Abstract
Purpose
Alkylating agents with or without ionizing radiation are frequently used in pre-transplant conditioning regimens. Damage induced by these agents is commonly repaired by the base excision repair (BER) pathway. Hence, we hypothesized that genetic polymorphisms in the BER pathway will be associated with post hematopoietic cell transplant (HCT) outcomes.
Patients and Methods
We evaluated the association between single nucleotide polymorphisms (SNPs) (n= 179) in the BER pathway with transplant related mortality at one year (TRM) and disease relapse in a cohort of 470 recipients who underwent allogeneic HCT for treatment of hematologic malignancies at the University of Minnesota.
Results
After adjustment for age at transplant, donor type, race, and conditioning regimen, four SNPs in OGGI, LIG3 and MUTYH genes (rs159153, rs3135974, rs3219463, rs3219476) were associated with increased risk of TRM whereas two SNPs in TDG gene (rs167715, rs2374327) were associated with decreased risk of TRM at one year (p≤0.01). Patients with increasing numbers of deleterious alleles in the BER pathway showed a higher cumulative incidence of TRM at one year (51% for ≥ 4 deleterious alleles vs. 14% for ≤ 1 deleterious allele; p<0.001). One SNP, rs3135974, in LIG3 gene was associated with decreased risk of disease relapse (p<0.001) post HCT.
Conclusions
These findings suggest that SNPs in the BER pathway can be used as genetic biomarkers to predict individuals at high risk for TRM and disease relapse. Modulation of pre-transplant conditioning may alter risk in these patients.
Keywords: Hematopoietic cell transplant, base excision repair, single nucleotide polymorphisms, transplant related mortality, disease relapse
Introduction
Despite rapid advances in hematopoietic cell transplant (HCT) techniques, the procedure continues to be associated with a high morbidity and mortality with transplant related mortality (TRM) ranging from 15% to as high as 50% 1-3. In those who survive the acute toxicity, disease relapse remains a major cause of death 3. High doses of ionizing radiation and alkylating agents such as cyclophosphamide and mephalan, induce significant DNA damage 4 and are commonly used during HCT 3. Single nucleotide polymorphisms (SNPs) in DNA repair genes may potentially influence the effectiveness of various chemotherapeutic regimens used in HCT and have been associated with treatment outcomes and overall survival in patients undergoing chemotherapy for a variety of hematological malignancies 5,6. One study has also shown an association between two polymorphisms in the DNA repair genes and overall survival in patients undergoing autologous transplants for treatment of multiple myeloma 7.
The base excision repair (BER) pathway is the primary DNA repair pathway that corrects base lesions that arise due to oxidative, alkylation, deamination, and depurination/depyrimidination damage 8 and plays an important role in repairing the damage caused by ionizing radiation and chemotherapeutic agents 4. There are at least 8 specialized DNA damage proteins that recognize specific types of DNA damage, which create abasic or apurinic/apyrimidinic (AP) sites 9,10. The DNA at the AP site is incised by a AP lyase (DNA polymerase beta) and repair of a single nucleotide is completed by DNA ligase III (short patch repair pathway) 8. Alternatively, the DNA at the AP site can be cleaved by an AP endonuclease (APEX1) and repair proceeds via a PCNA dependent manner and repair of two or more nucleotides is completed by DNA ligase I (long patch repair pathway) 8.
No study has yet systematically evaluated the association between SNPs in the BER pathway and outcomes after allogeneic HCT. We evaluated the association between SNPs in the BER pathway, TRM at one year and disease relapse in a cohort of patients who received an allogeneic HCT for hematologic malignancy at the University of Minnesota. We hypothesized that SNPs in BER genes would be independently associated with TRM at one year and disease relapse.
Material and Methods
Patient Selection
All patients (pediatric and adult) who underwent an allogeneic HCT from an HLA-identical sibling donor, matched or mismatched URD or single umbilical cord blood (UCB) transplant for treatment of hematological malignancies from January 1, 1998 to December 31, 2007 were eligible for study (n=587). All patients received a conditioning regimen that included an alkylating agent (e.g. cyclophosphamide) and/or ionizing radiation. After excluding patients for whom DNA samples were not available, 480 (82%) patients were included in this study. This study was approved by the Institutional Review Board at the University of Minnesota and waiver of informed consent from individual patients was obtained for this study.
Clinical data and collection of DNA samples
All clinical data was obtained using the University of Minnesota Blood and Marrow Transplant Database and supplemented by chart reviews where necessary. Demographic and transplant characteristics, which included patient and donor age at transplant, patient and donor gender, type of donor (related, unrelated donor (URD) or umbilical cord blood (UCB)), graft source (bone marrow versus peripheral blood stem cell versus cord blood), race, patient's underlying disease, disease status at transplant, HLA matching between donor and recipient, conditioning intensity (myeloablative or reduced intensity), conditioning regimen used for transplant, graft versus host disease (GVHD) prophylaxis, cytomegalovirus (CMV) serological status of recipient and donor prior to transplant and survival at time of last follow up were collected. HLA- matching status for URD transplants was categorized as well matched, partially matched or mismatched based on classification proposed by Weisdorf et al 11. HLA-matching status for cord blood transplants was based on antigen level HLA-A, B and allele level HLA-DRB1 typing.
Transplant related mortality (TRM) was defined as mortality due to all causes other than disease relapse or progression. Causes of death were carefully evaluated only to exclude patients with accidental death unrelated to HCT (n=1). Disease relapse or progression was defined by first evidence of hematologic, cytogenetic, molecular or radiological evidence of relapse or progression of the malignancy.
DNA samples from patients were obtained from peripheral blood samples collected prior to HCT. All patients with acute leukemias had no evidence of leukemic blasts in peripheral blood at the time of obtaining blood for DNA extraction. DNA was extracted from buffy coat using either the Puregene DNA extraction method (for samples collected prior to 2001) (Gentra Systems, Minneapolis, MN) or the Qiagen Mini Blood kit (all samples collected during or after 2001) (Qiagen Inc., San Jose, CA) as per manufacturer recommendations. All DNA samples were collected and stored at 4°C in the Molecular Diagnostics Laboratory at the University of Minnesota Medical Center, Fairview prior to use in the study.
SNP selection and genotyping
We used data from Environmental Genome Project 10,12 to identify and select Linkage Disequilibrium (LD) tagSNPs that had a frequency of ≥ 5% in the Caucasian population in 27 BER genes. If genes did not have tagSNPs information in the Environmental Genome Project, we used HapMap data for Caucasian populations to identify tagSNPs that had ≥5% frequency in Caucasian populations using the Genome Variation Server software (http://gvs.gs.washington.edu/GVS/). We then supplemented this list with non-synonymous SNPs that were identified to be functionally important using three software programs; PolyPhen 13, SIFT 14 and SNP3D 15. This approach allowed us to select 263 SNPs (210 tagSNPs and 53 SNPs that may be functionally important) in 27 genes with known function in the BER pathway. All DNA samples were genotyped using the Sequenom iPLEX system at the Biomedical Genomics Center at the University of Minnesota. All primers and probes were designed using Sequenom primer design software. We were able to design primers and probes for 239 SNPs.
Statistical Analysis
All statistical analyses were performed using SAS software version 9.1 (SAS Institute, Inc., Cary, NC) and R Statistical software version 2.4.1 (http://cran.opensourceresources.org/) 16. Minor allele frequencies (MAF), Hardy Weinberg proportions and linkage disequilibrium (LD) among SNPs genotyped were estimated. Assuming an additive model for the SNPs, the association between each of these SNPs and transplant related mortality (TRM) at 1 year was evaluated using a proportional hazards model with disease relapse as the competing risk as described by Fine and Gray 17 after adjustment for covariates. Covariates included age at transplant as a continuous variable, race, type of donor (sibling, URD or UCB), and conditioning regimen (myeloablative or reduced intensity). Other covariates such as clinical diagnosis, disease status at transplant, gender mismatch, and CMV status at time of transplant were neither independent predictors of TRM at one year in this dataset nor did they significantly change the observed associations between SNPs and TRM at one year and were excluded from the final model. A p value ≤ 0.01 was considered to be significant for this study and individual SNPs that were significant were evaluated in a backward stepwise regression model after adjustment for clinical covariates. Only SNPs that remained significantly associated with TRM in the multivariate model were combined to create a “SNP score” in which each individual received a point for each allele associated with increased incidence of TRM (i.e.) “deleterious alleles”. We then evaluated the association between the SNP score and cumulative incidence of TRM at one year using the Fine and Gray proportional hazards model with competing risks. Cumulative incidence estimates were calculated for TRM with disease relapse as the competing risk. Incidence of TRM at 1 year was plotted by categories of the SNP score. Gray's test 18 was used to compare the incidence rates across time by categories of the SNP score.
We also evaluated the association between BER SNPs and disease relapse where TRM was used as the competing risk for disease relapse using methods described above. Covariates considered when evaluating the association between SNPs and disease relapse included age at transplant, race, disease status at transplant, clinical diagnosis, and conditioning regimen.
Results
The baseline characteristics of the study population are described in table 1. Briefly, a majority of the patients underwent myeloablative HCT from an HLA-identical sibling donor (n=231), 72% of the cohort were ≥ 21years at the time of transplant and 86% of the cohort were Caucasian. Acute leukemia (acute myeloid leukemia and acute lymphoid leukemia) (52%) was the most common diagnosis. Among 75 URD recipients, 27 (36%) of recipients received a transplant from a well matched donor, 36 (48%) from a partially matched donor and 10 (13%) from a mismatched donor. Insufficient data was available to categorize two patients. Among 99 UCB recipients, 47% were mismatched at a single locus (5/6), 43% were mismatched at 2 loci (4/6) and 10% received a 6/6 matched transplant. Cumulative incidence of TRM at 1 year was 25% (95% CI 21%-29%) and the cumulative incidence of disease relapse/progression at 2 years was 26% (95% CI 22%-30%).
Table 1.
Demographic and transplant characteristics of patients who underwent allogeneic hematopoietic stem cell transplant at the University of Minnesota: 1998-2007
| Variable | N (%) |
|---|---|
| N | 470 (100) |
| Recipient Age at Transplant (Years) | |
| <20 | 133 (28) |
| 21 – 40 | 109 (23) |
| >40 | 228 (49) |
| Median age (range), years | 35.7 (0.7 - 69.8) |
| Donor Age at Transplant (Years) | |
| <20 | 113 (24) |
| 21-40 | 121 (25) |
| >40 | 195 (42) |
| Not known | 41 (9) |
| Median age (range), years | 37.7 (0.3 - 71.7) |
| Race | |
| African American | 14(3) |
| Asian | 18(4) |
| Caucasian | 404(86) |
| Hispanic | 13(3) |
| Native American | 4(1) |
| Unknown | 10(2) |
| Mixed | 7(1) |
| Diagnosis | |
| Acute lymphoid leukemia | 92 (20) |
| Acute myeloid leukemia | 151 (31) |
| Chronic myeloid leukemia | 73 (16) |
| Lymphoma | 78 (16) |
| Myelodysplastic syndrome | 40 (9) |
| Others | 36 (8) |
| Disease Status at Transplant | |
| Complete response | 288 (62) |
| Partial response | 24(5) |
| Relapse / primary induction failure | 118 (25) |
| Myelodysplastic syndrome | 38 (8) |
| Missing | 2 (<1) |
| CMV Serological status | |
| Recipient positive / donor positive | 101 (22) |
| Recipient positive / donor negative | 146 (31) |
| Recipient negative / donor positive | 62 (13) |
| Recipient negative / donor negative | 161 (34) |
| Conditioning | |
| Myeloablative | 378 (80) |
| Reduced Intensity | 92 (20) |
| Donor Type | |
| Sibling | 296 (63) |
| Unrelated donor | 75 (16) |
| Cord blood | 99 (21) |
| Stem Cell Source | |
| Marrow | 145 (31) |
| PBSC | 226 (48) |
| Cord blood | 99 (21) |
| GVHD Prophylaxis | |
| CSA/MMF | 125 (27) |
| CSA/MTX | 244 (52) |
| T-cell depletion | 43 (9) |
| Other | 58 (12) |
| Acute Graft versus host disease | |
| No AGVHD | 234 (50) |
| Grade I | 49 (10) |
| Grade II | 121 (26) |
| Grade III/IV | 66 (14) |
| Chronic graft versus host disease | 145 (31) |
| Follow up in years (Survivors), median (range) | 3.3 (0.9-8.6) |
Abbreviations CMV: cytomegalovirus; PBSC: peripheral blood stem cell; CSA: cyclosporine; MMF: mycophenolate mofetil; MTX: methotrexate
Genotypes for these candidate SNPs were available on 98% (n=470) of all patients for whom DNA was available (n=480). Among SNPs that were successfully genotyped (n=179) 80% were tagSNPs (n=143) and 20% were functionally important SNPs (n=36). Among SNPs that could not be genotyped (n=60) 85% were tagSNPs (n=51) and 15% were functionally important SNPs (n=9). Poor completion rates (<80%) was the most common reason for inability to genotype SNPs (n=57) while genotypes for 3 SNPs were not analyzed due to poor concordance (<95%) in blinded duplicate samples. Detailed description of biological function of genes, number of SNPs genotyped within individual genes and number of samples genotyped is given in online supplement 1.
Association between BER SNPs and one year Transplant Related Mortality
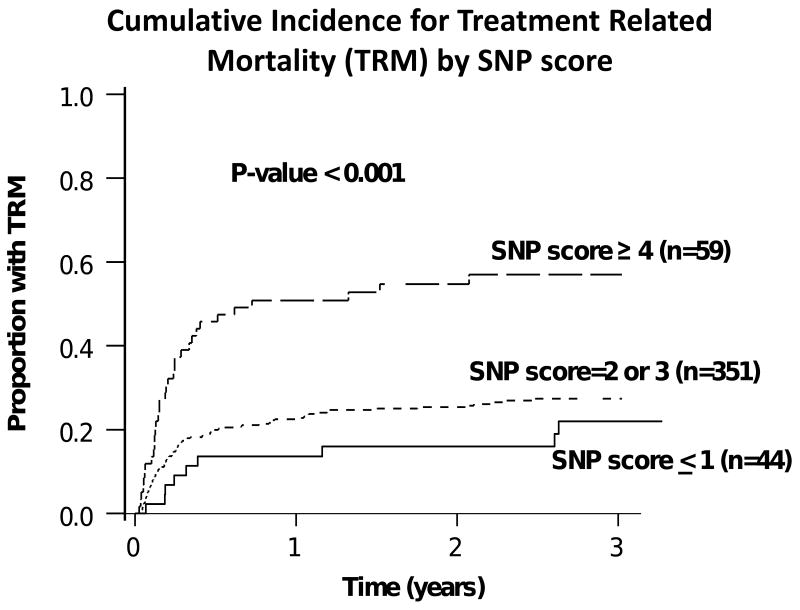
After adjustment for age at transplant, donor type, race, and conditioning regimen, 6 SNPs (rs159153, rs167715, rs2374327, rs3135974, rs3219463, rs3219476) were associated with 1 year TRM (p≤0.01) (Table 2). Three of these SNPs (rs167715, rs3219463 and 3219476) remained significantly associated with TRM when the analyses were restricted to a more homogeneous cohort of patients who underwent myeloablative allogeneic sibling transplants (Table 2). In multiple regression analysis of the entire cohort, three SNPs (rs167715, rs3135974, and rs3219463) remained significantly and independently associated with one year TRM (p≤0.01) after adjustment for other SNPs and clinical covariates (Table 3). There were no significant gene-gene interactions (all p>0.3). These three SNPs were combined to create the “SNP score” variable where each individual received 1 point for alleles that increased TRM risk in any of the three SNPs (rs167715, rs3135974, and rs3219463). Thus, individuals received 0 points if they had the T/T genotype for rs167715 or G/G genotype for SNPs rs3135974, and rs3219463, 1 point if they had T/C genotype for rs167715 or G/A genotype for SNPs rs3135974, and rs3219463 and 2 points if they had the C/C genotype for rs167715 or A/A genotype for SNPs rs3135974, and rs3219463. Thus the minimum SNP score for any given individual could be 0 and the maximum SNP score could be 6. In this study there were no individuals who were homozygous mutant for all 3 SNPs and hence the SNP score ranged from 0 to 5. The SNP score was found to be positively associated with one year TRM [HR, 95% CI): 1.84 (1.50, 2.25); p < 0.001]. The cumulative incidence of TRM at 1 year for patients with SNP score of ≥ 4 was 51% (95% CI: 37%-65%) as compared to 14% (95% CI:4%-24%) for those with SNP score ≤ 1 (p <0.001) (Figure 1). None of the interactions between the SNP score and the clinical covariates were statistically significant (all p>0.1).
Table 2.
Association between individual SNPs in BER genes and one year transplant related mortality after hematopoietic cell transplant: Univariate analysis of each SNP adjusted for clinical covariates
| SNPs | Gene (Location within gene) | Alleles | Minor allele Frequency | Hazard Ratio (95% CI); p-value | |
|---|---|---|---|---|---|
| Entire cohort* (n=470) | +Myeloablative transplants with sibling donors+ (n=231) | ||||
| rs159153 | OGG1 (5′ region near gene) | T/C | 0.33 | 1.41 (1.08, 1.85);p=0.013 | 1.28 (0.87, 1.89);p=0.211 |
| rs167715 | TDG (intron 2) | T/C | 0.10 | 0.44 (0.25, 0.78);p=0.004 | 0.34 (0.14, 0.81);p=0.015 |
| rs2374327 | TDG (intron 1) | A/T | 0.25 | 0.62 (0.45, 0.87);p=0.006 | 0.65 (0.42, 1.01);p=0.053 |
| rs3135974 | LIG3 (intron 3) | G/A | 0.10 | 1.80 (1.24, 2.59);p=0.002 | 1.63 (0.98, 2.73);p=0.061 |
| rs3219463 | MUTYH (3′ non coding region) | G/A | 0.26 | 1.62 (1.24, 2.13);p<0.001 | 1.88 (1.35, 2.62);p<0.001 |
| rs3219476 | MUTYH (3′ non coding region) | G/T | 0.34 | 1.66 (1.26, 2.18);p<0.001 | 1.70 (1.19, 2.44);p=0.004 |
Shown are SNPs significant at p ≤0.01 level in univariate analysis in the entire cohort.
This model was adjusted for age at transplant, race, type of donor and conditioning regimen.
Restricted to patients who underwent myeloablative transplants from sibling donors.
Table 3.
Association between individual SNPs in BER genes and one year transplant related mortality after hematopoietic cell transplant: multiple regression analysis
| Variables | Hazard ratio (95% CI); p value Entire cohort (n= 454) |
|---|---|
| rs167715 | 0.44 (0.25, 0.78); p=0.005 |
| rs3135974 | 1.79 (1.24, 2.60); p=0.002 |
| rs3219463 | 1.78 (1.35, 2.36); p<0.001 |
| Age at transplant | 1.05 (1.03,1.06); p<0.001 |
| Donor type | |
| Sibling (reference) | 1.00 |
| Unrelated | 1.97 (1.21, 3.18); p=0.006 |
| Cord Blood | 2.06 (1.20, 3.55); p=0.009 |
| Reduced intensity conditioning | 0.32 (0.18, 0.55); p<0.001 |
| Caucasian race | 1.25 (0.72, 2.17); p=0.419 |
None of the gene-gene interactions were significant (all p >0.3)
Figure 1.
Cumulative incidence estimates for treatment related mortality (TRM) according to different categories of SNP score (p<0.001). Disease relapse was used as the competing risk in this model. The SNPs rs167715, rs3135974, and rs3219463 were used to create the SNP score variable. Each individual received one point for each allele associated with increased incidence of TRM (i.e.) “deleterious alleles”. Since the mutant alleles for SNPs rs3135974 (A allele)and rs3219463 (A allele) were associated with increased TRM risk, individuals received 0 points for being homozygous for the wild type allele (i.e. G/G genotype), 1 point for being heterozygous (i.e. G/A genotype) and 2 points for being homozygous for the mutant allele (i.e. A/A genotype) for each of these SNPs. Since the mutant allele for rs167715 was associated with decreased TRM risk individuals received 2 points if they had the normal genotype (i.e. T/T genotype), 1 point for being heterozygous (i.e. T/C genotype) and 0 points for being homozygous for the mutant allele for rs167715 (i.e. C/C genotype).
Association between BER SNPs and disease relapse
After adjustment for age at transplant, race, disease status at transplant, diagnosis, and conditioning regimen, the GA genotype for rs3135974 in LIG3 gene was significantly associated with reduced risk of disease relapse [HR (95% CI): 0.39 (0.22, 0.69); p = 0.001] as compared to the GG genotype. The cumulative incidence of disease relapse at 2 years was 29% (95% CI: 24%-34%) for those with the GG genotype as compared to 15% (95% CI: 7%-23%) with the G/A genotype while there was no disease relapse (0%) among those with the A/A genotype (p=0.007) (Figure 2). The results were similar when the analyses were restricted to patients who underwent a myeloablative transplant from a sibling donor [HR (95% CI): 0.43 (0.19, 0.99); p=0.047]. There were no significant gene-clinical covariate interactions (all p>0.1). Specifically, there was no interaction between clinical factors such as disease status, diagnosis, conditioning regimen, recipient age at transplant; potential risk factors for disease relapse; and rs3135974 (p>0.1).
Figure 2.
Cumulative incidence estimates for disease relapse according to rs3135974 (p=0.007). Treatment related mortality was used as the competing risk in this model.
Discussion
This study found six SNPs in four BER genes to be individually associated with one year TRM and one SNP to be associated with disease relapse among patients undergoing allogeneic HCT after adjustment for clinical covariates. We also showed that increasing numbers of deleterious alleles in the BER pathway led to a higher cumulative incidence of one year TRM.
Previous studies in non-transplant populations did not find an association between specific SNPs in BER genes (rs3136820 in APEX1 gene and rs25487, rs25489, and rs1799782 in XRCC1) and treatment response to AML or NHL 5,6. One previous report examining patients undergoing autologous transplantation for multiple myeloma evaluated four SNPs in three DNA repair genes (related to the nucleotide excision repair and double strand DNA repair pathway) and found a significant association between one SNP in ERCC2 (rs13181), one SNP in XRCC3 (rs861539) and median time to treatment failure, but not overall survival 7. However, this study did not evaluate any of the SNPs evaluated in the present study 7.
This is the first study to evaluate the role of BER in predicting treatment response after allogeneic HCT. Five of the six SNPs associated with TRM were located in 3 DNA damage recognition genes (OGG1, MUTYH and TDG) indicating that variation in recognition of DNA damage may be an important determinant of the DNA repair response. The 8-oxoguanine DNA glycosylase (OGG1) and mutY homolog (MUTYH) both encode enzymes involved in oxidative DNA damage repair 19. The OGG1 gene is responsible for the excision of 8-oxoguanine, a mutagenic base byproduct which occurs as a result of exposure to reactive oxygen 19,20. The SNP, rs159153, in the OGG1 gene is located in the 5′ region of the gene upstream from the known promoter regions for this gene. The mutY homolog (MUTYH) gene excises adenine bases from the DNA backbone at sites where adenine is inappropriately paired with guanine, cytosine, or 8-oxo-7, 8-dihydroguanine, a major oxidatively damaged DNA lesion 19,20. Both SNPs, rs3219463 and rs3219476, are located in the non coding 3′ region of the MUTYH gene. The thymine-DNA glycosylase (TDG) removes thymine moieties from G/T mismatches, C/T and T/T mispairings by hydrolyzing the carbon-nitrogen bond between the sugar-phosphate backbone of DNA and the mispaired thymine 21. This enzyme plays a central role in cellular defense against genetic mutation caused by the spontaneous deamination of 5-methylcytosine and cytosine 21. Both SNPs, rs167715 and rs2374327, are located in intronic regions (introns 2 and 1 respectively) of the gene. DNA damage caused by chemo- and radiotherapeutic agents is thought to be repaired primarily by the short patch repair 4. The remaining SNP, rs3135974, located in intron 3 of the LIG3 gene, a member of the DNA ligase family that catalyzes the join of DNA ends and is involved in short patch repair 20, was associated with increased risk of TRM and decreased risk disease relapse. Though the biological relevance of rs3135974 is unknown, the association of rs3135974 with both increased TRM risk and decreased disease relapse supports the hypothesis that rs3135974 may decrease BER activity. Lower BER activity would increase damage to normal tissues, resulting in higher cell death in normal tissues and increasing the risk of TRM. In contrast, tumor cells with lower BER would not be able to efficiently repair DNA damage due to chemotherapeutic agents, leading to cell death and decreasing risk of disease relapse. None of the six SNPs found to be associated with TRM have been evaluated in previous studies.
The association between 3 SNPs, rs167715, rs3219463 and rs 3219476 and TRM, found in univariate analyses after adjustment for all clinical covariates in a broad HCT population, was also observed after restricting to a homogenous population (recipients of HLA-identical sibling donor myeloablative transplant). Strengths of this study include high quality genotyping, (only SNPs that had a concordance rate of 95% or greater using 61 pairs of blinded duplicate samples were selected for further analysis) and use of a LD tagSNP approach to comprehensively evaluate the association between genetic variation in the base excision repair pathway and treatment outcome after allogeneic HCT. Limitations of this study include the lack of information regarding the biological significance of these SNPs and inability to examine somatic mutations in BER genes within tumor cells that may also influence treatment outcomes following allogeneic HCT. To control for multiple comparisons, we have used a stringent p-value of ≤ 0.01, but we recognize that our findings need to be validated in independent datasets. Finally, TRM, itself includes several causes of death (after excluding disease relapse). Limited sample size in this study prevented subgroup analyses to evaluate to study the association of BER SNPs with individual causes of TRM.
This study provides support to the hypothesis that common genetic variants in BER are likely associated with outcomes after allogeneic HCT. If confirmed, polymorphisms in BER genes can be used as genetic biomarkers to identify groups of patients who would be at high risk for TRM and disease relapse. Pre-transplant conditioning regimens can then be modified in these high risk individuals to improve transplant outcomes.
Footnotes
Financial Disclosure Statement: The authors have no financial interests to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bredeson CN, Zhang MJ, Agovi MA, et al. Outcomes following HSCT using fludarabine, busulfan, and thymoglobulin: a matched comparison to allogeneic transplants conditioned with busulfan and cyclophosphamide. Biol Blood Marrow Transplant. 2008;14:993–1003. doi: 10.1016/j.bbmt.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canals C, Martino R, Sureda A, et al. Strategies to reduce transplant-related mortality after allogeneic stem cell transplantation in elderly patients: Comparison of reduced-intensity conditioning and unmanipulated peripheral blood stem cells vs a myeloablative regimen and CD34+ cell selection. Exp Hematol. 2003;31:1039–43. doi: 10.1016/s0301-472x(03)00223-6. [DOI] [PubMed] [Google Scholar]
- 3.Schattenberg AV, Levenga TH. Differences between the different conditioning regimens for allogeneic stem cell transplantation. Curr Opin Oncol. 2006;18:667–70. doi: 10.1097/01.cco.0000245318.90015.72. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem. 2008;15:360–7. doi: 10.2174/092986708783497328. [DOI] [PubMed] [Google Scholar]
- 5.Kuptsova N, Kopecky KJ, Godwin J, et al. Polymorphisms in DNA repair genes and therapeutic outcomes of AML patients from SWOG clinical trials. Blood. 2007;109:3936–44. doi: 10.1182/blood-2006-05-022111. [DOI] [PubMed] [Google Scholar]
- 6.Wang SS, Maurer MJ, Morton LM, et al. Polymorphisms in DNA repair and one-carbon metabolism genes and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. Leukemia. 2009;23:596–602. doi: 10.1038/leu.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vangsted A, Gimsing P, Klausen TW, et al. Polymorphisms in the genes ERCC2, XRCC3 and CD3EAP influence treatment outcome in multiple myeloma patients undergoing autologous bone marrow transplantation. Int J Cancer. 2007;120:1036–45. doi: 10.1002/ijc.22411. [DOI] [PubMed] [Google Scholar]
- 8.Robertson AB, Klungland A, Rognes T, et al. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–93. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto Y. Molecular mechanism of PCNA-dependent base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:129–38. doi: 10.1016/s0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- 10.NIEHSSNPs. NIEHS Environmental Genome Project. University of Washington; Seattle, WA: Jan, 2008. URL: http://egp.gs.washington.edu. [Google Scholar]
- 11.Weisdorf D, Spellman S, Haagenson M, et al. Classification of HLA-matching for retrospective analysis of unrelated donor transplantation: revised definitions to predict survival. Biol Blood Marrow Transplant. 2008;14:748–58. doi: 10.1016/j.bbmt.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.http://egp.gs.washington.edu/ber.html
- 13.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–46. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R D, Core, Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: Foundation for Statistical Computing; 2008. [Google Scholar]
- 17.Fine J, Gray R. A proportional hazards model for the subdistribution of a competing risk. Journal of The American Statistical Association. 1999;94:496–509. [Google Scholar]
- 18.Gray R. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Annals of Statistics. 1988;16:1140–54. [Google Scholar]
- 19.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–50. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood RD, Mitchell M, Sgouros J, et al. Human DNA repair genes. Science. 2001;291:1284–9. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 21.Cortazar D, Kunz C, Saito Y, et al. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Mehta PA, Alonzo TA, Gerbing RB, et al. XPD Lys751Gln polymorphism in the etiology and outcome of childhood acute myeloid leukemia: a Children's Oncology Group report. Blood. 2006;107:39–45. doi: 10.1182/blood-2005-06-2305. [DOI] [PubMed] [Google Scholar]