Abstract
The major barrier to treating or preventing Alzheimer’s disease (AD) is its unknown etiology/pathogenesis. Although increasing evidence supports a role for mitochondrial dysfunction in the pathogenesis of AD, there have been few studies that simultaneously evaluate changes in multiple mitochondrial proteins. To evaluate changes in suites of potentially interacting mitochondrial proteins, we applied 2-dimensional liquid chromatography coupled with tandem mass spectrometry (LC/MS/MS) and the isotope coded affinity tag (ICAT) method to identify and quantify proteins in mitochondrial enriched fractions isolated from short postmortem interval temporal pole specimens from subjects with mild cognitive impairment (MCI, 4 subjects pooled), early Alzheimer’s disease (EAD, 4 subjects pooled), late-stage AD (LAD, 8 subjects pooled) and age-matched normal control (NC, 7 subjects pooled) subjects. A total of 112 unique, non-redundant proteins were identified and quantified in common to all three stages of disease progression. Overall, patterns of protein change suggest activation of mitochondrial pathways that include proteins responsible for transport and utilization of ATP. These proteins include adenine nucleotide translocase (ADT1), voltage dependent anion channels (VDACs), hexokinase (HXK1) and creatine kinase (KCRU). Comparison of protein changes throughout the progression of AD suggests the most pronounced changes occur in EAD mitochondria.
1. Introduction
Alzheimer’s disease (AD), the fourth leading cause of death in the United States, currently affects ∼ 4 million Americans and may affect nine million by the year 2040 unless preventive strategies are found [1]. The major barrier to treating and eventually preventing AD is the lack of a complete understanding of the etiology and pathogenesis of neuron degeneration and loss. Numerous pathogenic/etiologic mechanisms have been suggested for AD including mitochondrial dysfunction and altered energy metabolism [2].
Alzheimer’s disease is characterized clinically by a progressive decline in multiple cognitive functions and is thought to begin with amnestic mild cognitive impairment (MCI), widely considered to be a transition between normal aging and dementia. As the disease progresses patients are classified as early AD (EAD and ultimately as late stage AD (LAD) patients. The mean length of life following diagnosis is 8.5 years with a range of 1 – 25 years [3].
Mitochondria are critical subcellular organelles responsible for energy production through coupling of respiration to the generation of ATP. Mitochondria possess their own DNA (mtDNA) that code for only 13 proteins in the electron transport chain. The remaining ∼1500 proteins required for mitochondrial function are generated by nuclear transcription processes and cytoplasmic synthesis machinery and are selectively transported to mitochondria for utilization. Mitochondria communicate protein requirements to the nucleus via a complex system of retrograde communication that is not completely understood at this point. Mitochondria also play a pivotal role in several cellular processes such as the effector phase of apoptosis (programmed cell death) [4, 5], through release of cytochrome C or the apoptosis-inducing factor. Release of cytochrome C is considered to be the key event in caspase (cysteine protease) activation promoting degradation of the cell [4].
Previous studies of mitochondrial function and AD showed altered oxidative metabolism in AD brain that appeared to precede development of AD pathology [2]. Other studies showed reduced activities of metabolic and energy metabolism related enzymes in AD brain including decreases in the pyruvate dehydrogenase complex (PDHC) and various components of cytochrome C oxidase (COX) (reviewed [2]). In vivo imaging studies showed reduced cerebral glucose metabolism in adjacent parietal and temporal cortices [6] and increased oxidative utilization compared with glucose utilization in AD [7] that appeared to precede atrophy [7, 8] and functional impairment on neuropsychologic testing (reviewed [9]). A recent study of frontal and temporal cortex showed a significant 25% decrease in normal mitochondria in AD subjects compared to controls [10].
Although several studies show deficiencies of select mitochondrial enzymes (COX, α-ketogluatarate dehydrogenase complex [KGDHC] and PDHC) associated with AD, these studies have been limited to the analysis of protein levels by immunochemical methods, functional activity assays, or by the measurement of mRNA and have only addressed a small fraction of the total mitochondrial proteome through the use of proteomics, simultaneous analysis of multiple proteins in complex samples is now possible.
Two dimensional liquid chromatography coupled with data dependent tandem mass spectrometry (2D-LC/MS/MS) provides an excellent means to analyze complex protein mixtures. By using strong cation exchange and an orthogonal reversed phase separation, remarkable increases in peptide peak capacity can be obtained. Data-dependent tandem mass spectrometry takes advantage of this increased peak capacity by sorting through peptide precursor ions and automatically performing tandem mass spectrometry for peptide amino acid sequence information. Relative quantification of peptides can be obtained simultaneously by incorporating stable isotope labeling. Development of the cleavable isotope coded affinity tag (cICAT) method that uses light (12C9) or heavy (13C9) reagents to selectively label proteins through alkylation of cysteines [11–13], provides one approach to obtain relative quantification.
In this pilot study, we applied 2D-LC/MS/MS and cICAT labeling to measure relative abundances of proteins in enriched mitochondrial fractions prepared from temporal pole (TP) brain specimens of MCI, EAD, LAD and age-matched control subjects.
2. Materials and Methods
2.1 Subjects
Mitochondria were isolated from TP specimens from MCI, EAD, LAD and age-matched NC subjects. These TP specimens were chosen for initial study because relatively large specimens were available for mitochondrial isolation and because TP demonstrates significant AD pathology. The brains of subjects used in these studies were obtained at autopsy through the University of Kentucky-Alzheimer’s Disease Center (UK-ADC) following UK IRB approved protocols. All subjects were followed longitudinally with annual neuropsychological testing, physical and neurological examinations. All LAD patients demonstrated progressive intellectual decline and met NINCDS-ADRDA Workgroup criteria [14] for the clinical diagnosis of probable AD. All LAD patients met accepted criteria for the histopathologic diagnosis of AD [15, 16], typically demonstrated Braak staging scores of VI, and met high likelihood NIA/Reagan criteria for the neuropathologic diagnosis elements. Normal control subjects were derived from the control population followed longitudinally by the UK-ADC. Control subjects demonstrated Braak scores of I or II, and met NIA/Reagan low likelihood criteria for the histopathologic diagnosis of AD. Subjects with amnestic MCI were derived from the control group and were followed longitudinally in the UK-ADC clinic. MCI patients were normal on enrollment into the longitudinal study and developed MCI during follow-up. The clinical criteria for diagnosis of amnestic MCI are those of Petersen et al. [17]. Subjects with EAD were also derived from the normal control group and were normal on enrollment but progressed to EAD on follow-up. The clinical criteria for EAD are a) a decline in cognitive function from a previous higher level, b) declines in one or more areas of cognition in addition to memory, c) a clinical dementia rating scale score of 0.5 to 1, d) impaired ADLs, and e) a clinical evaluation that excludes other causes of dementia. All subjects studied had neuropathological examination of multiple sections of neocortex, hippocampus, entorhinal cortex, amygdala, basal ganglia, nucleus basalis of Meynert, midbrain, pons, medulla, and cerebellum using the modified Bielschowsky stain, hematoxylin and eosin stain, and Aβ and α-synuclein immunostains Braak staging [18] scores were determined using the Gallyas stain on sections of entorhinal cortex, hippocampus, and amygdala and the Bielschowsky stain on neocortex. Subject demographic data are shown in Table 1.
Table 1.
Subject demographic data.
| Mean ± SEM Age (y) | Sex | Mean ± SEM PMI (hr) |
Median Braak Staging Score |
|
|---|---|---|---|---|
| Control | 83.3 ± 2.3 | 4M/3F | 3.4 ± 0.4 | I |
| MCI | 89.8 ± 1.4† | 2M/2F | 4.2 ± 1.8 | IV* |
| EAD | 89.5 ± 2.7 | 1M/3F | 2.8 ± 0.7 | V* |
| LAD | 81.6 ± 2.0‡ | 4M/4F | 3.1 ± 0.1 | VI* |
MCI subjects were significantly older than normal controls (p < 0.05).
LAD subjects were significantly younger than MCI and EAD subjects (p < 0.05) but not younger than normal controls.
Median Braak staging scores were significantly higher (p < 0.05) in MCI (median = IV), EAD (median = V) and LAD subjects (median = VI) compared to NC subjects (median = I).
2.2 Isolation of Mitochondria
Specimens of TP obtained from short postmortem interval (PMI) autopsies were immediately frozen in liquid nitrogen and subsequently stored at −80°C until used for analysis. Preparation of an enriched mitochondria fraction was as previously described [19, 20] with slight modification [21]. Brain specimens (0.5 – 1 g) were homogenized on ice in 10 ml MSB-Ca2+ buffer [22] consisting of 0.21 M mannitol, 0.07 M sucrose, 0.5 M Tris HCl and 3 mM CaCl2 (pH 7.5) using a motor driven Teflon coated Dounce homogenizer. Following homogenization, 1/10 volume of 0.1 M Na2EDTA was added to the solution and it was centrifuged at 1,500 × g and 4° C for 20 min. Following centrifugation the supernatant was removed and centrifuged at 20,000 × g and 4 °C for 20 min. The resulting pellet was washed three times in MSB-Ca2+ followed by centrifugation. Because this crude pellet was often contaminated with Golgi and cytosolic proteins, the pellet was resuspended in 2 ml 50/50 Percoll/MSB-Ca2+ and centrifuged at 50,000 × g for 1 h. Following centrifugation, enriched mitochondria were isolated at a density of ∼1.035 g/ml, pelleted, resuspended in Percoll/MSB-Ca2+ and centrifuged through a second Percoll gradient. The resulting enriched mitochondrial pellet was then rinsed three times in PBS and freeze dried. In general, yields of mitochondria enriched through two Percoll gradients approached ∼ 200 to 300 µg/g tissue.
2.3 Verification of mitochondrial purity
To verify mitochondrial fractions were relatively free of other subcellular proteins, 50 µg samples of enriched mitochondria, nuclei, and cytosol were separated on a 10 to 20% gradient SDS-PAGE gel and transferred to nitrocellulose. The blots were blocked in 5% dry milk in tris buffered saline containing 1% Tween-20 (TTBS) for 2 h and incubated at 4 °C overnight in a polyclonal antibody that recognizes isoforms 1 and 3 of human voltage dependent anion channel (VDAC ) (Oncogene, San Diego, CA; 1:1000) as a marker specific for mitochondria, rabbit anti-lamin B (Santa Cruz Biotechnology, Santa Cruz, CA; 1:1000) as a specific marker of nuclei, or rabbit anti-MAP-2 (Sigma, St. Louis, MO; 1:1000) as a cytosolic marker. The blots were rinsed 3 to 5 times in TTBS and incubated 2 h in horseradish peroxidase labeled anti-rabbit IgG (Vector Laboratories, Burlingham, CA). Following 3 −5 washes in TTBS the bands were observed using the enhanced chemiluminescence (ECL) method per manufacturer’s instructions. The choice of proteins for verification of each subcellular fraction was based on the availability of suitable commercially available antibodies.
2.4 ICAT Labeling
For proteomic analysis, mitochondrial pellets were resuspended in 300 µl 80% 50 mM ammonium bicarbonate/20% acetonitrile and passed through a 26 gauge needle ten times to disrupt mitochondria. Protein concentration was determined using the Pierce BCA method and equal aliquots of each MCI, EAD, LAD and NC sample were pooled to generate representative samples for each group. The pooled samples were divided into triplicate 100 µg samples (MCI, EAD, LAD) for ICAT labeling. Pooled NC specimens were aliquoted into 3 triplicate sets for comparison to MCI, EAD and LAD mitochondria.
ICAT analysis of mitochondrial proteins was carried out using the commercially available cleavable ICAT Reagent Kit (Applied Biosystems, Foster City, CA) as previously described [13, 23] with modification. Briefly, 100 µg samples of MCI, EAD, LAD and NC mitochondria were solubilized in 80 µl Tris-SDS denaturing buffer by passing the sample through a 26 gauge needle 10 times and heating in a boiling water bath for 10 min with tris(2-carboxyethyl)phosphine (TCEP) to reduce protein disulfide bonds. After cooling to room temperature, NC mitochondrial proteins were reacted with light (12C9-labeled) ICAT reagent, whereas MCI, EAD and LAD proteins were reacted with heavy (13C9–labeled) ICAT reagent for 2 h at 37° C. Labeled protein samples were combined and digested with sequencing grade trypsin (Promega Madison, WI) (1:40 protease:protein) at 37°C for 16 h. The resulting peptide mixture was passed through a cation-exchange cartridge to remove TCEP, SDS, and unreacted ICAT reagents. ICAT labeled peptides in the eluent were isolated on an avidin cartridge and eluted with 30% acetonitrile/70% aqueous trifluoroacetic acid (TFA)(0.4%). The isolated labeled peptides were evaporated to dryness, resuspended in TFA and incubated for two hours at 37° C to cleave the biotin portion of the ICAT tags. Solutions containing ICAT-labeled peptides were evaporated to dryness, reconstituted in 10 µl 5% acetonitrile/95% aqueous formic acid (0.1%) prior to 2D-LC/MS/MS.
2.5 Chromatography
Peptides were separated using a laboratory constructed [24, 25] 2D HPLC capillary column (350 µm i.d.) containing 5 cm of strong cation exchange resin (Partisil 10 µm, Alltech, Deerfield, IL) packed on top of 15 cm of reversed phase C18 resin (Macrosphere 300 5 µm, Alltech, Deerfield, IL). Peptide separations used ammonium acetate steps from 0 to 300 mM to elute peptides from the cation exchange phase. Each salt step was subjected to a complete reversed phase gradient from 5% acetonitrile (ACN)/95% aqueous formic acid (0.1%) to 70% ACN/aqueous 30% formic acid (0.1%) over 110 min. Salt and organic gradients were generated using an LC Packings Ultimate HPLC pump (Dionex, Sunnyvale, CA) at a solvent flow of 4µL/min. LC/MS/MS spectra were acquired on a ThermoFinnigan LCQ Deca quadrupole ion trap mass spectrometer (ThermoScientific, San Jose, CA). Tandem mass spectra were acquired in a data dependent mode. Three spectra were averaged to generate the data dependent full scan spectrum with the most intense ion subjected to tandem mass spectrometry with five spectra averaged to produce the MS/MS spectrum. Masses subjected to MS/MS were excluded from further mass spectrometry for 2 min.
2.6 Mass spectrometric data analysis
Acquired ICAT MS/MS spectral files were converted to a mzXML format using Thermo2mzXML (Version 1, ThermoElectron, San Jose, CA ) subjected to protein database searches using Sorcerer-Sequest (Sorcerer PE version V2.5.4, Sage-N Research, Milpitas, CA). A human sub-database was constructed from the SwissProt non-redundant protein database (Release 50.0) using the FASTA Database Utility found in Bioworks 3.1 (ThermoElectron, San Jose, CA). The Sequest parameters were modified to accommodate the ICAT protocol by requiring a static modification of 227.13 u (light ICAT label) and a differential modification of 9.03 u (heavy ICAT label) for all cysteine containing peptides. Thus every light and heavy labeled cysteine residue was expected to have masses at 330.136 and 339.166 u, respectively. The raw database search results were input into the Trans-Proteomic Pipeline (TPP) (Version 2.7, MIST Rev. 2, Institute for Systems Biology, Seattle, WA). Database results were validated and filtered using the Peptide- [26] and ProteinProphet [27] modules (4.0(TPP v2.9 GALE rev.4, Build 200610111250) of the TPP. Protein quantification was accomplished within the TPP using the Xpress module [28]. Data from the TPP were combined and parsed using the Ions 1.0 (Adaptive Bioinformatics, Lexington, KY) for each analytical replicate and converted to an Excel 2003 xls spreadsheet (Microsoft, Redmond, WA). Spreadsheets from individual analytical replicates were combined using the Pivot table module to produce a disease stage protein summary. Evaluation of false positive identifications was accomplished by appending a decoy database (a sequence reversed version of the database) to the SwissProt human sub-database using a Perl script available from Matrix Science ( London, UK ). The original mzXML files were searched against this new database using X!TANDEM2 (version 2007.07.01.2, www.theGPM.org).
2.7 Western Blot Analysis
Verification of proteomic data was carried out using Western blot analysis of representative proteins. For analysis, 20 µg aliquots of mitochondrial protein from 4 individual LAD and 4 individual NC subjects (subsets of subjects used for the proteomic study) were separated on 8 to 16% linear gradient gels and transferred to nitrocellulose. The blots were probed for voltage dependent anion channel (VDAC) using a 1:100 dilution of mouse anti-VDAC (Calbiochem; La Jolla, CA), stripped and re-probed for hexokinase using a 1:100 dilution of rabbit anti-hexokinase (Abgent; San Diego, CA). Protein loading efficiency was verified by probing the gel for 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNP ) using a 1:500 dilution of mouse anti-CNP (Chemicon, Temecula, CA). Western blots were then scanned, cropped using ADOBE Photoshop (ver. 6.0) and band intensities measured using Scion Image Analysis (NIH). Results are expressed as mean ± SEM % control staining.
2.8 Statistical Analysis
Protein ICAT values were evaluated statistically using a 2-tailed t-test and the commercially available ABSTAT software (rel. 1.96). Results were considered significant for p≤0.05, trending toward significant for 0.06≤p≤0.1 and insignificant for p>0.1
3. Results
To determine which proteins were significantly altered in AD mitochondria, we isolated enriched mitochondrial fractions from TP specimens of 4 MCI, 4 EAD, 8 LAD, and 7 age-matched control subjects. To reduce variability associated with disease/control pairwise comparisons, we followed the lead of Montine and co-workers [29] and pooled the enriched mitochondrial samples according to their stage of disease and analyzed each pool in triplicate. Aliquots (100 µg each) were subjected to quantitative proteomic analysis using 2D-HPLC coupled with tandem mass spectrometry and ICAT quantification. The subject demographic data is shown in Table 1. There were no significant differences in age between control, EAD, or LAD subjects, but MCI subjects were significantly (p < 0.05) older than NC subjects. In addition, LAD subjects were significantly (p < 0.05) younger than both the MCI and EAD subjects. Median Braak staging scores, a measure of NFT pathology, were significantly higher (p < 0.05) in MCI (median = IV), EAD (median = V) and LAD subjects (median = VI) compared to NC subjects (median = I). There were no significant differences in PMI for any of the subjects. The mitochondria isolated using protocols described here are derived from a mixture of cell types including normal and degenerating neurons, glia and astrocytes and represent a global mitochondrial pool. Therefore, these quantitative results represent average proteomic changes from the global mitochondrial pool and thus minimized the likelihood of observing gross changes in protein concentrations. Although gliosis is often associated with AD [30], analysis of sections of TP from representative LAD subjects that were immunostained for glial fibrillary acidic protein (GFAP, a marker of glia) and visualized by confocal microscopy, did not show evidence of an excess number of glia compared to age-matched controls (data not shown).
To characterize changes in the mitochondrial proteins in the progression of AD, it was necessary to verify that our preparations were sufficiently free of contamination by other organelles. Western blot analysis of mitochondrial, nuclear, and cytosolic fractions for VDAC, a mitochondrial protein, lamin-B, a nuclear specific protein, and MAP-2, a cytosolic marker, showed minimal cross contamination of the isolated fractions (data not shown). Although we used two Percoll gradients and thoroughly rinsed the mitochondrial pellet with PBS to ensure that the mitochondrial samples were free of cytosolic proteins, we still observed the presence of proteins not generally considered to be mitochondrial proteins. However, several of these proteins are physically associated with mitochondria in the cell and likely isolated along with mitochondria due to interactions with the outer membrane.
Two dimensional liquid chromatography tandem mass spectrometric analyses of ICAT-labeled samples identified and quantified 112 unique, non-redundant proteins in at least two of the three analytical replicates for all three disease stages (Supplementary Table 1). Supplementary Table 2 through 13 show details of the protein identification including protein name (SwissProt), TPP probability of each protein identification, percent coverage of the protein, mean and standard deviations for Xpress ratio (disease/control values), the number of peptides used for Xpress calculations, the precursor ion charge, peptide sequence identified and the number of instances each peptide was observed for MCI, EAD and LAD subjects.
False positive identifications were evaluated by searching the mass spectral data against a database composed of both proper human protein sequences and corresponding reverse (or nonsense) sequences (decoy database). Using this approach, false positive identifications ranged from 4 to 6% for any individual analytical replicate. However, requiring consensus identification in least two to the three analytical replicates dropped the false positive identifications to less than 1%. By requiring that a protein must be observed in all three disease states, no false positives appeared in the 112 reported proteins.
Utilization of three analytical replicates permitted an analysis of the coefficient of variance for the 112 quantified proteins. The mean coefficient of variance for the entire study was determined to be 24.9%.
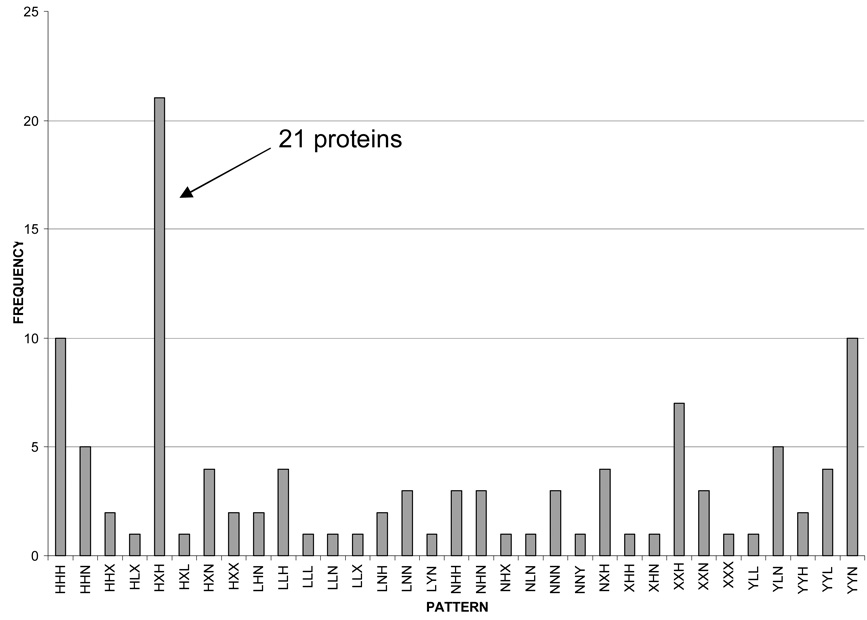
Because of the extensive amount of data generated from this pilot study, a pattern recognition approach was developed to segregate proteins into similar groups. To accomplish this, five arbitrary levels of change in expression were used to describe individual proteins as a function of disease progression. Proteins with ICAT ratios between 0.91 and 1.10 were labeled N, ratios between 1.11 and 1.50 were labeled H, ratios greater than 1.50 were labeled X. Equally distanced in the less than 1 category, ratios between 0.90 and 0.66 were labeled L and rations less than 0.66 were labeled Y. Application of this algorithm produced the frequency distribution plot shown in Figure 1. Interestingly, 21 proteins displayed a pattern of HXH in the progression from MCI to EAD to LAD which means that the MCI to control ratio was between 1.10 and 1.50, the EAD to control ratio was greater than 1.50 and the LAD to control ratio was between 1.10 and 1.50, thus producing an arc of protein levels as a function of disease progression. Table 2 lists the proteins, their mean ICAT ratios, standard error of the means (SEM) and the number of times each protein was observed in each disease category. Of the 21 proteins with an HXH pattern, 3 belong to the tricarboxylic acid cycle (ACON, CISY and MDHM)[31], 7 proteins are part of the electron transport chain and oxidative phosphorylation (ATP8, ATPG, COX5B, CY1, NUIM, SCOT and UQCR1)[31], 4 proteins are participate in ATP transport/utilization (HXK1, VDAC1, ADT1and KRCU)[32–35], 2 proteins are involved in anaplerotic reactions (AATM and GLSK)[36], 1 protein functions as a chaperone (CH60)[31], 1 protein is involved in mitochondrial protein biosynthesis (EFTU), 1 protein in lipid metabolism (ECHM)[31] and 1 protein in the regulation of the electron transport/oxidative phosphorylation pathways (CJ070)[37].
Figure 1.
A frequency distribution histogram resulting from the protein clustering algorithm showing that 21 of the 112 proteins were clustered into the HXH category.
Table 2.
Twenty-one proteins exhibiting a similar disease related pattern of protein change. The data column shows the mean, standard error of the mean (SEM ) and the number of instances the protein was observed in each disease category (Count)
| Protein Description | Data | MCI | EAD | LAD |
|---|---|---|---|---|
| AATM_HUMAN (P00505) Aspartate aminotransferase, mitochondrial precursor (EC 2.6.1.1) (Glutamate oxaloacetate transaminase 2) [MASS=47475] |
Mean | 1.43 | 1.70 | 1.31 |
| SEM | 0.07 | 0.09 | 0.09 | |
| Count | 3.00 | 3.00 | 3.00 | |
| ACON_HUMAN (Q99798) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) (Aconitase) [MASS=85425] |
Mean | 1.24 | 1.61 | 1.19 |
| SEM | 0.05 | 0.17 | 0.18 | |
| Count | 3.00 | 2.00 | 3.00 | |
| ADT1_HUMAN (P12235) ADP/ATP translocase 1 (Adenine nucleotide translocator 1) (ANT 1) (ADP,ATP carrier protein 1) [MASS=32933] |
Mean | 1.22 | 1.79 | 1.35 |
| SEM | 0.10 | 0.05 | 0.13 | |
| Count | 3.00 | 3.00 | 3.00 | |
| ANXA2_HUMAN (P07355) Annexin A2 (Annexin II) (Lipocortin II) (Calpactin I heavy chain) (Chromobindin-8) (p36) [MASS=38473] |
Mean | 1.40 | 1.64 | 1.28 |
| SEM | 0.14 | 0.01 | 0.27 | |
| Count | 3.00 | 2.00 | 2.00 | |
| ATP8_HUMAN (P03928) ATP synthase protein 8 (EC 3.6.3.14) (ATPase subunit 8) (A6L) [MASS=7992] |
Mean | 1.23 | 1.88 | 1.26 |
| SEM | 0.15 | 0.06 | 0.08 | |
| Count | 3.00 | 2.00 | 3.00 | |
| ATPG_HUMAN (P36542) ATP synthase gamma chain, mitochondrial precursor (EC 3.6.3.14) [MASS=32996] |
Mean | 1.33 | 1.95 | 1.25 |
| SEM | 0.07 | 0.07 | 0.13 | |
| Count | 3.00 | 3.00 | 3.00 | |
| CH60_HUMAN (P10809) 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (Mitochondrial matrix protein P1) [MASS=61054] |
Mean | 1.29 | 1.67 | 1.23 |
| SEM | 0.05 | 0.10 | 0.02 | |
| Count | 3.00 | 3.00 | 3.00 | |
| CISY_HUMAN (O75390) Citrate synthase, mitochondrial precursor (EC 2.3.3.1) [MASS=51712] |
Mean | 1.46 | 1.69 | 1.30 |
| SEM | 0.07 | 0.19 | 0.05 | |
| Count | 3.00 | 3.00 | 3.00 | |
| CJ070_HUMAN (Q9NZ45) Protein C10orf70 [MASS=12199] |
Mean | 1.16 | 1.61 | 1.24 |
| SEM | 0.08 | 0.12 | 0.04 | |
| Count | 3.00 | 3.00 | 3.00 | |
| COX5B_HUMAN (P10606) Cytochrome c oxidase polypeptide Vb, mitochondrial precursor (EC 1.9.3.1) [MASS=13696] |
Mean | 1.44 | 1.87 | 1.35 |
| SEM | 0.10 | 0.41 | 0.03 | |
| Count | 3.00 | 2.00 | 3.00 | |
| CY1_HUMAN (P08574) Cytochrome c1, heme protein, mitochondrial precursor (Cytochrome c-1) [MASS=35390] |
Mean | 1.18 | 1.81 | 1.26 |
| SEM | 0.10 | 0.18 | 0.05 | |
| Count | 3.00 | 3.00 | 3.00 | |
| ECHM_HUMAN (P30084) Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Enoyl-CoA hydratase 1) [MASS=31387] |
Mean | 1.31 | 1.95 | 1.31 |
| SEM | 0.23 | 0.14 | 0.23 | |
| Count | 2.00 | 3.00 | 2.00 | |
| EFTU_HUMAN (P49411) Elongation factor Tu, mitochondrial precursor (EF-Tu) (P43) [MASS=49541] |
Mean | 1.36 | 1.57 | 1.19 |
| SEM | 0.06 | 0.22 | 0.03 | |
| Count | 3.00 | 2.00 | 3.00 | |
| GLSK_HUMAN (O94925) Glutaminase kidney isoform, mitochondrial precursor (EC 3.5.1.2) (L-glutamine amidohydrolase) [MASS=73461] |
Mean | 1.30 | 1.61 | 1.44 |
| SEM | 0.09 | 0.11 | 0.08 | |
| Count | 3.00 | 3.00 | 3.00 | |
| HXK1_HUMAN (P19367) Hexokinase-1 (EC 2.7.1.1) (Hexokinase type I) (HK I) (Brain form hexokinase) [MASS=102484] |
Mean | 1.24 | 1.61 | 1.33 |
| SEM | 0.04 | 0.11 | 0.09 | |
| Count | 3.00 | 3.00 | 3.00 | |
| KCRU_HUMAN (P12532) Creatine kinase, ubiquitous mitochondrial precursor (EC 2.7.3.2) (U-MtCK) (Acidic- typemitochondrial creatine kinase) [MASS=47036] |
Mean | 1.20 | 1.51 | 1.27 |
| SEM | 0.01 | 0.20 | 0.05 | |
| Count | 3.00 | 3.00 | 3.00 | |
| MDHM_HUMAN (P40926) Malate dehydrogenase, mitochondrial precursor (EC 1.1.1.37) [MASS=35531] |
Mean | 1.25 | 1.79 | 1.35 |
| SEM | 0.08 | 0.09 | 0.02 | |
| Count | 3.00 | 3.00 | 3.00 | |
| NUIM_HUMAN (O00217) NADH-ubiquinone oxidoreductase 23 kDa subunit, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (Complex I-23KD) [MASS=23705] |
Mean | 1.49 | 2.39 | 1.19 |
| SEM | 0.22 | 0.34 | 0.26 | |
| Count | 2.00 | 2.00 | 2.00 | |
| SCOT_HUMAN (P55809) Succinyl-CoA:3-ketoacid- coenzyme A transferase 1, mitochondrial precursor (EC 2.8.3.5) [MASS=56157] |
Mean | 1.28 | 1.66 | 1.36 |
| SEM | 0.10 | 0.20 | 0.19 | |
| Count | 3.00 | 3.00 | 3.00 | |
| UQCR1_HUMAN (P31930) Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor (EC 1.10.2.2) [MASS=52645] |
Mean | 1.25 | 1.76 | 1.13 |
| SEM | 0.12 | 0.20 | 0.11 | |
| Count | 3.00 | 3.00 | 3.00 | |
| VDAC1_HUMAN (P21796) Voltage-dependent anion- selective channel protein 1 (VDAC-1) (hVDAC1) (Outer mitochondrial membrane protein porin 1) [MASS=30641] |
Mean | 1.12 | 1.87 | 1.43 |
| SEM | 0.07 | 0.19 | 0.37 | |
| Count | 3.00 | 3.00 | 3.00 |
Twenty of the 21 proteins identified with an HXH pattern of expression represent several important mitochondrial protein groups/pathways including the electron transport chain, tricaboxylic acid pathway, chaperon proteins, and proteins involved in ATP transport and utilization. Only one protein, annexin A2 (ANXA2_Human) does not appear to be primarily mitochondrial. Annexin A2 is a plasma membrane protein that binds phospholipids under calcium dependent control [38] and is likely co-precipitated with actin during mitochondrial isolation [39].
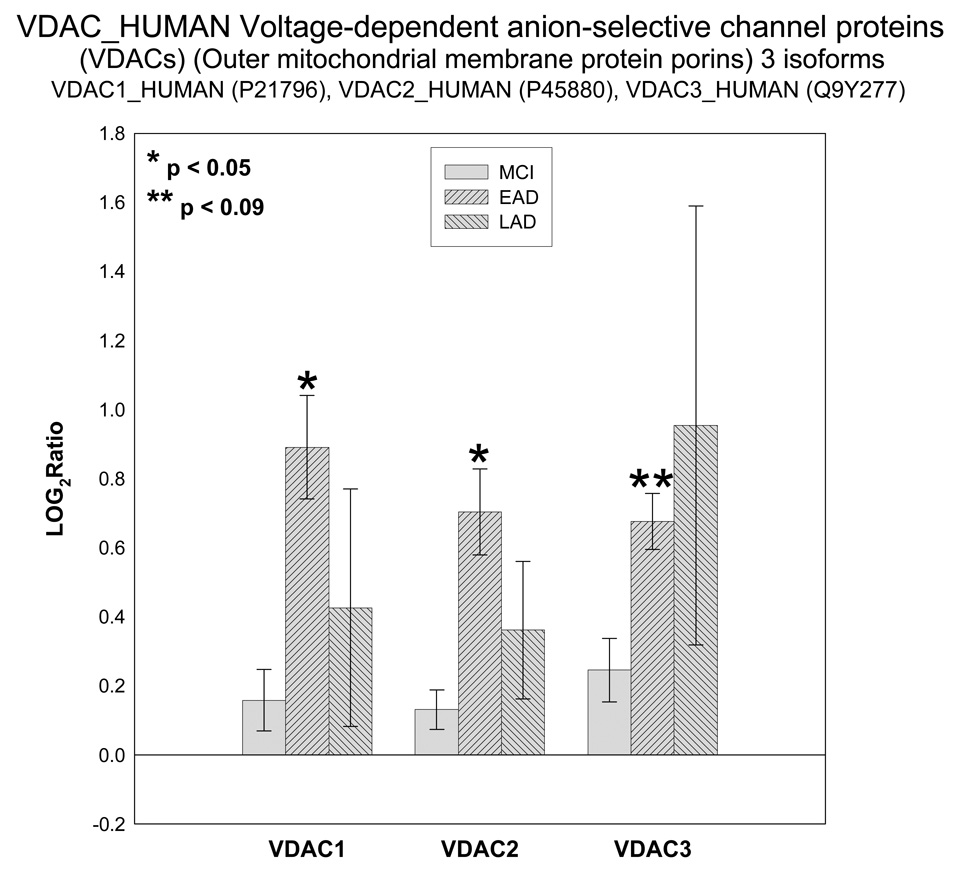
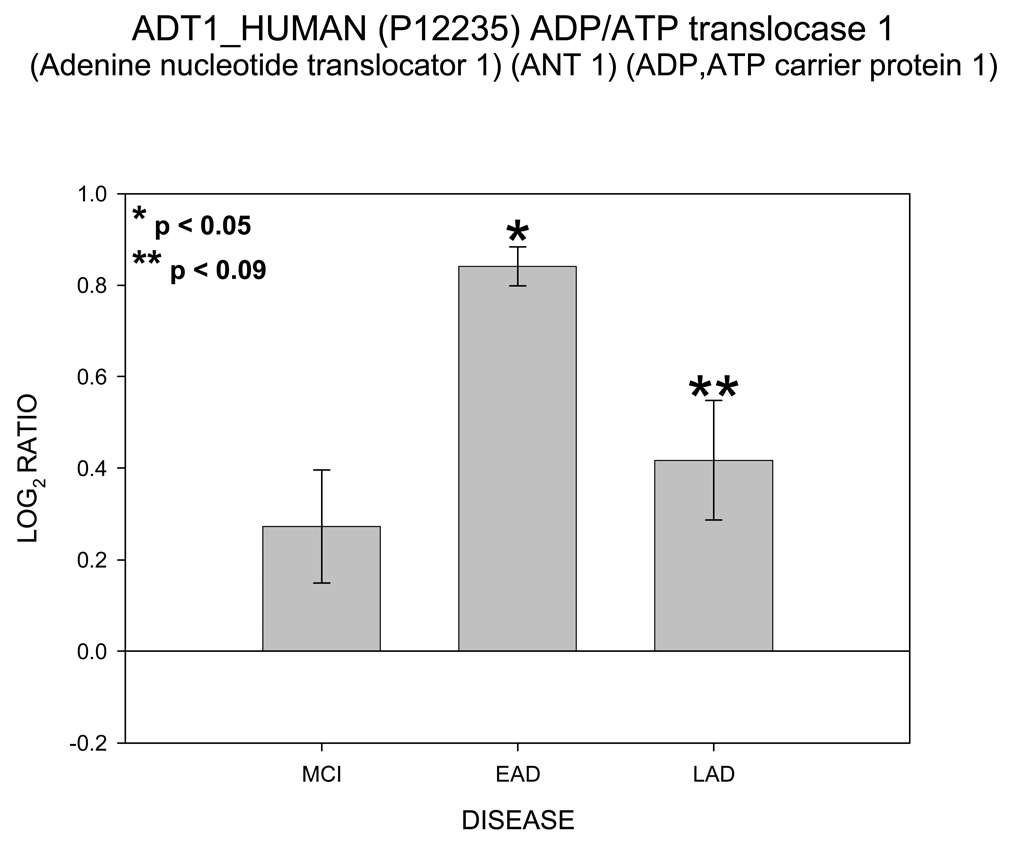
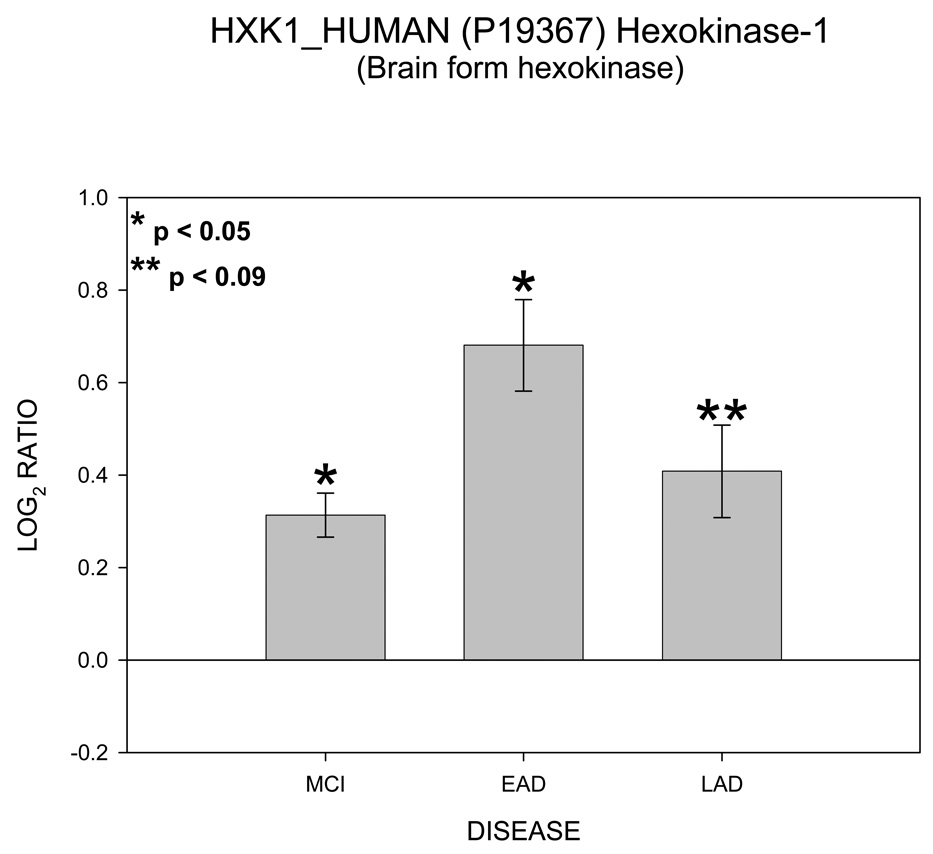
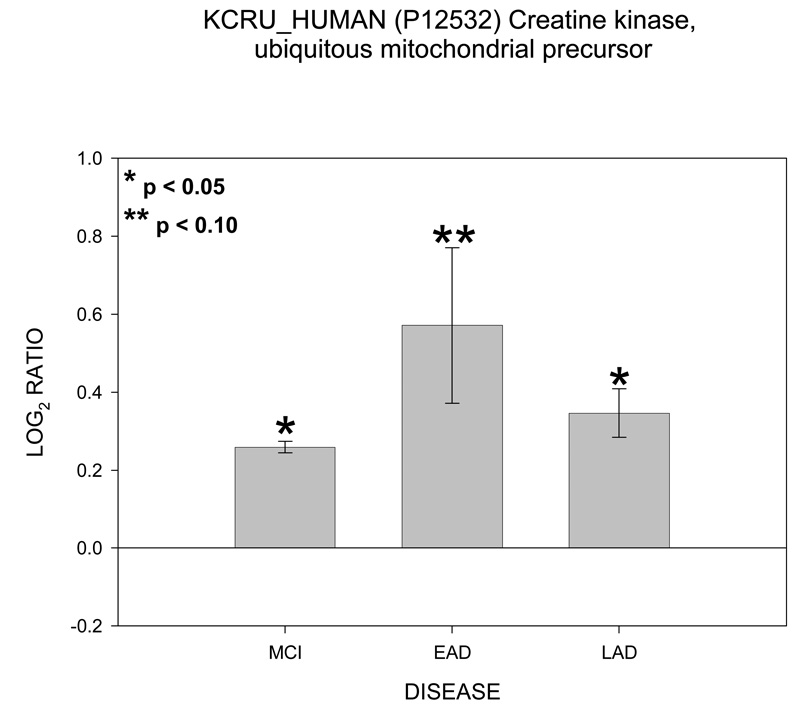
Of particular interest are the appearance of four proteins involved in ATP transport and utilization. These proteins are hexokinase 1 (HXK1), voltage dependent anion channel (VDAC 1), ATP/ADP translocase 1 (ADT1) and mitochondrial creatine kinase (KCRU). Figure 1 shows mean ± SEM levels of HXK1 in MCI, EAD and LAD mitochondria relative to control levels. The ICAT ratios were converted to log base 2 values for two reasons; 1) this approach results in similar output values as those obtained from gene chip arrays and thus may be more easily interpreted; and, 2) log base 2 values result in a linear scale of increased and decreased ratios. For example, an ICAT ratio of 1 equals 0 (log2), however an ICAT ratio of 0.5 and 2 are equally displaced in log2 space (log2(0.5) = −1 and log2(2) = 1). For HXK1, statistically significant (p < 0.05) increases were observed for MCI and EAD mitochondria and were trending toward significance (p = 0.06) in LAD relative to NC mitochondria. Figure 2 shows levels of the 3 isoforms of VDAC. It should be noted that only VDAC1 followed the HXH pattern of change whereas VDAC2 followed a LXH pattern and VDAC3 followed an HXX pattern. Figure 3 shows levels of ADT1. The EAD to control ratio was statistically significantly (p < 0.05) and the LAD to control ratio was trending toward significance (p = 0.09). Finally, Figure 4 shows a histogram plot for KCRU where significant changes were observed for MCI and LAD relative to control (p < 0.05)and EAD was trending toward significance (p = 0.1).
Figure 2.
Histogram plot of the log base 2 ratio data for the VDAC isoforms as a function of the three disease states, MCI, EAD and LAD.
Figure 3.
Histogram plot of the log base 2 ratio data for ADT1 as a function of the three disease states, MCI, EAD and LAD.
Figure 4.
Histogram plot of the log base 2 ratio data for HXK1 as a function of the three disease states, MCI, EAD and LAD.
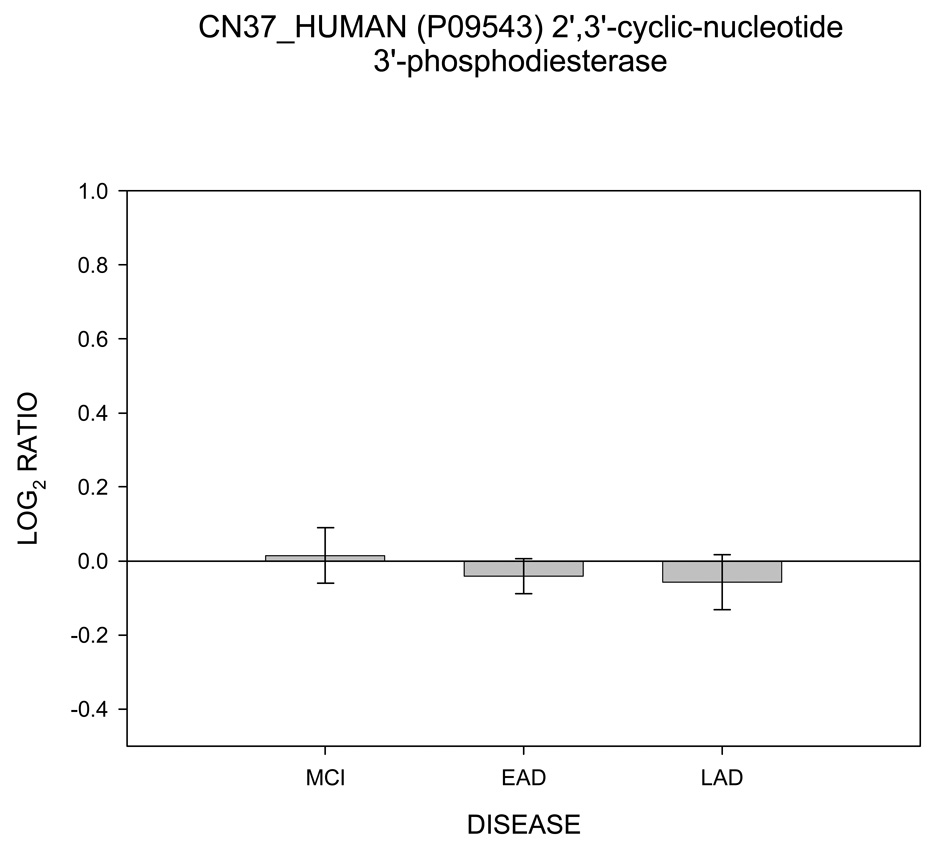
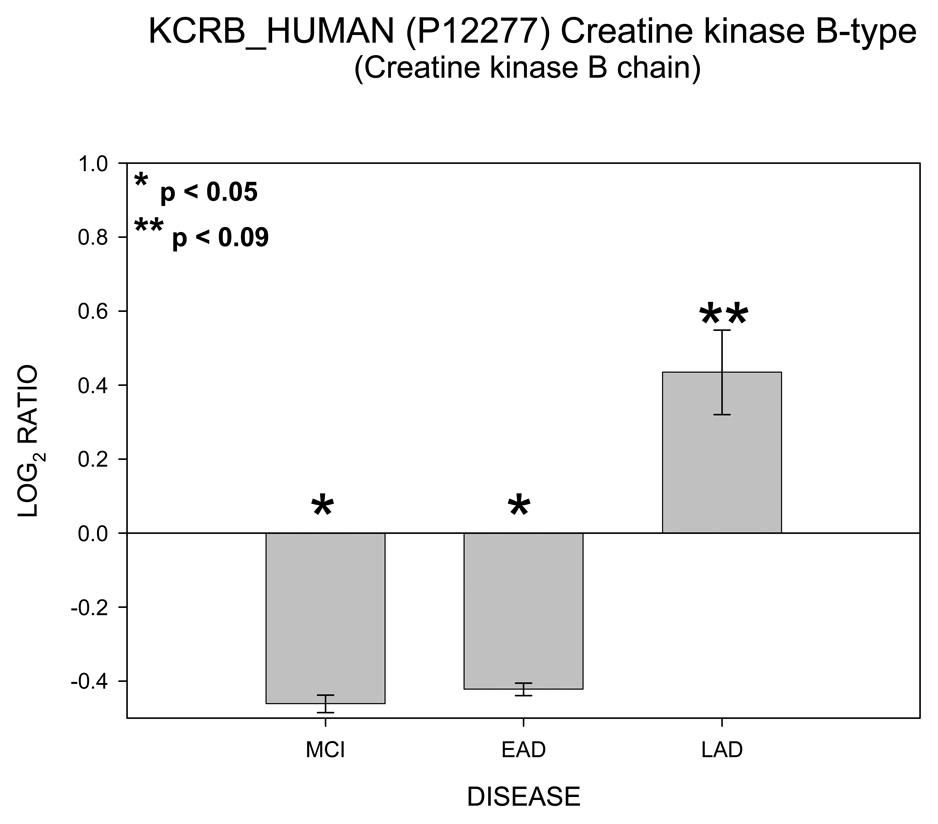
In contrast to Figure 1 – Figure 5, Figure 6 shows no change in log2 ICAT ratio for 2’,3’-cyclic-nucleotide 3’-phosphodiesterase (CN37) as a function of disease progression and therefore was categorized as NNN. Results for creatine kinase (KCRB) are shown in Figure 7. For KCRB, statistically significant negative log2 values for MCI and EAD (p < 0.05) indicate a reduction in levels of this protein relative to control. This trend reverses for LAD mitochondria which showed a positive value that trended toward significance (p = 0.06). Both CN37 and KCRB are not considered mitochondrial proteins but were co-isolated by our isolation protocol.
Figure 5.
Histogram plot of the log base 2 ratio data for KRCU as a function of the three disease states, MCI, EAD and LAD.
Figure 6.
Histogram plot of the log base 2 ratio data for the CN37 (CNP) as a function of the three disease states, MCI, EAD and LAD showing no change in protein levels.
Figure 7.
Histogram plot of the log base 2 ratio data for KRCB as a function of the three disease states, MCI, EAD and LAD showing a completely different pattern of protein change..
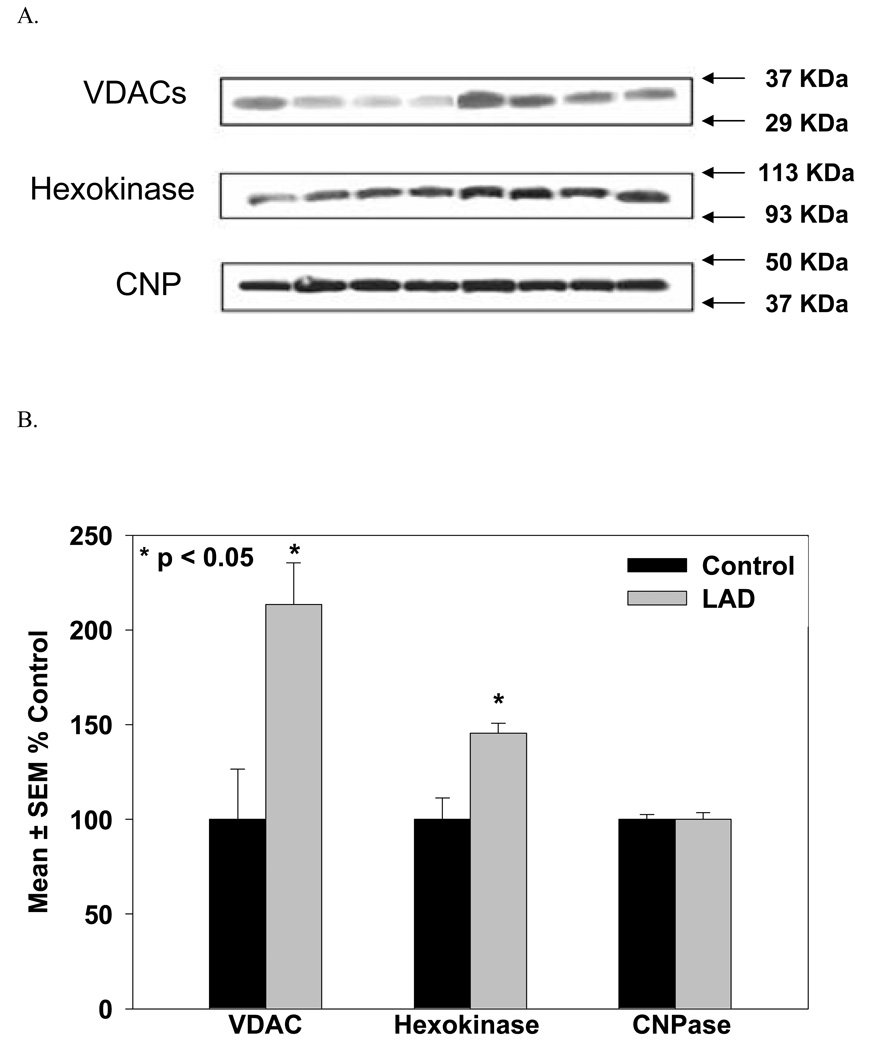
Western blot validation of proteomic data was carried out on 4 control and 4 LAD subjects for total VDAC and HXK1. VDAC and hexokinase were chosen for comparison with the ICAT results in light of our previous findings using rat primary neurons [40] and because of the pivotal roles played by VDAC and hexokinase in ATP transport/utilization and mitochondrial triggered apoptosis. Protein loading efficiencies in the Western blot analyses were verified using an antibody against CN37 which showed no significant differences between AD and control subjects using ICAT analyses. Results from these Western blots are shown in Figure 8. Consistent with proteomic analysis, Western blots showed total VDAC and HXK1 were significantly different from NC specimens. Western blot analysis of CN37 showed no significant differences between LAD and NC specimens also consistent with proteomic observations.
Figure 8.
8a. Western blot images for VDACs, HXK1 and CN37 (CNP) where the first four lanes are control and the next four lanes are LAD. 8b. Histogram plots for VDACs, HXK1 and CN37 resulting from quantitative analysis of the Western blot images (expressed as percent of control).
4. Discussion
The basic rationale for this study was to screen for potential changes in the mitochondrial proteome as a function of disease progression. Proteins identified in this pilot study will be the focus of future in-depth investigations. As with every discovery phase study, certain compromises must be made to facilitate the work. Due to the relatively limited nature of the samples, we chose to pool mitochondria within a disease category. Pooling mitochondrial samples not only extended the amount of material available for the study but also provided a means to characterize analytical variation. Data for three analytical replicates across all three disease states indicated less than a 25% coefficient of variance. Additionally, replicates facilitated by pooling were used as a means of filtering out false positive hits in the proteomic results. By requiring that a protein be observed in two of the three analytical replicates and in all three disease state before it was reported, no false positive identifications contributed to the final data set as evidenced by our decoy database test. Because the ICAT protocol used in this study selects for labeled cysteine containing peptides, we compared cysteine containing proteins in the mitochondrial database to the entire human proteome [41]. A comparison of the Mitoproteome fasta database [42] with the SwissProt human sub-database for cysteine containing tryptic peptides in a range from m/z 400 to 3500 indicated that 90% of mitochondrial proteins contain at least one cysteine containing tryptic peptide compared to 95% from the human sub-database. This result suggested that the ICAT approach did not underestimate proteins present in the sample. However, the percentage of proteins with at least two cysteine containing tryptic peptides decreases to 43% for mitochondria and 62% for the human sub-database respectively, perhaps indicating a potential limitation for the ICAT approach when multiple unique peptides are required for identification.
In this pilot study, we identified and quantified 112 unique, non-redundant proteins in at least two of the three analytical replicates common to all three disease stages. Twenty-one of 112 showed an unusual pattern of disease related change (HXH). Twenty of these proteins were components of several important mitochondrial protein groups/pathways including the electron transport chain, tricaboxylic acid pathway, chaperon proteins, and proteins involved in ATP transport and utilization. Since mitochondria are the sole producers of ATP, changes in proteins responsible for transport and utilization of ATP would be of great consequence to the host cell [43]. Our data show alterations in four proteins directly involved in transport and utilization of ATP; namely the VDACs, ADT1, HXK1 and KCRU.
VDACs are pore forming proteins that provide selective transport of anions across the mitochondrial outer membrane [32]. VDACs are also a major constituent of the mitochondrial membrane permeability transition pore [44]. These VDAC proteins exist in three isoforms, VDAC1, VDAC2 and VDAC3 [32]. It is currently unclear why three VDAC isoforms exist although it has been suggested that two of the isoforms are evolutionary vestiges [45]. Alternatively, multiple VDAC isoforms may serve different functions [46].
The ADT1 protein is a transmembrane protein located in the inner mitochondrial membrane. When VDAC couples with ANT, these two proteins produce a channel that spans the inter- and outer-membrane facilitating efflux of ATP and influx of ADP [32, 47, 48]. Hexokinase, an extra-mitochondrial protein that strongly associates with VDAC during certain metabolic sequences, directly utilizes the ATP delivered by VDAC to phosphorylated glucose in the first stage of glycolysis [33, 49]. Therefore, VDAC, ANT, and hexokinase play pivotal roles in aerobic glycolysis and glucose utilization [50]. Several studies show a reduction in glucose utilization in the AD brain [8, 51, 52], thus changes in the abundance of the three-protein-complex could play a role in the progression of AD. Hexokinase is tightly associated with VDAC while glucose-6-phosphate production is active. However, under alosteric control, hexokinase dissociates from VDAC when levels of G-6-P increase [49]. After hexokinase dissociation, VDAC closes which blocks the efflux of ATP from the mitochondria [32]. To prevent disruption of oxidative phosphorlyation, creatine kinase in the inter-membrane space utilizes ATP to phosphorylate creatine [35]. The resulting phosphocreatine can be transported to other systems in the cell requiring energy [53]. Our data show mitochondrial creatine kinase levels were elevated and followed the same pattern observed for other members of the ATP utilization pathway (VDAC, ADT1 and hexokinase). In contrast, cytoplasmic creatine kinase (KCRB) co-isolated with the mitochondrial fractions did not show this characteristic pattern of change.
Results from 21 proteins in this study show an unusual pattern of mitochondrial changes as a function of disease progression. Slight increases in protein abundance observed for MCI specimens lead to significant alterations in EAD. As the disease progresses to LAD, the abundance changes observed for EAD subjects tend to moderate. This arc of mitochondrial protein changes was rather surprising and difficult to explain. It has been suggested that glial proliferation during LAD may account for some of this moderation [30, 54]. Because we isolated a total mitochondrial pool (including glial mitochondria), gliosis could lead to a proteomic profile of mitochondria that is more representative of the increasing glial pool. However, immunohistochemical staining for GFAP did not support this hypothesis suggesting that contributions from glial mitochondria would be minimal.
Overall, this pilot study was the first to employ 2D-LC/MS/MS and ICAT quantification for the analysis of multiple samples of mitochondria prepared from MCI, EAD, LAD, and age-matched NC subjects. In contrast to previous studies, this approach allows simultaneous analysis of a variety of proteins and provides information regarding changes in levels of potentially inter-related proteins. It must be emphasized that this pilot study utilized a limited number of subjects to construct the disease state pools. Therefore caution should be used when interpreting the biological significance of these results. However, our results suggest that mitochondrial proteins associated with ATP transport and utilization exhibited protein abundance changes as a function of disease progression. Increased protein levels associated with energy metabolism perhaps suggest attempts by the AD brain to maintain ATP levels in presence of declining mitochondrial membrane potentials that occur following oxidative stress [55]. Our results are consistent with those of Johnson et al. [56] who showed increased expression of proteins associated with ATP production in primary mouse neurons undergoing p53-mediated cell death. We are currently evaluating mitochondrial protein levels in specimens of hippocampus from AD and control subjects to determine if the patterns observed in TP are also present in a brain region with more advanced pathology.
Supplementary Material
Acknowledgements
The authors express their gratitude to Drs. David Wekstein, Frederick Schmidt, Charles Smith, Gregory Cooper and Richard Krysico for patient procurement and patient data. We also express our gratitude to Dr. Jack Goodman and William Nelson for assistance with the TPP and Dr. Shuling Xiong for assistance with Western blots. Finally we wish to thank Paula Thomason for editorial assistance and Sonya Anderson for subject demographic data. This work was supported by NIH grants 5-P305AG028383 and 5-R01-AG25403.
Abbreviations
- MCI
Mild Cognitive Impairment
- EAD
Early Alzheimer’s disease
- LAD
Late Alzheimer’s disease
- VDAC
voltage dependent anion channel
- PMI
postmortem interval
- TP
temporal pole
- PTP
permeability transition pore
References
- 1.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow RH, Kish SJ. Mitochondria in Alzheimer's disease. Int Rev Neurobiol. 2002;53:341–385. doi: 10.1016/s0074-7742(02)53013-0. [DOI] [PubMed] [Google Scholar]
- 3.Jost BC, Grossberg GT. The natural history of Alzheimer's disease: a brain bank study. J Am Geriatr Soc. 1995;43:1248–1255. doi: 10.1111/j.1532-5415.1995.tb07401.x. [DOI] [PubMed] [Google Scholar]
- 4.Regula KM, Ens K, Kirshenbaum LA. Mitochondria-assisted cell suicide: a license to kill. J. Molecular and Cellular Cardiology. 2003;35:559–567. doi: 10.1016/s0022-2828(03)00118-4. [DOI] [PubMed] [Google Scholar]
- 5.Pollack M, Leeuwaenburgh Apoptosis and aging: Role of the mitochondria. J. Gerontology. 2001;56A:B475–B482. doi: 10.1093/gerona/56.11.b475. [DOI] [PubMed] [Google Scholar]
- 6.Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 7.Hoyer S. Intermediary metabolism disturbance in AD/SDAT and its relation to molecular events. Prog Neuropsychopharmacol Biol Psychiatry. 1993;17:199–228. doi: 10.1016/0278-5846(93)90043-r. [DOI] [PubMed] [Google Scholar]
- 8.Fukuyama H, Ogawa M, Yamauchi H, Yamaguchi S, Kimura J, Yonekura Y, Konishi J. Altered cerebral energy metabolism in Alzheimer's disease: a PET study. J Nucl Med. 1994;35:1–6. [PubMed] [Google Scholar]
- 9.Blass JP. The mitochondrial spiral. An adequate cause of dementia in the Alzheimer’s syndrome. Ann N Y Acad Sci. 2000;924:170–183. doi: 10.1111/j.1749-6632.2000.tb05576.x. [DOI] [PubMed] [Google Scholar]
- 10.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu Y, Sousa EA, Hewick RM, Wang JH. Acid-labile isotope-coded extractants: a class of reagents for quantitative mass spectrometric analysis of complex protein mixtures. Anal Chem. 2002;74:4969–4979. doi: 10.1021/ac0256437. [DOI] [PubMed] [Google Scholar]
- 12.Tao WA, Aebersold R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr Opin Biotechnol. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 13.Hansen KC, Schmitt-Ulms G, Chalkley RJ, Hirsch J, Baldwin MA, Burlingame AL. Mass spectrometric analysis of protein mixtures at low levels using cleavable 13C–isotope-coded affinity tag and multidimensional chromatography. Mol Cell Proteomics. 2003;2:299–314. doi: 10.1074/mcp.M300021-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 15.Consensus recommendations for the postmortem diagnosis of Alzheimer's disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer's Disease. Neurobiol Aging. 1997;18:S1–S2. [PubMed] [Google Scholar]
- 16.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 17.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 18.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 19.Mecocci P, MacGarvey U, Kaufman AE, Koontz D, Shoffner JM, Wallace DC, Beal MF. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 20.Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease. Ann Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer's disease. J Neurochem. 2005;93:953–962. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- 22.Poon HF, Frasier M, Shreve N, Calabrese V, Wolozin B, Butterfield DA. Mitochondrial associated metabolic proteins are selectively oxidized in A30P alpha-synuclein transgenic mice - a model of familial Parkinson's disease. Neurobiology of Disease. 2005;18:492–498. doi: 10.1016/j.nbd.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J Proteome Res. 2002;1:47–54. doi: 10.1021/pr015509n. [DOI] [PubMed] [Google Scholar]
- 24.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 25.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 26.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 27.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 28.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Goodlett DR, Quinn JF, Peskind E, Kaye JA, Zhou Y, Pan C, Yi E, Eng J, Wang Q, Aebersold RH, Montine TJ. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J Alzheimers Dis. 2005;7:125–133. doi: 10.3233/jad-2005-7205. discussion 173–180. [DOI] [PubMed] [Google Scholar]
- 30.Masters CL, Cappai R, Barnham KJ, Villemagne VL. Molecular mechanisms for Alzheimer's disease: implications for neuroimaging and therapeutics. J Neurochem. 2006;97:1700–1725. doi: 10.1111/j.1471-4159.2006.03989.x. [DOI] [PubMed] [Google Scholar]
- 31.www.expasy.org, Accessed
- 32.Lemasters JJ, Holmuhamedov E. Voltage-dependent anion channel (VDAC) as mitochondrial governator--thinking outside the box. Biochim Biophys Acta. 2006;1762:181–190. doi: 10.1016/j.bbadis.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Robey RB, Hay N. Mitochondrial hexokinases: guardians of the mitochondria. Cell Cycle. 2005;4:654–658. doi: 10.4161/cc.4.5.1678. [DOI] [PubMed] [Google Scholar]
- 34.Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim Biophys Acta. 2003;1604:77–94. doi: 10.1016/s0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- 35.Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim Biophys Acta. 2006;1762:164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 37.Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc Natl Acad Sci U S A. 2007;104:5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diakonova M, Gerke V, Ernst J, Liautard JP, van der Vusse G, Griffiths G. Localization of five annexins in J774 macrophages and on isolated phagosomes. J Cell Sci. 1997;110(Pt 10):1199–1213. doi: 10.1242/jcs.110.10.1199. [DOI] [PubMed] [Google Scholar]
- 39.Yoshii K, Sugimoto K, Tai Y, Konishi R, Tokuda M. Purification, identification and phosphorylation of annexin I from rat liver mitochondria. Acta Med Okayama. 2000;54:57–65. doi: 10.18926/AMO/32286. [DOI] [PubMed] [Google Scholar]
- 40.Lovell MA, Xiong S, Markesbery WR, Lynn BC. Quantitative proteomic analysis of mitochondria from primary neuron cultures treated with amyloid beta peptide. Neurochem Res. 2005;30:113–122. doi: 10.1007/s11064-004-9692-5. [DOI] [PubMed] [Google Scholar]
- 41.Cagney G, Amiri S, Premawaradena T, Lindo M, Emili A. In silico proteome analysis to facilitate proteomics experiments using mass spectrometry. Proteome Sci. 2003;1:5. doi: 10.1186/1477-5956-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.www.mitoproteome.org, Accessed
- 43.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 44.Tsujimoto Y, Nakagawa T, Shimizu S. Mitochondrial membrane permeability transition and cell death. Biochim Biophys Acta. 2006;1757:1297–1300. doi: 10.1016/j.bbabio.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 46.De Pinto V, Messina A, Accardi R, Aiello R, Guarino F, Tomasello MF, Tommasino M, Tasco G, Casadio R, Benz R, De Giorgi F, Ichas F, Baker M, Lawen A. New functions of an old protein: the eukaryotic porin or voltage dependent anion selective channel (VDAC) Ital J Biochem. 2003;52:17–24. [PubMed] [Google Scholar]
- 47.Vyssokikh M, Brdiczka D. VDAC and peripheral channelling complexes in health and disease. Mol Cell Biochem. 2004;256–257:117–126. doi: 10.1023/b:mcbi.0000009863.69249.d9. [DOI] [PubMed] [Google Scholar]
- 48.Buck CR, Jurynec MJ, Gupta DK, Law AK, Bilger J, Wallace DC, McKeon RJ. Increased adenine nucleotide translocator 1 in reactive astrocytes facilitates glutamate transport. Exp Neurol. 2003;181:149–158. doi: 10.1016/s0014-4886(03)00043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skaff DA, Kim CS, Tsai HJ, Honzatko RB, Fromm HJ. Glucose 6-phosphate release of wild-type and mutant human brain hexokinases from mitochondria. J Biol Chem. 2005;280:38403–38409. doi: 10.1074/jbc.M506943200. [DOI] [PubMed] [Google Scholar]
- 50.Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- 51.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bigl M, Bruckner MK, Arendt T, Bigl V, Eschrich K. Activities of key glycolytic enzymes in the brains of patients with Alzheimer's disease. J Neural Transm. 1999;106:499–511. doi: 10.1007/s007020050174. [DOI] [PubMed] [Google Scholar]
- 53.Adhihetty PJ, Beal MF. Creatine and its potential therapeutic value for targeting cellular energy impairment in neurodegenerative diseases. Neuromolecular Med. 2008;10:275–290. doi: 10.1007/s12017-008-8053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugaya K. [Glial activation and brain aging] Nippon Yakurigaku Zasshi. 2001;118:251–257. doi: 10.1254/fpj.118.251. [DOI] [PubMed] [Google Scholar]
- 55.Atlante A, Calissano P, Bobba A, Giannattasio S, Marra E, Passarella S. Glutamate neurotoxicity, oxidative stress and mitochondria. FEBS Lett. 2001;497:1–5. doi: 10.1016/s0014-5793(01)02437-1. [DOI] [PubMed] [Google Scholar]
- 56.Johnson MD, Yu LR, Conrads TP, Kinoshita Y, Uo T, Matthews JD, Lee SW, Smith RD, Veenstra TD, Morrison RS. Proteome analysis of DNA damage-induced neuronal death using high throughput mass spectrometry. J Biol Chem. 2004;279:26685–26697. doi: 10.1074/jbc.M401274200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.