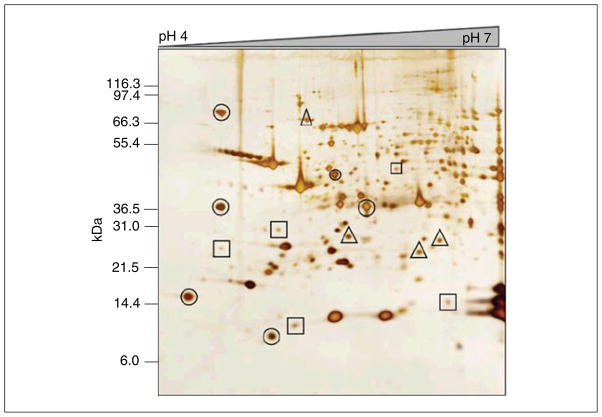
For Basic Protocol 1, a common issue is that insufficient protein is present in the gel spot or band for protein identification by MS. When this happens, it is important to first rule out that there was a technical issue with the steps involved with the digestion and extraction of the peptides. To ensure that processing was successful, it is helpful to include a positive control. This means that one should process one of the proteins from the molecular weight markers or another high-abundance protein from the same gel as the spot/band of interest. At the same time, one should include a negative control, such as a piece of empty gel (i.e., a piece with no protein present), to determine any contamination that may have occurred during sample processing. As a general rule, if a gel band/spot is visible using traditional Coomassie stain (TCS), there should be sufficient material for a confident identification. Lower-abundance spots/bands can also be successfully analyzed by MS. To maximize the quantity of peptides obtained from any protein spot or band, the following should be considered: (1) the largest spot or band should be used, which usually means that it must be excised manually; and (2) one should combine the equivalent spots/bands from multiple gels into the same tube and process this as if it were a single sample. Finally, it is the authors’ experience that excising the spot or band from a dried gel increases the signal-to-noise ratio in the MS data, presumably due to the removal of chemical contaminants normally present in the gel. Drying the gel also extends the “shelf life,” as spots/bands can be excised, successfully digested, and used for MS analysis apparently indefinitely. Figure 10.25.3 shows a 2-DE gel, which illustrates those proteins that will likely result in an identification from a single gel compared to spots which may need to be pooled from multiple gels.
Figure 10.25.3.
200 μg of whole-tissue extract from mouse myocardium run on pH 4 to 7 two-dimensional gel and visualized with an MS-compatible silver stain. Protein spots that will likely provide reliable protein identification from a single gel (circled), as well as protein spots that will likely need to be pooled from multiple gels (squares), are indicated. Also shown are protein spots (triangles) that may or may not contain sufficient protein for identification, depending upon the MS instrumentation used.
For Basic Protocols 2 and 3, one of the most important factors that can affect the efficiency of a trypsin digestion is pH. If digestion is not complete, double check that the digestion solution pH is ~8 before adding trypsin, using a small drop applied to pH paper. Another important consideration is the solubility of the protein and the access of trypsin to the digestion sites within the protein sequence. If protein solubilization is a problem, the addition of MS-compatible surfactants—e.g., low concentrations of Progenta (Protea Biosciences), Rapigest (Waters), PPS (ProteinDiscovery), or acetonitrile may increase the effectiveness of the enzymatic digestion. It may be helpful to examine aliquots of the digestion for undigested protein using SDS-PAGE (UNIT 10.2A) to determine the appropriate digestion times and/or enzyme concentrations.
For Basic Protocol 3, it may be necessary to optimize the elution conditions for releasing the protein from the affinity matrix. Sequential extraction using increasingly stringent conditions can be helpful to indicate the efficiency of a given condition. For example, after eluting with 0.1 M glycine, pH 2.5, the peptide or protein can be re-eluted from the affinity matrix with reducing SDS-PAGE sample buffer (UNIT 10.2A).
For Basic Protocol 4, it is advisable to test the protocol with a series of proteins that contain known sites of phosphorylation. It is important to have fresh buffers with the correct pH. TiO2 beads are light sensitive, and the efficiency of TiO2 beads to select for phosphorylated peptides will decrease over time. 2,5-Dihydroxybenzoic acid (DHB) will require higher laser intensities than other widely used MALDI-MS matrices (e.g., α-cyano-4-hydroxycinnamic acid).
For Basic Protocol 5, if no peptides are recovered, try re-eluting with a higher concentration of acetonitrile (e.g., 90%). Also, if it is suspected that the peptides did not initially bind to the resin (e.g., fewer peptides observed than expected), save the non-bound fraction and reprocess it with a new column. It is important to consider that the sample should contain 0.1% TFA and low organic (e.g., <10% acetonitrile) to ensure maximum binding to the column.
Anticipated Results
Improvements in each step of the proteome technical pipeline illustrated in Figure 10.25.1 will result in increased efficiency, confidence in protein identifications, and improved protein characterization. Intact protein separation prior to digestion should help to reduce sample complexity and thus assist in identifying more of the proteome of interest. Selective enrichment of peptides and proteins by affinity chromatography will increase the likelihood of identifying any particular modifications and/or characterizing specific proteins of interest.
Time Considerations
Basic Protocol 1 will take ~1 day, while other protocols can typically be performed within a few hours.
Acknowledgments
This work was supported by an NIH Pathway to Independence Award K99-L094708-01 (RLG), a C.J. Martin Postdoctoral Research Fellowship, The National Health and Medical Research Foundation (464905) of Australia (MYW), American Heart Association Pre-Doctoral Fellowships 0715247U (LAK), and 0815145E (CIM), and the NHLBI Proteomics Innovation Contract N01-HV-28180 (JEV).
Footnotes
Key References
Thingholm et al., 2008. See above.
This is a landmark paper in which both IMAC and TiO2 are combined in a sequential manner to allow expanded phosphopeptide enrichment.
Shevchenko et al., 1996. See above.
This is the most commonly used silver staining and peptide extraction method for MS compatible samples.
Internet Resources
Two-dimensional polyacrylamide gel electrophoresis database.
Database of proteomic analysis tools which include cleavage site predictions, modification predictions, topology prediction, and visualization software, among others.
Database of useful protocols for preparing proteomic samples, as well as tutorials and principles of chromatography and mass spectrometry.
Comprehensive database of protein modifications and their masses for mass spectrometry.
http://www.cbs.dtu.dk/services/NetPhos/
Predicts sites of phosphorylation on eukaryotic proteins.
http://www.proteomecenter.org/course.php
Excellent repository of lecture notes for proteomics, mass spectrometry, and bioinformatics courses designed for beginners, taught at the Institute for Systems Biology at the Seattle Proteome Center.
Literature Cited
- Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marban E, Van Eyk JE. Proteomic analysis of pharmacological preconditioning: Novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Chrambach A, Reisfeld RA, Wyckoff M, Zaccari J. A procedure for rapid and sensitive staining of protein fractionated by polyacrylamide gel electrophoresis. Anal Biochem. 1967;20:150–154. doi: 10.1016/0003-2697(67)90272-2. [DOI] [PubMed] [Google Scholar]
- Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: Principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath J. Immobilized metal ion affinity chromatography. Protein Expr Purif. 1992;3:263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. J Mass Spectrom. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- Smith JC, Figeys D. Recent developments in mass spectrometry-based quantitative phosphoproteomics. Biochem Cell Biol. 2008;86:137–148. doi: 10.1139/O08-007. [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Larsen MR. The use of titanium dioxide micro-columns to selectively isolate phosphopeptides from proteolytic digests. Methods Mol Biol. 2009;527:57–66. doi: 10.1007/978-1-60327-834-8_5. [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics. 2008;7:661–671. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Larsen MR. Enrichment and separation of mono- and multiply phosphorylated peptides using sequential elution from IMAC prior to mass spectrometric analysis. Methods Mol Biol. 2009;527:67–78. doi: 10.1007/978-1-60327-834-8_6. [DOI] [PubMed] [Google Scholar]
- Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr Opin Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]