Abstract
Background and rationale
Hepatic stellate cell (HSC) activation is an essential event during liver fibrogenesis. Methionine adenosyltransferase (MAT) catalyzes biosynthesis of S-adenosylmethionine (SAMe), the principle methyl donor. SAMe metabolism generates two methylation inhibitors, methylthioadenosine (MTA) and S-adenosylhomocysteine (SAH). Liver cell proliferation is associated with induction of two non-liver specific MATs, MAT2A that encodes the catalytic subunit α2 and MAT2β , which encodes a regulatory subunit β that modulates the activity of the MAT2A-encoded isoenzyme MATII. We reported that MAT2A and MAT2β genes are required for liver cancer cell growth that is induced by the profibrogenic factor, leptin. Also, MAT2β regulates leptin signaling. The strong association of MAT genes with proliferation and leptin signaling in liver cells led us to examine the role of these genes during HSC activation.
Results
MAT2A and MAT2β are induced in culture-activated primary rat HSCs and HSCs from 10-day bile duct ligated (BDL) rat livers. HSC activation led to a decline in intracellular SAMe and MTA levels, a drop in the SAMe/SAH ratio and global DNA hypomethylation. The decrease in SAMe levels was associated with lower MATII activity during activation. MAT2A silencing in primary HSCs and MAT2A or MAT2β silencing in the human stellate cell line LX-2, resulted in decreased collagen and alpha-smooth muscle actin (α-SMA) expression and cell growth and increased apoptosis. MAT2A knockdown decreased intracellular SAMe levels in LX-2 cells. Activation of extracellular signal-regulated kinase and phosphatidylinositol-3-kinase signaling in LX-2 cells required the expression of MAT2β but not that of MAT2A.
Conclusion
MAT2A and MAT2β genes are induced during HSC activation and are essential for this process. SAMe level falls resulting in global DNA hypomethylation.
Keywords: Fibrosis, Fibrogenesis, Collagen, Bile duct ligation, DNA Methylation
INTRODUCTION
The hepatic stellate cell (HSC) is now well established as the key cellular element involved in the development of hepatic fibrosis. Because of its importance in the fibrotic process, there is considerable interest in establishing the molecular events that trigger and perpetuate HSC activation. Development of liver fibrosis entails major alterations in the quantity and quality of hepatic extracellular matrix (ECM) and there is overwhelming evidence that activated HSCs are the major producers of the fibrotic neomatrix (1). In normal liver, HSCs are the major storage sites of vitamin A, stored in the cytoplasm as retinyl esters. Following chronic liver injury, HSCs proliferate, lose their vitamin A and undergo a major phenotypical transformation to α-smooth muscle actin (α-SMA) positive activated HSCs, which produce a wide variety of collagenous and non-collagenous ECM proteins (1). The profibrogenic potential of activated HSCs is due to their capacity to synthesize fibrotic matrix proteins and components that inhibit fibrosis degradation. Among the large number of factors identified as activators of matrix production are transforming growth factor-β (2), connective tissue growth factor (3), leptin (4) and platelet-derived growth factor (PDGF) (5). Activation of HSCs is mediated by various cytokines and reactive oxygen species released from damaged hepatocytes and activated Kupffer cells (6). Hence, inhibition of HSC activation and its related events such as ECM formation and cellular proliferation are important targets for therapeutic intervention.
Quiescent primary HSCs undergo spontaneous activation when plated on uncoated plastic and attain a myofibroblast-like phenotype with loss of vitamin A and increased expression of α-SMA and collagen (7, 8). In vivo activated HSCs can be obtained from livers of animals undergoing experimentally induced biliary fibrosis resulting from bile duct ligation (BDL) (9). In this animal model, the number of activated HSCs increase during liver injury (10). The BDL model has been used extensively to study the pathogenesis of liver injury, HSC activation and to test the efficiency of potential antifibrotic drugs (11).
Methionine adenosyltransferase (MAT) is a critical enzyme required for biosynthesis of S-adenosylmethionine (SAMe), the principle biological methyl donor (12). SAMe also donates propylamine moiety for polyamine biosynthesis and in the process generates methylthioadenosine (MTA), which is an inhibitor of methylation (13). Transmethylation reactions of SAMe result in its conversion to another potent methylation inhibitor, S-adenosylhomocysteine (SAH) (14). Mammalian cells express two genes MAT1A and MAT2A that encode the two MAT catalytic subunits, α1 and α2, and a third gene MAT2β , that encodes the regulatory subunit β that regulates the activity of MAT2A-encoded isoenzyme MAT II by lowering the inhibition constant (Ki) for SAMe and Michaeli’s constant (Km) for methionine (15, 16). MAT1A is expressed mainly in hepatocytes and maintains the differentiated state of these cells (12). MAT2A is expressed in all extrahepatic tissues and is induced in liver during active growth and de-differentiation (12, 17, 18). The MAT2β gene is induced during liver cirrhosis and hepatocellular carcinoma (HCC) (19). Hepatic stellate cells do not express MAT1A (20). MAT2A is the only enzyme responsible for SAMe biosynthesis in these cells. Our recent work in liver cancer cells showed that induction of MAT2A and MAT2β genes is required for cell growth that is induced by leptin (21), an adipokine that plays a pivotal role in liver fibrogenesis and carcinogenesis (4, 22). Furthermore, leptin signaling in the liver cancer cell line, HepG2 requires the expression of MAT2β gene but not that of MAT2A. Knockdown of MAT2β inhibits upstream events like leptin-mediated signal transducers and activators of transcription 3 (STAT3) activation as well as downstream events like extracellular signal-regulated kinase (ERK) and phosphatidylinositol-3-kinase (PI3-K) activation (21). Since leptin is a potent pro-fibrogenic growth factor regulated by MAT gene expression and MAT genes are associated with cellular proliferation, we investigated the hypothesis that MAT2A and MAT2β genes may play important roles in the activation of HSCs. Our results indicate dramatic changes in MAT genes and SAMe homeostasis during activation of HSCs and provide evidence that activation of the MAT genes is an essential event during fibrogenesis.
MATERIALS AND METHODS
Materials
All reagents used in this study were of analytical grade and obtained from commercial sources.
HSC isolation and cell culture
The use of animals in this study was approved by the InstitutionalAnimal Care and Use Committee (IACUC) of the University of Southern California (USC). HSCs were isolated from normal male Wister rats or 10-day BDL rats by the Non-Parenchymal Liver Cell Core of the Research Center for AlcoholicLiver and Pancreatic Diseases and Cirrhosis as previously described (23). The purity of isolated HSCs was examined by ultraviolet-excited fluorescence microscopy and viability was determined by trypan blue exclusion. Normal rat HSCs were cultured in low glucose DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics for 1, 3, 5 or 7 days to examine culture activation. HSC isolated from BDL and sham-operated rats were cultured for 16 hours in low glucose DMEM with 3% FBS before analysis. LX-2, a human stellate cell line that expresses key components in fibrogenic response like PDGF receptor, leptin receptor and α-SMA (24), was cultured in DMEM with 10% FBS and antibiotics.
Real-time PCR analysis
Total RNA was subjected to reverse transcription (RT) by using M-MLV Reverse transcriptase (Invitrogen, CarlsBad, CA). Two μl of RT product was subjected to real-time PCR analysis. The primers and TaqMan probes for rat or human MAT2A, MAT2β, alpha2(1) collagen (Col1A2), α-SMA and the Universal PCR Master Mix were purchased from ABI (Foster City, CA). Hypoxanthine phosphoribosyl-transferase 1 (HPRT1) was used as housekeeping gene as described (25). The thermal profile consisted of initial denaturation at 95°C for 15 minutes followed by 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute. The cycle threshold (Ct value) of the target genes was normalized to that of HPRT1 to obtain the delta Ct (ΔCt). The ΔCt was used to find the relative expression of target genes according to the formula: relative expression= 2−ΔΔCt, where ΔΔCt= ΔCt of target genes in experimental condition – ΔCt of target gene under control condition.
Western blot analysis
Total cellular protein from primary HSCs or LX-2 cell line was extracted following standard protocols (Amersham BioSciences, Piscataway, NJ) and resolved on 12% SDS-polyacrylamide gels for MAT2A, MAT2β , α-SMA, phospho-ERK and phospho-AK strain transforming (AKT) or on 7.5% gels for type I collagen protein. Western blotting was performed using primary antibodies for MAT2A (GenWay Biotech Inc., San Diego, CA), MAT2β (Novus Biologicals, Littleton, CO), type I collagen, phospho and total ERK, phospho and total AKT, β-actin (Cell Signaling, Danvers, MA), α-SMA (Abcam, Cambridge, MA) and horse radish peroxidase (HRP)-conjugated secondary antibodies. Membranes were developed by chemiluminescence using the ECL detection system (Amersham BioSciences, Piscataway, NJ). Quantitation of blots was done using the Quantity One densitometry program (Bio-Rad laboratories, Hercules, CA).
Determination of SAMe, MTA and SAH levels
Cellular SAMe, MTA and SAH levels were measured in 3x106 culture-activated or BDL HSCs and human LX-2 cells by high performance liquid chromatography (HPLC) as described (26).
Measurement of global DNA methylation
The cytosine extension assay was used to evaluate global DNA methylation as previously described (27) using 3x106 HSCs in 10 cm dishes. Briefly, 1 μg of genomic DNA extracted using Qiagen mini-columns (Qiagen, Valencia, CA) was digested for 16–18 h with 20 U of HpaII (New England Biolabs, Beverly, MA, USA). A second DNA aliquot served as background control and was incubated without addition of the restriction enzyme. The single nucleotide extension reaction was performed in a 25 μL reaction mixture containing 0.5 μg of DNA, 1X PCR buffer, 1.5 mM magnesium chloride, 0.25 U of Choice Taq DNA polymerase (Denville Scientific Inc., NJ), 0.1 μL of [3H]-dCTP (47.7 Ci/mmol; Perkin Elmer, Waltham, MA), incubated at 56°C for 1 hour and then placed on ice. Duplicate 10 μL aliquots from each reaction were applied to a Whatman DE-81 ion exchange filter, washed three times with sodium phosphate buffer, pH 7.0, dried and processed for scintillation counting. Background radioactivity in untreated samples was subtracted from enzyme-treated samples. An increase in [3H]-dCTP incorporation (higher dpm values) indicated that DNA was hypomethylated.
Measurement of MATII enzyme activity in primary HSCs
Primary HSCs (3x106 cells) were cultured on 10 cm dishes and cytosolic protein was tested for MATII enzyme activity using 20 μM L-methionine (Sigma, St. Louis, MO) as described previously (17).
RNA interference (RNAi) analysis
RNAi experiments in primary rat HSCs were performed by forward transfection in day 2 cultured HSCs (1x106 cells per 6 cm dish) using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s protocol. For LX-2 cells, reverse transfection with RNAiMax was done as previously described (21). For phospho-ERK and phospho-AKT immunoblotting, LX-2 cells were cultured in serum-free DMEM for 14 hours and then subjected to reverse transfection with RNAiMax in 10% FBS-containing DMEM. Small interfering RNA (siRNA) oligonucleotides against MAT genes or scrambled sequences were synthesized by the USC Norris Comprehensive Cancer Center Microchemical Core Laboratory and annealed to form duplexes. The following siRNA sequences were used: si-MAT2A (human and rat), 5’-GUGAGAGAGAGCUAUUAGATT-3’ (sense) and5’-UCUAAUAGCUCUCUCUCACTC-3’(antisense); si-MAT2β (human), 5 ’-GAAUGCUGGAUCCAUCAAUTT-3’ (sense) and 5’-AUUGAUGGAUCCAGCAUUCTC-3’ (antisense); si-control with scrambled sequence (negative control siRNA having no perfect matches to known human or rat genes), 5’-UUCUCCGAACGUGUCACAUdTdT-3’(sense) and 5’-AUGUGACACGUUCGGAGAAdTdT-3’ (antisense). Transfection was allowed to proceed for various times and cells were processed for different assays. The siRNA transfection efficiency of Lipofectamine RNAiMax in cells was determined by the BLOCK-iT Alexa FluorR Red Fluorescent Oligo protocol (Invitrogen).
Cell proliferation assay
To assay for cell proliferation, primary HSCs or LX-2 cells were plated at a density of 1x104 per well of a 96-well plate under different knockdown conditions. Bromodeoxyuridine (BrDU) was added to each well at a dilution of 1:2000 during the last 16 hours of knockdown and its incorporation into DNA (a measure of growth) was measured using the BrDU Cell Proliferation assay kit (CalBiochem, San Diego, CA).
Apoptosis assay
LX-2 cells were grown in 6-well dishes and apoptosis in control or RNAi-treated cells was measured at different times using Hoechst staining as described previously (28).
Statistical analysis
Data are represented as Mean±SE. Statistical analysis was performed using ANOVA followed by Students t-test. For changes in mRNA or protein levels, ratios of mRNA (relative expression) and protein (densitometric values) to respective housekeeping controls were compared. Significance was defined as p<0.05.
RESULTS
Expression of MAT genes during HSC activation
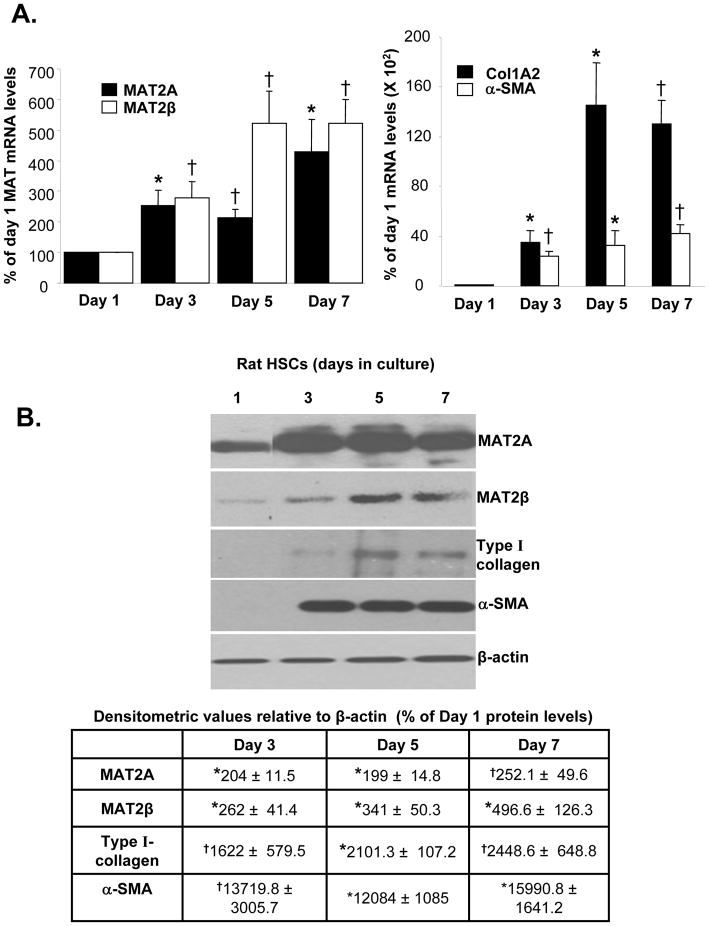
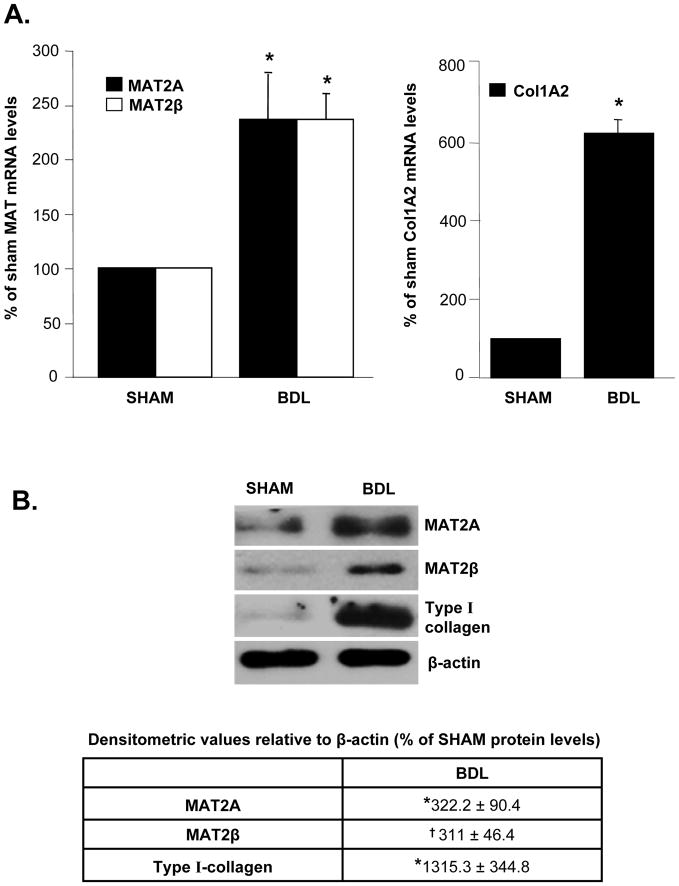
The steady state mRNA levels of MAT2A and MAT2β were induced in culture-activated HSCs at day 3, 5 and 7 compared to quiescent HSCs (day 1) along with the induction of Col1A2 and α-SMA mRNA, markers of HSC activation (Fig. 1A). MAT2A and MAT2β proteins were induced by 250 and 496% respectively by day 7 compared to day 1. MAT2A was maximally induced by day 3, while the expression of MAT2β continued to increase from day 3 to 5. This corresponded to a progressive induction in type I collagen and α-SMA protein expression from day 3 to 5 (Fig. 1B). To make sure that changes in MAT genes during culture activation of HSCs also occur in vivo, expression of MAT genes was examined in HSCs isolated from BDL rats. Expression of MAT2A and MAT2β mRNA and protein was also induced in in vivo activated HSCs isolated from 10-day BDL rat livers compared to sham control livers (Fig. 2). HSC activation in BDL livers was demonstrated by induction of Col1A2 mRNA and type I collagen protein (Fig. 2).
Fig. 1. Expression of MAT2A and MAT2β genes is induced during in vitro HSC activation.
Quiescent HSCs were isolated from rat liver as described in Methods. HSCs (1x106 cells) were cultured on 6 cm plastic dishes and allowed to activate till day 7. (A) RNA was isolated from HSCs at different days in culture and the expression of MAT2A, MAT2β , Col1A2 and α-SMA mRNA was assessed by real-time RT-PCR. Results represent Mean±SE from four HSC preparations; *p<0.05, †p<0.005 vs. day 1. (B) Total cellular protein was extracted from HSCs as described in Methods and subjected to Western blotting for detection of MAT2A, MAT2β , type I collagen and α-SMA. Representative images and densitometric analysis (Mean±SE) from 3 to 4 HSC preparations is shown; *p<0.005, †p<0.05 vs. day 1.
Fig. 2. Expression of MAT2A and MAT2β genes is induced during in vivo HSC activation.
Rats were subjected to BDL surgery or sham operation for a period of 10 days. HSCs were isolated from BDL or sham control rat livers and 1x106 cells were plated on 6 cm plastic dishes for 16 hours. (A) RNA was isolated from BDL or sham HSCs and the expression of MAT2A, MAT2β and Col1A2 mRNA was assessed by real-time RT-PCR. Results represent Mean±SE from four BDL or four sham HSC preparations; *p<0.005 vs. sham control. (B) Total cellular protein was extracted from BDL or sham HSCs as described in Methods and subjected to Western blotting for detection of MAT2A, MAT2β and type I collagen. Representative images and densitometric analysis (Mean±SE) from four BDL or sham preparations is shown; *p<0.05, †p<0.005 vs. sham control.
Changes in SAMe homeostasis and global DNA methylation during HSC activation
As indicated in Table I, a 70 to 75% decrease in intracellular SAMe levels was observed in culture-activated HSCs at day 3, 5 and 7 compared to day 1. A slight decrease in intracellular MTA and SAH levels was also observed by day 7. HSC activation resulted in a two-fold reduction in global DNA methylation. Concomitant to activation-induced DNA hypomethylation in HSCs, SAMe/SAH ratio, an indicator of cellular methylation capacity (29), was also reduced. Similar to results obtained from culture-activated HSCs, the intracellular level of SAMe was also lower in HSCs from BDL livers compared to sham controls (Table II). A moderate reduction of MTA and SAH levels was observed along with decreased SAMe/SAH ratio (Table II).
Table I.
Changes in SAMe, MTA, SAH levels and [3H]-dCTP incorporation (DNA methylation) during in vitro HSC activation
| Days in culture | SAMe | MTA | SAH | SAMe/SAH ratio | DNA methylation (X 100) |
|---|---|---|---|---|---|
| 1 | 1.94 ± 0.28 | 0.41 ±0.1 | 0.17 ± 0.02 | 11.3 ± 1.6 | 106.2 ± 16.5 |
| 3 | 0.6 ± 0.04* | 0.46 ± 0.001* | 0.19 ± 0.01 | 3.1 ± 0.2 | 225.4 ± 48.7* |
| 5 | 0.59 ± 0.2* | 0.30 ± 0.06 | 0.15 ± 0.02 | 2.5 ± 0.7 | 210.2 ± 44.2* |
| 7 | 0.47 ± 0.18* | 0.14 ± 0.01* | 0.14 ± 0.003* | 2.4 ± 0.5 | 186.9 ± 28.1* |
Rat HSCs were grown in culture till day 7 as described in methods and HPLC was performed for detection of SAMe, MTA and SAH levels. The unit for these metabolites is nmol/mg protein. The results represent Mean±S.E from three HSC preparations. As a measure of genome-wide methylation, the incorporation of [3H]-dCTP into DNA was assessed after treatment with HpaII endonuclease that cleaves non-methylated sites and compared to undigested control DNA. The units for DNA methylation are disintegrations per minute (dpm)/μg of DNA. Results represent Mean±S.E from four HSC preparations.
p<0.05 vs. day 1.
Table II.
Changes in SAMe, MTA, SAH levels during in vivo HSC activation
| Rat HSCs | SAMe | MTA | SAH | SAMe/SAH ratio |
|---|---|---|---|---|
| Sham | 2.17 ± 0.36 | 0.32 ± 0.02 | 0.28 ± 0.09 | 8.99 ± 1.82 |
| BDL | 0.91 ± 0.13* | 0.21 ± 0.02† | 0.17 ± 0.04 | 5.85 ± 1.07 |
Rat HSCs were isolated from 10-day BDL livers or corresponding sham control livers as described in methods. SAMe, MTA and SAH levels were assessed by HPLC analysis of HSCs from each group. The unit for these metabolites is nmol/mg protein. The results represent Mean±S.E from four HSC preparations;
p<0.05,
p<0.005 vs. sham control.
MATII-specific enzyme activity during HSC activation
Our results showed an up-regulation of MAT2A and MAT2β expression during HSC activation. However, despite induction of MAT2A, the SAMe synthesizing enzyme in HSCs, the intracellular level of SAMe decreased during activation. In order to examine what factors were responsible for this drop in SAMe level, we measured the activity of the MATII enzyme during HSC activation. Interestingly, we found that there was a 40% inhibition of MATII activity at day 3, 5 and 7 compared to day 1 (Table III). Concurrent with the in vitro findings, we noticed a 50% inhibition of MATII activity during in vivo HSC activation in BDL rats (Table III). These results indicate that the low SAMe levels in activated HSCs is due at least in part to lower MATII activity in these cells.
Table III.
Changes in MATII enzyme activity during HSC activation
| Rat HSCs | MAT activity (pmol/μg/min) |
|---|---|
| Day 1 | 2.12 ± 0.14 |
| Day 3 | 1.44 ± 0.06* |
| Day 5 | 1.60 ± 0.12* |
| Day 7 | 1.23 ± 0.22* |
| Sham HSCs | 4.14 ± 1.24 |
| BDL HSCs | 2.11 ± 0.41† |
Rat HSCs were grown in culture till day 7 or isolated from 10-day BDL rats or corresponding sham controls and MATII enzyme activity was measured in cytosolic fraction as described in methods. The results represent Mean±S.E. from three HSC preparations in duplicates.
p<0.05 vs. day 1,
p<0.005 vs. sham control.
Role of MAT genes during HSC activation
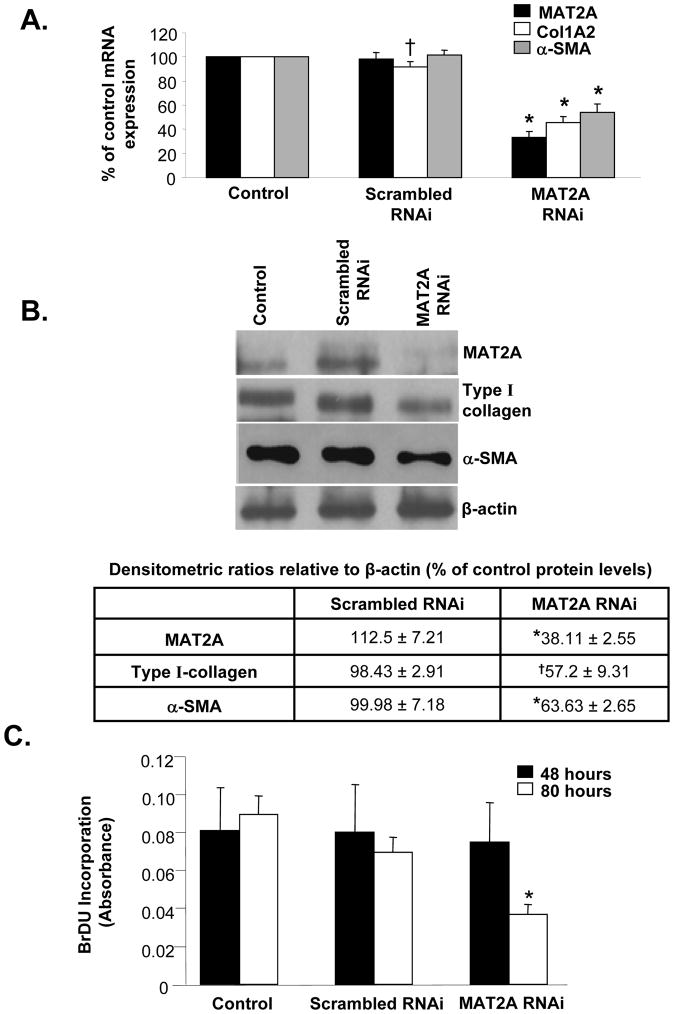
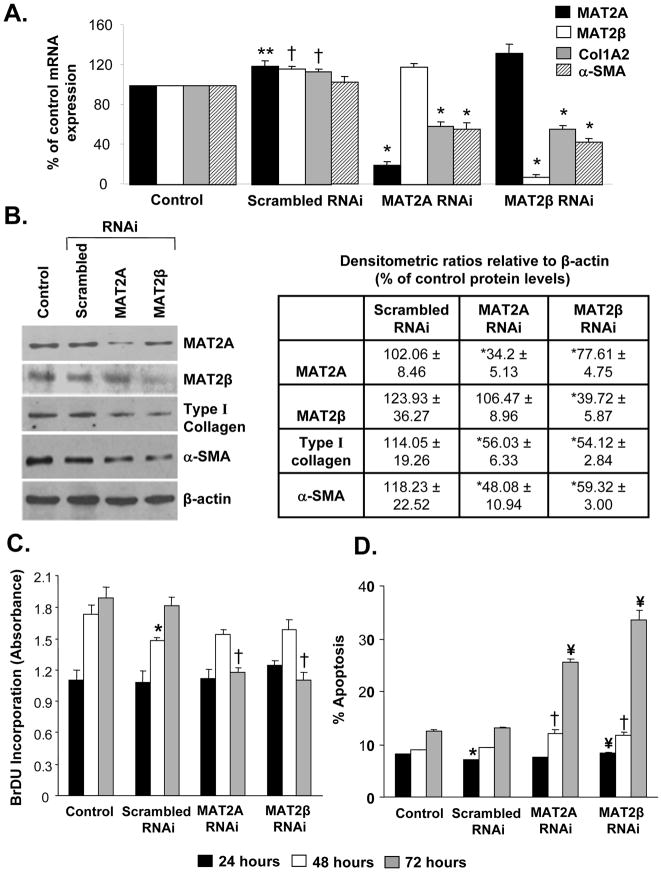
We used an RNAi-based knockdown approach to understand whether MAT genes play a role during HSC activation and proliferation. As shown in Figures 3A and B, knockdown of MAT2A for 48 hours in primary rat HSCs resulted in decreased HSC activation as measured by the lower levels of Col1A2 mRNA and type I collagen protein, respectively, compared to scrambled RNAi controls. A similar decrease in α-SMA mRNA and protein was also observed after MAT2A silencing. MAT2A gene silencing did not cause any change in cellular proliferation at this time, thereby indicating a lack of any toxicity at the 48 hour time point during which gene expression changes were measured (Fig. 3C). However, longer periods of MAT2A knockdown (80 hours) resulted in decreased BrDU incorporation in HSCs indicative of suppressed cell growth (Fig. 3C). Despite several attempts, we were unable to get sufficient knockdown of the MAT2β gene in primary rat HSCs. In order to examine the role of MAT2β in HSC activation, we performed gene silencing studies in the easily transfectable, human LX-2 cell line. As shown in Figure 4A, knockdown of either MAT2A by 80% or MAT2β by 93% inhibited expression of Col1A2 and α-SMA mRNA by 50% as compared to scrambled RNAi-treated cells. These results were further confirmed at the protein level (Fig. 4B). Concurrent with the findings in primary HSCs (Fig. 3), knockdown of MAT2A or MAT2β in LX-2 cells did not significantly affect cell growth up to 48 hours but there was decreased proliferation at the 72-hour time point (Fig. 4C). Toxicity effects of MAT gene knockdown were also evaluated by performing apoptosis assays in LX-2 cells. As shown in Figure 4D, knockdown of either MAT2A or MAT2β up to 48 hours did not significantly affect the number of apoptotic cells compared to scrambled RNAi controls. However, at 72 hours, there was a significant increase in the percent apoptosis when compared to scrambled RNAi.
Fig. 3. Effect of MAT2A gene silencing on HSC activation and proliferation.
Knockdown of MAT2A gene in primary rat HSCs was performed as described in Methods. (A) Total RNA from MAT2A knockdown HSCs was subjected to real-time PCR and expression of MAT2A, Col1A2 and α-SMA was compared to control and scrambled RNAi. Results represent Mean±SE from five HSC preparations; *p<0.005, †p<0.05 vs. scrambled RNAi. (B) Total cellular protein from MAT2A knockdown HSCs was subjected to Western blotting for detection of MAT2A, type I collagen and α-SMA and compared to control and scrambled RNAi. Representative images and densitometric analysis (Mean±SE) from four HSC preparations is shown; *p<0.005, †p<0.05 vs. scrambled RNAi. (C) Knockdown of MAT2A was done for 48 or 80 hours and BrDU incorporation in RNAi-treated cells was compared to control or scrambled RNAi. Mean±SE of two HSC preparations in quadruplets is shown; *p<0.005 vs. scrambled RNAi.
Fig. 4. Effect of MAT2A and MAT2β knockdown on activation, proliferation and apoptosis in human LX-2 cells.
Knockdown of MAT2A and MAT2β genes in LX-2 cells was performed as described in Methods. (A) Total RNA isolated from MAT2A or MAT2β knockdown cells was subjected to real-time RT-PCR analysis and expression of MAT2A, MAT2β , Col1A2 and α-SMA was compared to control or scrambled RNAi. Results represent Mean±SE from three experiments in duplicates for MAT2A, MAT2β , Col1A2 and four experiments for α-SMA; **p<0.05, †p<0.005 vs. control, *p<0.005 vs. scrambled RNAi. (B) Total cellular protein from MAT2A or MAT2β knockdown cells was subjected to Western blotting for detection of MAT2A, MAT2β, type I collagen and α-SMA and compared to control and scrambled RNAi. Representative images and densitometric analysis (Mean±SE) from three experiments is shown; *p<0.05 vs. scrambled RNAi. (C) Knockdown of MAT2A or MAT2β was done for 24, 48 or 72 hours and BrDU incorporation in RNAi-treated cells was compared to control or scrambled RNAi-treated cells. Results represent Mean±SE of three experiments in duplicates; *p<0.05 vs. control, †p<0.005 vs. scrambled RNAi. (D) Apoptotic cells in control or RNAi-treated samples were detected after 24, 48 or 72 hours using Hoechst staining as described in methods. Result represent Mean±SE of two experiments in duplicates; *p<0.005 vs. control, †p<0.05, ¥p<0.005 vs. scrambled RNAi.
Mechanisms underlying the influence of MAT2A or MAT2β on cell proliferation and apoptosis in LX-2 cells
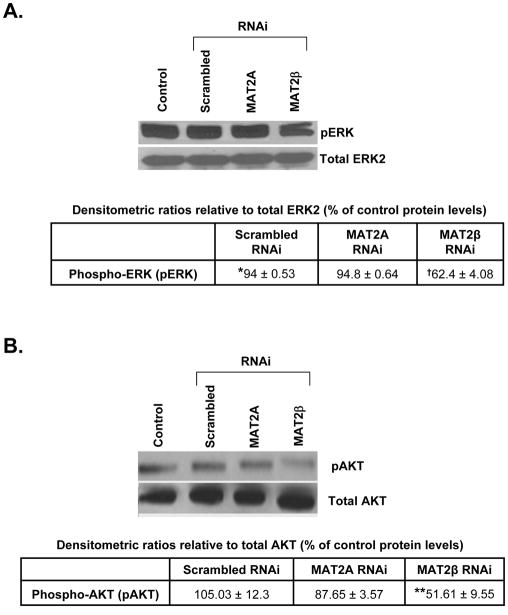
Since MAT2A or MAT2β silencing affects growth in LX-2 cells, we investigated the mechanism by which these genes influence cell proliferation and apoptosis. Our results showed that MAT2A knockdown lowered intracellular SAMe levels by 75% compared to control or scrambled RNAi-treated cells (Table IV). We also examined the effect of MAT2A or MAT2β silencing on the survival signaling pathways, ERK and PI-3K, in LX-2 cells. In Figure 5 we showed that knockdown of MAT2A did not significantly influence phosphorylation of ERK or AKT (a measure of PI-3K signaling) but knockdown of MAT2β prevented activation of both ERK and AKT in LX-2 cells, thereby indicating a role of this gene in signal transduction pathways associated with HSC activation.
Table IV.
Effect of MAT2A knockdown on intracellular SAMe levels in human LX-2 cells
| Treatment condition | SAMe (nmol per mg protein) |
|---|---|
| Control | 0.73 ± 0.08 |
| Scrambled RNAi | 0.66 ± 0.07 |
| MAT2A RNAi | 0.17 ± 0.03* |
Human LX-2 cells were transfected with scrambled or MAT2A RNAi as described in methods. SAMe levels were assessed by HPLC analysis of cells from each treatment condition. The results represent Mean±S.E from four experiments;
p<0.005 vs. scrambled RNAi.
Fig. 5. Effect of MAT2A and MAT2β knockdown on signaling in LX-2 cells.
Silencing of MAT2A and MAT2β genes in LX-2 cells was performed and phosphorylation of ERK1/2 (A) or AKT (for PI-3K signaling) (B) was checked by Western blotting as described in Methods. Subsequently the membranes were stripped and reprobed with antibodies against total ERK2 or AKT to verify protein levels. Representative images from 3 to 4 independent experiments are shown and results of densitometric analysis (Mean±SE) are shown below each blot; *p<0.005 vs. control, †p<0.005, **p<0.05 vs. scrambled RNAi.
DISCUSSION
The expression of MAT genes in the liver is a determinant of cell proliferation and differentiation. While MAT1A is a marker of differentiation, MAT2A and MAT2β are markers of growth and de-differentiation (12). Recently we showed in liver cancer cells that MAT2A and MAT2β genes are required for the mitogenic effect of leptin (21), a potent pro-fibrogenic growth factor that also induces activation of HSCs (22). We showed that MAT2A regulates leptin-mediated growth by changes in intracellular SAMe levels and identified the MAT2β gene as a novel entity that could regulate leptin signaling in liver cancer cells at multiple steps such as STAT3, ERK and PI-3K activation (21). Since leptin also influences these signaling pathways in HSCs (30), we sought to examine possible functions of MAT genes during HSC activation.
MAT2A and MAT2β genes were induced in HSCs undergoing activation both in vitro and in the BDL model of liver injury and their expression correlated strongly with the activation process as measured by induction of collagen and α-SMA, known markers of activation (1, 7, 8, 31). Both of these genes are markedly up-regulated during cellular proliferation caused by liver injury (12, 17, 18, 19, 32). Since HSCs are activated during liver injury, MAT2A and MAT2β signaling in these cells may be an essential mechanism during fibrogenesis. Consistent with this is the observation that when either one of these genes is knocked down in HSCs, collagen and α-SMA gene expression and cell proliferation is reduced.
The MAT2A-encoded protein is the only SAMe-synthesizing enzyme in HSCs because the liver-specific MAT1A-encoded isoenzymes that are expressed in hepatocytes are absent in HSCs (20). Since MAT2A is induced during the shift of HSCs from quiescence to activation, we expected an increase in intracellular SAMe levels during this process. However, our results showed that the intracellular SAMe levels were markedly decreased during HSC activation. One possible explanation is that SAMe is being consumed for polyamine biosynthesis. Another possible explanation is the up-regulation of MAT2β , a regulatory subunit of MAT2A, during HSC activation. The β subunit lowers the Km for methionine and the Ki for SAMe, making MATII more susceptible to feedback inhibition (16). With higher β expression, the steady state SAMe level would be lower due to this regulation. Even though both MAT2A and MAT2β are induced to similar extents in in vitro and in vivo activated HSCs, the ratio of the β to α2 subunit in HSCs may be such that the effect of the β subunit is more apparent. Consistent with this is the fact that the MATII enzyme activity decreased progressively during HSC activation. These results are also in agreement with the work of Shimizu-Saito et al who reported a decrease in MATII enzyme activity in HSCs from rats treated with carbon tetrachloride to induce liver fibrosis (20).
It is interesting to point out that while MAT2β induction occurs during de-differentiation and growth of hepatocytes and HSCs, the opposite occurs for lymphocyte activation (33). During T-lymphocyte activation, MAT2A expression increases while MAT2β disappears, allowing the steady state SAMe level to rise (33). If this is blocked, lymphocyte activation is blocked (34). Thus, the role of the MAT2β gene clearly differs in different cell types.
HSCs are similar to hepatocytes in that a lower SAMe level correlates with growth. In the case of hepatocytes, SAMe can exert an inhibitory effect on mitogens (12). In HSCs, exogenous SAMe has been reported by several groups to inhibit HSC activation and carbon tetrachloride-induced fibrosis (35). While the inhibitory effect of exogenous SAMe on fibrogenesis is well-known, the fact that SAMe level falls during HSC activation has not been reported to our knowledge. The levels of SAMe metabolites, MTA and SAH, exhibited less variation early on but both also fell modestly by day 7. These changes culminated in a dramatic decrease in the SAMe/SAH ratio, which is known to be a major determinant of transmethylation reactions (29). The fall in SAMe level and the SAMe/SAH ratio resulted in global DNA hypomethylation. Mann and colleagues have reported that treatment of quiescent HSCs with DNA methylation inhibitor, 5-aza-2’-deoxycytidine (5-azadC) blocks transdifferentiation and induces the expression of peroxisome proliferator-activated receptor gamma and inhibitor of kappaB-alpha. They showed that DNA methylation exerts epigenetic control over myofibroblast transdifferentiation (36). These findings seem to be at odds with our results on global DNA hypomethylation in activated HSCs. However, we have only examined global CpG methylation changes and do not provide evidence of any gene-specific methylation pattern in HSCs that relates to the activation process. This situation is somewhat similar to those observed in many human cancers where there is global DNA hypomethyation but certain ‘hotspots’ are hypermethylated (37). It is also interesting to note that loss of DNA methylation has been reported by Jiang and colleagues in gastric cancer stromal myofibroblasts in culture (38). Our findings in activated HSCs are consistent with this report.
Silencing of MAT2A in primary HSCs inhibited activation as detected by the decrease in collagen and α-SMA expression. This also led to inhibition of cell growth during extended periods of MAT2A knockdown. MAT2A silencing might have prevented SAMe biosynthesis in HSCs and hence inhibited activation and growth. To clearly establish whether MAT2A silencing affects SAMe levels, we studied these changes in the LX-2 cell line because for SAMe measurement under knockdown conditions, very large amounts of cells are required which is difficult to achieve with primary HSCs. Knockdown of MAT2A severely depleted intracellular SAMe pools in LX-2 cells and this led to decreased cell proliferation and increased apoptosis after extended periods of knockdown. These findings are supported by previous observations showing that SAMe depletion invoked by cycloleucine, a chemical inhibitor of MAT, led to apoptosis in rat hepatocytes (39). Our results thereby suggest that a certain physiological level of SAMe is required for HSC activation and entry into cell cycle. Our inability to achieve efficient knockdown of the MAT2β gene in primary HSCs led us to study this gene in human LX-2. Interestingly, knockdown of MAT2β inhibited both collagen and α-SMA expression in these cells. Similar to the results with MAT2A, MAT2β silencing also decreased growth and increased apoptosis after extended periods of knockdown. Our previous work showed that MAT2β influenced leptin signaling in liver cancer cells and this involved regulation of ERK and PI-3K pathways (21). In this work, we have examined the effect of MAT2β silencing on these two pathways because they are well known survival mechanisms in HSCs and are essential components of pro-fibrogenic response (30). Consistent with our previous findings on the effect of this gene on ERK and PI-3K (21), here we show that MAT2β knockdown also inhibited activation of these signaling components in LX-2 cells. This function was specific to MAT2β because MAT2A did not influence these signaling pathways. Apart from its role in regulating MATII activity and SAMe homeostasis, MAT2β also plays an important part in regulating signaling in activated HSCs.
In summary we have demonstrated that MAT2A and MAT2β genes, the sole regulators of SAMe homeostasis in HSCs, are induced during in vitro and in vivo activation. A drop in MATII enzyme activity and intracellular SAMe levels occur during HSC activation along with a fall in global DNA methylation. Silencing of MAT2A or MAT2β gene inhibits collagen and α-SMA expression and cell growth, markers of HSC activation. MAT2β gene affects HSC activation by influencing ERK and PI-3K survival signal mechanisms in HSCs whereas MAT2A affects growth by changes in intracellular SAMe levels. These findings have important implications regarding epigenetic changes during HSC activation as well as provide novel therapeutic targets against fibrosis.
Acknowledgments
Financial support.: This work was supported by NIH grants DK51719 (SC Lu), K99AA017774 (K Ramani), and R01AT1576 (SC Lu and JM Mato) and Plan Nacional of I+D SAF 2005-00855, and HEPADIP-EULSHM-CT-205 (JM Mato). Hepatic stellate cells were provided by the Non-Parenchymal Cell Core of the USC-UCLA Research Center for Alcoholic Liver and Pancreatic Diseases and Cirrhosis (R24 AA12885-07).
Abbreviations
- α-SMA
alpha-smooth muscle actin
- AKT
AK strain transforming
- BDL
bile duct ligation
- BrDU
bromodeoxyuridine
- Col1A2
alpha2(1) collagen mRNA
- ECM
extracellular matrix
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- HPLC
high performance liquid chromatography
- HPRT1
hypoxanthine phosphoribosyl-transferase 1
- HSC
hepatic stellate cell
- MAT
methionine adenosyltransferase
- MTA
methylthioadenosine
- RT
reverse transcription
- SAH
S-adenosylhomocysteine
- SAMe
S-adenosylmethionine
- siRNA
short interfering RNA
- RNAi
RNA interference
- STAT
signal transducers and activators of transcription
References
- 1.Burt AD. Cellular and molecular aspects of hepatic fibrosis. J Pathol. 1993;70:105–114. doi: 10.1002/path.1711700203. [DOI] [PubMed] [Google Scholar]
- 2.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 3.Paradis V, Dargere D, Bonvoust F, Vidaud M, Segarini P, Bedossa P. Effects and regulation of connective tissue growth factor on hepatic stellate cells. Lab Invest. 2002;82:767–774. doi: 10.1097/01.lab.0000017365.18894.d3. [DOI] [PubMed] [Google Scholar]
- 4.Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, et al. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67:2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamphorst BE, Stoll D, Gressner AM, Weiskirchen R. Inhibitory effect of soluble PDGF-beta receptor in culture-activated hepatic stellate cells. Biochem Biophys Res Commun. 2004;317:451–462. doi: 10.1016/j.bbrc.2004.03.064. [DOI] [PubMed] [Google Scholar]
- 6.Iredale JP, Benyon RC, Arthur MJP, et al. Tissue inhibitor of metalloproteinase-1 messenger RNA expression is enhanced relative to interstitial collagenase messenger RNA in experimental liver injury and fibrosis. Hepatology. 1996;24:176–184. doi: 10.1002/hep.510240129. [DOI] [PubMed] [Google Scholar]
- 7.Maher JJ, McGuire RF. Extracellular matrix gene expression increases preferentially in rat lipocytes and sinusoidal endothelial cells during hepatic fibrosis in vivo. J Clin Invest. 1990;86:1641–1648. doi: 10.1172/JCI114886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutiérrez-Ruiz MC, Gómez-Quiroz LE. Liver fibrosis: searching for cell model answers. Liver International. 2007;27:434–439. doi: 10.1111/j.1478-3231.2007.01469.x. [DOI] [PubMed] [Google Scholar]
- 9.Lotersztajn S, Julien B, Clerc FT, Grenard P, Mallat AH. Hepatic fibrosis: Molecular mechanisms and drug targets. Annu Rev Pharmacol Toxicol. 2005;45:605–628. doi: 10.1146/annurev.pharmtox.45.120403.095906. [DOI] [PubMed] [Google Scholar]
- 10.Kinnman N, Goria O, Wendum D, Gendron M, Rey C, Poupon R, et al. Hepatic Stellate Cell Proliferation is an Early Platelet-Derived Growth Factor-Mediated Cellular Event in Rat Cholestatic Liver Injury. Laboratory Investigation. 2001;81:1709–2001. doi: 10.1038/labinvest.3780384. [DOI] [PubMed] [Google Scholar]
- 11.Greupink R, Bakker HI, Bouma W, Reker-Smit C, Meijer DKF, Beljaars L. The Antiproliferative Drug Doxorubicin Inhibits Liver Fibrosis in Bile Duct-Ligated Rats and Can Be Selectively Delivered to Hepatic Stellate Cells in Vivo. J Pharmacol Exp Ther. 2006;317:514–521. doi: 10.1124/jpet.105.099499. [DOI] [PubMed] [Google Scholar]
- 12.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45:1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 13.Clarke SG. Inhibition of mammalian protein methyltransferases by 5’-methylthioadenosine (MTA): a mechanism of action of dietary SAMe? The Enzymes. 2006;24:467–493. doi: 10.1016/S1874-6047(06)80018-1. [DOI] [PubMed] [Google Scholar]
- 14.Lu SC, Tsukamoto H, Mato JM. Role of abnormal methionine metabolism in alcoholic liver injury. Alcohol. 2002;27:155–162. doi: 10.1016/s0741-8329(02)00226-4. [DOI] [PubMed] [Google Scholar]
- 15.Kotb M, Mudd SH, Mato JM, Geller AM, Kredich NM, Chou JY, et al. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13:51–62. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 16.Halim AB, Le Gros L, Geller AM, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- 17.Cai J, Sun W, Hwang JJ, Stain S, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: Molecular characterization and significance. Hepatology. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 18.Huang ZZ, Mao Z, Cai J, Lu SC. Changes in methionine adenosyltransferase during liver regeneration in the rat. Am J Physiol. 1998;275:G14–21. doi: 10.1152/ajpgi.1998.275.1.G14. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Chantar ML, García-Trevijano ER, Latasa MU, Martín-Duce A, Fortes P, Caballería J, et al. Methionine adenosyltransferase II β subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–948. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu-Saito K, Horikawa S, Kojima N, Shiga J, Senoo H, Tsukada K. Differential Expression of S-Adenosylmethionine Synthetase Isozymes in Different Cell Types of Rat Liver. Hepatology. 1997;26:424–431. doi: 10.1002/hep.510260224. [DOI] [PubMed] [Google Scholar]
- 21.Ramani K, Yang H, Xia M, Iglesias Ara A, Mato JM, Lu SC. Leptin’s Mitogenic Effect in Human Liver Cancer Cells Requires Induction of Both Methionine Adenosyltransferase 2A and 2β. Hepatology. 2008;47:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxena NK, Saliba G, Floyd JJ, Anania FA. Leptin induces increased alpha2(I) collagen gene expression in cultured rat hepatic stellate cells. J Cell Biochem. 2003;89:311–320. doi: 10.1002/jcb.10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukamoto H, Cheng S, Blaner WS. Effects of dietary polyunsaturated fat on ethanol-induced Ito cell activation. Am J Physiol Gastrointest Liver Physiol. 1996;270:G581–G586. doi: 10.1152/ajpgi.1996.270.4.G581. [DOI] [PubMed] [Google Scholar]
- 24.Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:34.1–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrar C, Clarke S. Altered levels of S-adenosylmethionine and S-adenosylhomocysteine in the brains of L-isoaspartyl (D-aspartyl) O-methyltransferase-deficient mice. J Biol Chem. 2002;277:27856–27863. doi: 10.1074/jbc.M203911200. [DOI] [PubMed] [Google Scholar]
- 27.Finnell RH, Spiegelstein O, Wlodarczyk B, Triplett A, Pogribny IP, Melnyk S, et al. DNA Methylation in Folbp1 Knockout Mice Supplemented with Folic Acid during Gestation. J Nutrition. 2002;132:2457S–2461S. doi: 10.1093/jn/132.8.2457S. [DOI] [PubMed] [Google Scholar]
- 28.Yang HP, Sadda MR, Li M, Zeng Y, Chen LX, Bae WJ, Ou X, Runnegar MT, Mato JM, Lu SC. S-Adenosylmethionine and its metabolite induce apoptosis in HepG2 cells: role of protein phosphatase 1 and Bcl-xS. Hepatology. 2004;40:221–231. doi: 10.1002/hep.20274. [DOI] [PubMed] [Google Scholar]
- 29.Lu SC, Huang ZZ, Yang H, Mato JM, Avila A, Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;279:G178–G185. doi: 10.1152/ajpgi.2000.279.1.G178. [DOI] [PubMed] [Google Scholar]
- 30.Saxena NK, Titus MA, Ding X, Floyd J, Srinivasan S, Sitaram SV, et al. Leptin as a novel profibrogenic cytokine in hepatic stellate cells: mitogenesis and inhibition of apoptosis mediated by extracellular regulated kinase (Erk) and Akt phosphorylation. FASEB J. 2004;18:1612–1614. doi: 10.1096/fj.04-1847fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner FR, Giambrone MA, Czaja MJ, Shah A, Annoni G, Takahashi S, et al. Ito-cell gene expression and collagen regulation. Hepatology. 1990;11:111–117. doi: 10.1002/hep.1840110119. [DOI] [PubMed] [Google Scholar]
- 32.Huang ZZ, Mato JM, Kanel G, Lu SC. Differential Effect of Thioacetamide on Hepatic Methionine Adenosyltransferase Expression in the Rat. Hepatology. 1999;29:1471–1478. doi: 10.1002/hep.510290525. [DOI] [PubMed] [Google Scholar]
- 33.Le Gros HL, Jr, Geller AM, Kotb M. Differential regulation of methionine adenosyltransferase in superantigen and mitogen stimulated human T lymphocytes. J Biol Chem. 1997;272:16040–16047. doi: 10.1074/jbc.272.25.16040. [DOI] [PubMed] [Google Scholar]
- 34.Kotb M, Dale JB, Beachey EH. Stimulation of S-adenosylmethionine synthetase in human lymphocytes by streptococcal M protein. J Immunol. 1987;139:202–206. [PubMed] [Google Scholar]
- 35.Nieto N, Cederbaum AI. S-Adenosylmethionine Blocks Collagen I Production by Preventing Transforming Growth Factor Induction of the COL1A2 Promoter. J Biol Chem. 2005;280:30963–30974. doi: 10.1074/jbc.M503569200. [DOI] [PubMed] [Google Scholar]
- 36.Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation fo myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- 37.Calvisi DF, Ladu S, Gorden A, Farina M, Lee J, Conner EA, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L, Gonda TA, Gamble MV, Salas M, Seshan V, Tu S, et al. Global Hypomethylation of Genomic DNA in Cancer-Associated Myofibroblasts. Cancer Res. 2008;68:9900–9908. doi: 10.1158/0008-5472.CAN-08-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuge J, Cederbaum AI. Depletion of S-adenosyl-L-methionine with cycloleucine potentiates cytochrome P450 2E1 toxicity in primary rat hepatocytes. Arch Biochem Biophys. 2007;466:177–185. doi: 10.1016/j.abb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]