Abstract
BACKGROUND
Candidate biomarkers discovered with high-throughput proteomic techniques (along with many biomarkers reported in the literature) must be rigorously validated. The simultaneous quantitative assessment of multiple potential biomarkers across large cohorts presents a major challenge to the field. Multiplex immunoassays represent a promising solution, with the potential to provide quantitative data via parallel analyses. These assays also require substantially less sample and reagents than the traditional ELISA (which is further limited by its ability to measure only a single antigen). We have measured the reproducibility, reliability, robustness, accuracy, and throughput of commercially available multiplex immunoassays to ascertain their suitability for serum biomarker analysis and validation.
METHODS
Assay platforms MULTI-ARRAY (Meso Scale Discovery), Bio-Plex (Bio-Rad Laboratories), A2 (Beckman Coulter), FAST Quant (Whatman Schleicher & Schuell BioScience), and FlowCytomix (Bender MedSystems) were selected as representative examples of technologies currently used for high-throughput immunoanalysis. All assays were performed according to protocols specified by the manufacturers and with the reagents (diluents, calibrators, blocking reagents, and detecting-antibody mixtures) included with their kits.
RESULTS
The quantifiable interval determined for each assay and antigen was based on precision (CV < 25%) and percentage recovery (measured concentration within 20% of the actual concentration). The MULTI-ARRAY and Bio-Plex assays had the best performance with the lowest limits of detection, and the MULTI-ARRAY system had the most linear signal output over the widest concentration range (105 to 106). Cytokine concentrations in unspiked and cytokine-spiked serum samples from healthy individuals were further investigated with the MULTI-ARRAY and Bio-Plex assays.
CONCLUSIONS
The MULTI-ARRAY and Bio-Plex multiplex immunoassay systems are the most suitable for biomarker analysis or quantification.
Clinical proteomic research is directed toward the identification of biomarkers that can aid in diagnosing disease, estimating prognosis, and monitoring treatment (1, 2). Large numbers of candidate biomarkers are typically identified in the discovery phase of a proteomic screen, and many candidate biomarkers have been reported in the literature (3). These candidates must be further evaluated to identify those markers that have a statistically significant association with disease (4, 5). In the biomarker verification/validation phase, the concentrations of candidate proteins must be measured across clinical cohorts (6, 7).
The concentrations of circulating proteins in plasma or serum can be measured with ELISAs (8). Although ELISAs provide precise and accurate results, they measure only a single antigen. Recent advances in mass spectroscopy–based protein identification have generated an avalanche of candidate biomarkers, thereby creating a market for high-throughput multiplex immunoassays that allow simultaneous quantification of many analytes. Two basic assay formats have been developed to facilitate simultaneous quantification of multiple antigens: planar array assays and microbead assays. In the first format, different capture antibodies are spotted at defined positions on a 2-dimensional array. In the second, the capture antibodies are conjugated to different populations of microbeads, which can be distinguished by their fluorescence intensity in a flow cytometer. In this study, we evaluated the performance of 3 planar array assays [MULTI-ARRAY (Meso Scale Discovery), A2 (Beckman Coulter), and FAST Quant (Whatman Schleicher & Schuell BioScience)] and 2 microbead assays [Bio-Plex (Bio-Rad Laboratories) and FlowCytomix (Bender MedSystems)]. For details about the various platform characteristics, see the Data Supplement that accompanies the online version of this Brief Communication at http://www.clinchem.org/content/vol56/issue2.
The accuracy of quantification for multiplexed immunoassays depends, as with all ELISAs, on the quality of the calibration curves, which is determined by the following: appropriate curve-fitting procedures, assay imprecision (CV), recoveries, and assay linearity (the limits of quantification). The performance characteristics of all the platforms were evaluated with respect to these parameters. For each assay system, duplicate or triplicate wells of supplier-provided calibrators were serially diluted as recommended by the manufacturer to produce calibration curves. We were unable to investigate cross-reactivity between analytes because the manufacturers provided the calibrators as pre-mixed solutions containing all of the analytes.
It is important to note that none of the cytokine assays on the 5 platforms have been cleared by the US Food and Drug Administration for clinical testing. They are to be used only in research applications.
The FlowCytomix assay was not further evaluated as a platform for clinical validation, owing to substantial losses of beads that might adversely affect performance. In this assay, multiplexing is achieved by coupling the detector antibodies to internally fluorescent beads of 2 different sizes (4.4 μm and 5.5 μm); however, we experienced substantial losses of the small beads when we assayed human serum samples. The loss of the small beads may have been due to the viscosity of the serum because, in contrast to the other kits, the manufacturer of the FlowCytomix kit did not supply a diluent specifically formulated for serum.
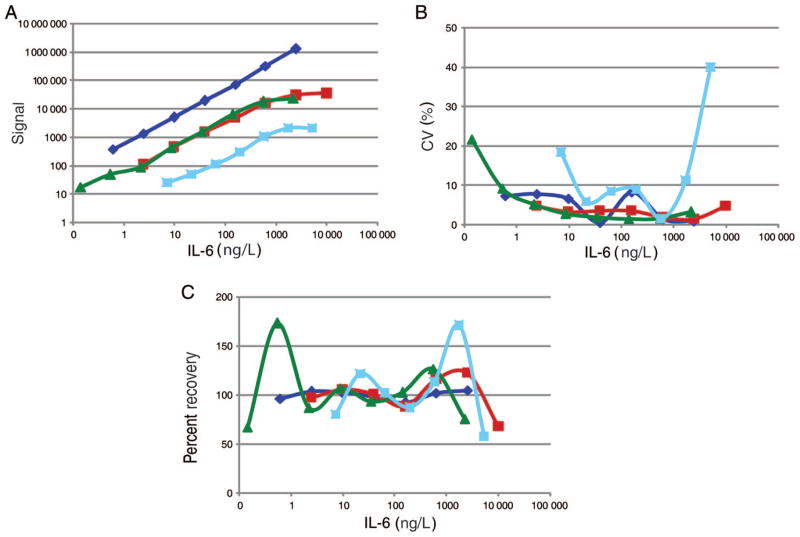
Most of the currently available multiplex immunoassays have been designed to quantify the concentrations of various cytokines, because cytokine concentrations can provide information about numerous diseases and inflammatory conditions (9, 10). Therefore, performance was analyzed for the remaining platforms, and the results were compared with data obtained for serial dilutions of 5 common cytokines [Fig. 1 for interleukin-6 (IL-6)4; Fig. 1 in the online Data Supplement for data for IL-12p70, IL-2, IL-10, and IL-1β]. All assays were performed on at least 3 different days.
Fig. 1. Calibration curve performance and comparison of MULTI-ARRAY (blue), Bio-Plex (green), A2 (light blue), and FAST Quant (red) multiplex assays.
The following cytokine kits were compared: Human Pro-Inflammatory 9-Plex MULTI-ARRAY kit [granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon γ (IFN-γ), IL-1β, IL-2, IL-6, IL-8, IL-10, IL-12p70, tumor necrosis factor-α (TNF-α)] from Meso Scale Discovery; Bio-Plex Human Cytokine 27-Plex Panel [IL-1β, IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17, eotaxin, basic fibroblast growth factor, granulocyte colony-stimulating factor, GM-CSF, IFN-γ, IFN-γ–induced protein 10 (IP-10), monocyte chemoattractant protein 1 (MCP-1/MCAF), macrophage inflammatory protein 1α (MIP-1α), MIP-1β, platelet-derived growth factor BB (PDGF-BB), RANTES (regulated upon activation, normal T cell expressed and secreted), TNF-α, VEGF (vascular endothelial growth factor)] from Bio-Rad Laboratories; A2 human 13-cytokine assay kit (GM-CSF, IFN-γ, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12p70, IL-13, TNF-α) from Beckman Coulter; FAST Quant Human II Kit 10-plex assay (IL-1β, IL-10, IL-2, IL-12p70, IL-4, GM-CSF, IL-6, RANTES, IL-8, MCP-1) from Whatman Schleicher & Schuell BioScience. (A), Log plots of raw IL-6 data (fluorescence signal intensity vs IL-6 concentration). (B), Corresponding plot of CV vs IL-6 concentration. The CV was calculated from 2 spots (2 wells in MULTI-ARRAY), 8 spots (2 pads in FAST Quant), 12 spots (3 wells in A2), and 2 wells (in Bio-Plex). (C), Plot of percentage calibrator recovery vs IL-6 concentration. A 4-parameter algorithm and a 1/y2 weighting function were used to fit (with Prism software) each data set from the MULTI-ARRAY, Bio-Plex, A2, and FAST Quant systems to a sigmoidal dose–response curve with a variable slope. Curve back-fitting is done by calculating the observed concentration of each calibrator and then comparing it with the original concentration (expected concentration) used in each analyte assay. For these tests, actual and expected concentrations were calculated, and the percentage recovery of calibrator was calculated as: [(observed concentration)/(expected concentration)] × 100.
The MULTI-ARRAY system had the greatest linear signal output, over the widest range (105 to 106), for every cytokine. By comparison, the signal output range was 103 to 104 for the Bio-Plex assay, 103 for the A2 assay, and 104 for the FAST Quant assay (Fig. 1A and Table 1; Fig. 1A in the online Data Supplement). Within each batch, CVs varied broadly, depending on the analytes: 0.4%–23% for the MULTI-ARRAY assay, 0.6%–40% for the Bio-Plex assay, 0.5%–39% for the A2 assay, and 0.8%–13% for the FAST Quant assay (Fig. 1B; Fig. 1B in the online Data Supplement).
Table 1.
Comparison of the performance of 4 multiplex immunoassay platforms.a
| MULTI-ARRAY | Bio-Plex | A2 | FAST Quant | |
|---|---|---|---|---|
| IL-12p70 | ||||
| Manufacturer’s calibrator interval, ng/Lb | 2500–0.6 | 4411–0.3 | 4800–6.5 | 50 000–12.2 |
| Quantifiable interval, ng/Lc | 2500–0.6 | 4411–0.27 | 577–7.1 | 625–2.4 |
| Signal intervald | ≅2 700 000–200 | ≅23 000–30 | ≅1000–25 | ≅16 000–100 |
| Mean CV within quantifiable intervale | 9.60% | 2.80% | 8.70% | 3.40% |
| IL-2 | ||||
| Manufacturer’s calibrator interval, ng/L | 2500–0.6 | 2161–0.1 | 6630–9.1 | 10 000–2.4 |
| Quantifiable interval, ng/L | 2500–2.4 | 540–2.1 | 245–9 | 10 000–2.4 |
| Signal interval | ≅600 000–200 | ≅18 000–100 | ≅4000–100 | ≅2000–30 |
| Mean CV within quantifiable interval | 5.00% | 5.90% | 10.00% | 3.20% |
| IL-6 | ||||
| Manufacturer’s calibrator interval, ng/L | 2500–0.6 | 2215–0.1 | 5200–7.1 | 10 000–2.4 |
| Quantifiable interval, ng/L | 2500–0.6 | 138–2.1 | 577–7.1 | 625–2.4 |
| Signal interval | ≅1 300 000–400 | ≅6000–80 | ≅1000–25 | ≅16 000–100 |
| Mean CV within quantifiable interval | 4.70% | 2.80% | 8.70% | 3.40% |
| IL-10 | ||||
| Manufacturer’s calibrator interval, ng/L | 2500–0.6 | 4311–0.3 | 4750–6.5 | 50 000–12.2 |
| Quantifiable interval, ng/L | 2500–0.6 | 269–1.05 | 175–6.5 | 50 000–195 |
| Signal interval | ≅500 000–300 | ≅70 000–20 | ≅3000–70 | ≅1500–300 |
| Mean CV within quantifiable interval | 5% | 8% | 6% | 4% |
| IL-1β | ||||
| Manufacturer’s calibrator interval, ng/L | 2500–0.6 | 3794–0.2 | 4840–6.6 | 10 000–2.4 |
| Quantifiable interval, ng/L | 2500–0.6 | 59–0.2 | 538–6.6 | 625–2.4 |
| Signal interval | ≅1 300 000–500 | ≅9000–50 | ≅3000–50 | ≅20 000–400 |
| Mean CV within quantifiable interval | 6.60% | 5.30% | 8.40% | 5.00% |
| Calibrators | ||||
| Manufacturer’s recommended standard dilution factorf | 1/4 Dilution | 1/4 Dilution | 1/3 Dilution | 1/4 Dilution |
| Calibration curve | 7 Concentrations | 8 Concentrations | 7 Concentrations | 7 Concentrations |
The manufacturers of the different multiplex platforms are Meso Scale Discovery (MULTI-ARRAY), Bio-Rad Laboratories (Bio-Plex), Beckman Coulter (A2), and Whatman Schleicher & Schuell (FAST Quant).
With the manufacturer’s recommended serial dilution of calibrators (see manufacturer’s recommended standard dilution factor).
The quantifiable interval of each analyte is defined as the concentration interval in which the CV is <25% and the percentage recovery is <100% ± 20% with the standard calibrators.
The signal interval of each analyte is defined as the signal output between upper and lower quantification limits with a CV <25% and a percentage recovery <100% ± 20% with the standard calibrators.
The mean CV within the quantifiable interval is calculated with a CV of <25% and a percentage recovery <100% ± 20% with the standard calibrators.
Serial dilution factors are expressed as the fraction of the concentration before dilution.
Because cytokines are present at low concentrations in nonpathologic sera, curve fitting has a direct effect on the accuracy of the results. Although each assay system includes software for curve fitting by nonlinear regression, differences among the algorithms and parameters embedded in these proprietary software programs made it impossible to compare the 4 different systems directly with regard to the quality of the results. To standardize the analytical process, we reanalyzed the data with Prism software (GraphPad Software). We used a 4-parameter algorithm and a 1/y2 weighting function to fit each data set to a sigmoidal dose–response curve with a variable slope.
To assess the quality of the fit at each concentration, we determined and compared actual and expected values to calculate percentage recovery (Fig. 1C; Fig. 1C in the online Data Supplement). Percentage recovery is an important measure for evaluating the accuracy of quantitative data from the lower limit of detection across the entire dynamic range of an assay (Fig. 1C; Fig. 1C in the online Data Supplement). We defined an upper limit of quantification (ULOQ) as the value below which the percentage recovery was 80%–120% and the CV was <25%. The lower limit of quantification (LLOQ) was defined as an analyte concentration that has (a) a response at least 3 times that of a blank sample, (b) a percentage recovery of 80%–120%, and (c) a CV <25%. We then calculated ULOQ and LLOQ values for the 5 cytokines on each assay platform (Table 1). Overall, the MULTI-ARRAY platform from Meso Scale Discovery had the largest quantifiable concentration interval (2.4–2500 ng/L).
Our assessments identified deficiencies in the A2 and FAST Quant platforms that may limit their overall suitability. The LLOQ was relatively poor on the A2 platform; similarly, the FAST Quant assay was not as sensitive as the other assays (Table 1). The FAST Quant software uses a point-to-point curve-fitting method and does not provide data on the back-fitting of a calibration curve (percentage recovery), making it difficult to evaluate the accuracy of the data.
The MULTI-ARRAY and Bio-Plex platforms demonstrated the best performance for quantifying purified cytokines. Although both systems were highly sensitive, the MULTI-ARRAY had a wider dynamic range because it had a higher ULOQ. On the other hand, the Bio-Plex system was able to quantify more analytes. We tested the ability of the MULTI-ARRAY and Bio-Plex systems to measure a common set of 9 cytokines in the complex milieu of human serum. Human serum samples from 12 healthy male individuals between 20 and 60 years of age were purchased from Bioreclamation. As expected, the concentrations of most cytokines in these healthy controls were very low, with many of the readings below the LLOQ (see Table 1 in the online Data Supplement). For samples of unspiked serum, the MULTI-ARRAY platform had better performance for 6 cytokines (IL-2, IL-10, IL-12p70, granulocyte-macrophage colony-stimulating factor, interferon γ, and tumor necrosis factor-α), and the Bio-Plex system had a better performance for 1 cytokine (IL-8). The 2 platforms performed similarly for IL-6. IL-1β could not be quantified with either platform. The inter- and intraplate CVs for duplicate assays were low for both platforms (mean interplate CVs of 8% and 10% for the MULTI-ARRAY and Bio-Plex platforms, respectively; mean intraplate CVs of 6% and 9% for the MULTI-ARRAY and Bio-Plex platforms, respectively).
To investigate the accuracy of cytokine quantification in human serum, we measured cytokine concentrations in serum samples supplemented with pure cytokines (see Fig. 2 in the online Data Supplement). Because we added each cytokine to a concentration of 50 ng/L, the difference between the measured cytokine concentrations in the supplemented and unspiked samples should have been 50 ng/L. On the MULTI-ARRAY platform, the differences varied from 53 ng/L to 70 ng/L. The Bio-Plex platform had larger differences (15–78 ng/L). One limitation of this study is that we did not evaluate potential effects related to pathophysiological changes in serum composition or the molecular form of the analytes. Thus, this study presents the best possible performance of the assays, and performance with real-world samples could be substantially worse.
The performance of multiplex assay platforms is analyte, sample type (matrix), and concentration dependent. Optimal assay performance depends on proprietary information about the antibody pair, the composition of diluents, and the software. Therefore, no single assay platform is suitable for all analytes. The MULTI-ARRAY and the Luminex-based Bio-Plex assays performed best overall, but their performance characteristics were not uniform (see Table 1 in the online Data Supplement). The 2 platforms performed similarly in detecting high concentrations (50 ng/L) of spiked cytokines (see Fig. 2 in the online Data Supplement). Although the MULTI-ARRAY platform was superior for quantifying cytokines in unspiked serum, it was severely limited by the small number of analytes that can be assayed with commercially available kits, which are provided exclusively by Meso Scale Discovery. In contrast, assays for a large selection of analytes are available from a network of Luminex partners. Luminex assays also have the potential to multiplex up to 100 analytes, whereas MULTI-ARRAY assays are limited to 10-plex.
In summary, the simultaneous analysis of multiple antigens makes it possible to identify combinations of biomarkers that have greater disease specificity and sensitivity than results obtained from the analysis of any single marker (11–15). We compared 5 commercial multiplex immunoassay platforms (representing both planar array and microbead assays) and found the MULTI-ARRAY and Bio-Plex systems to be the most suitable for biomarker verification and validation; however, the performance of even these systems was analyte dependent. Our results highlight the importance of an assay’s dynamic range, linearity, CV, and percentage recovery in obtaining accurate and reproducible assay results.
Acknowledgments
Research Funding: Funding was provided by grants from the National Heart, Lung, and Blood Institute (contract N0-HV-28180, grant PH SCCOR - 1 P50 HL 084946-01), the Daniel P. Amos Foundation, and an NIH Institutional Clinical and Translational Science Award to Johns Hopkins University (1U54RR023561–01A1).
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: IL-6, interleukin-6; ULOQ, upper limit of quantification; LLOQ, lower limit of quantification.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.Banks RE, Dunn MJ, Hochstrasser DF, Sanchez JC, Blackstock W, Pappin DJ, et al. Proteomics: new perspectives, new biomedical opportunities. Lancet. 2000;356:1749–56. doi: 10.1016/S0140-6736(00)03214-1. [DOI] [PubMed] [Google Scholar]
- 2.Patterson SD. Proteomics: beginning to realize its promise? Arthritis Rheum. 2004;50:3741–4. doi: 10.1002/art.20796. [DOI] [PubMed] [Google Scholar]
- 3.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5:310–20. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Q, Van Eyk JE. Proteomics and heart disease: identifying biomarkers of clinical utility. Expert Rev Proteomics. 2006;3:237–49. doi: 10.1586/14789450.3.2.237. [DOI] [PubMed] [Google Scholar]
- 5.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 6.Zolg JW, Langen H. How industry is approaching the search for new diagnostic markers and biomarkers. Mol Cell Proteomics. 2004;3:345–54. doi: 10.1074/mcp.M400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Rifai N, Gerszten RE. Biomarker discovery and validation. Clin Chem. 2006;52:1635–7. doi: 10.1373/clinchem.2006.074492. [DOI] [PubMed] [Google Scholar]
- 8.Price C, Newman D. Principles and practices of immunoassays. New York: Stockton Press; 1997. p. 750. [Google Scholar]
- 9.Sachdeva N, Asthana D. Cytokine quantitation: technologies and applications. Front Biosci. 2007;12:4682–95. doi: 10.2741/2418. [DOI] [PubMed] [Google Scholar]
- 10.Prabhakar U, Eirikis E, Miller BE, Davis HM. Multiplexed cytokine sandwich immunoassays: clinical applications. Methods Mol Med. 2005;114:223–32. doi: 10.1385/1-59259-923-0:223. [DOI] [PubMed] [Google Scholar]
- 11.Utz PJ. Multiplexed assays for identification of biomarkers and surrogate markers in systemic lupus erythematosus. Lupus. 2004;13:304–11. doi: 10.1191/0961203303lu1017oa. [DOI] [PubMed] [Google Scholar]
- 12.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359–65. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingsmore SF, Kennedy N, Halliday HL, Van Velkinburgh JC, Zhong S, Gabriel V, et al. Identification of diagnostic biomarkers for infection in premature neonates. Mol Cell Proteomics. 2008;7:1863–75. doi: 10.1074/mcp.M800175-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatine MS, Morrow DA, de Lemos JA, Gibson CM, Murphy SA, Rifai N, et al. Multimarker approach to risk stratification in non-ST elevation acute coronary syndromes: simultaneous assessment of troponin I, C-reactive protein, and B-type natriuretic peptide. Circulation. 2002;105:1760–3. doi: 10.1161/01.cir.0000015464.18023.0a. [DOI] [PubMed] [Google Scholar]
- 15.Ling MM, Ricks C, Lea P. Multiplexing molecular diagnostics and immunoassays using emerging microarray technologies. Expert Rev Mol Diagn. 2007;7:87–98. doi: 10.1586/14737159.7.1.87. [DOI] [PubMed] [Google Scholar]