Abstract
Hypoxia is a condition of low oxygen tension occurring in the tumor and negatively correlated with the progression of the disease. We studied the gene expression profiles of nine neuroblastoma cell lines grown under hypoxic conditions to define gene signatures that characterize hypoxic neuroblastoma. The l1-l2 regularization applied to the entire transcriptome identified a single signature of 11 probesets discriminating the hypoxic state. We demonstrate that new hypoxia signatures, with similar discriminatory power, can be generated by a prior knowledge-based filtering in which a much smaller number of probesets, characterizing hypoxia-related biochemical pathways, are analyzed. l1-l2 regularization identified novel and robust hypoxia signatures within apoptosis, glycolysis, and oxidative phosphorylation Gene Ontology classes. We conclude that the filtering approach overcomes the noisy nature of the microarray data and allows generating robust signatures suitable for biomarker discovery and patients risk assessment in a fraction of computer time.
1. Background
Neuroblastoma is the most common pediatric solid tumor, deriving from immature or precursor cells of the ganglionic lineage of the sympathetic nervous system [1, 2] endowed with remarkable heterogeneity with regard to histology and clinical behavior [3, 4]. The neuroblastoma cell lines derived from the fresh tumors show various degrees of differentiation, chromosomal alterations, and morphology and consequently, a great variability in the gene expression profile. We studied the transcriptional response of neuroblastoma cell lines to hypoxia by microarray analysis [5].
Hypoxia is a condition of low oxygen tension that characterizes many pathological tissues and that is a critical determinant of tumor cell growth, susceptibility to apoptosis, and resistance to radio and chemotherapy [6–8]. The general response to hypoxia involves activation of biochemical pathways leading to alternative ways to generate energy that becomes scant in low oxygen [9]. Hypoxia modulates gene expression through the activation of several transcription factors, among which the hypoxia-inducible transcription factor-1α (HIF-1α) [7, 10], and -2α (HIF-2α) [11] are the most studied. Rapidly expanding neuroblastoma tumors present areas of hypoxia [12] and it has been reported that HIF-2α expression correlates with poor prognosis [13, 14] suggesting a central role of hypoxia in tumor progression. HIFs transactivate the hypoxia-responsive element (HRE) present in the promoter or enhancer elements of many genes encoding angiogenic, metabolic, and metastatic factors [8, 15, 16]. However, neuroblastoma cell lines respond differently to hypoxia and the nature of the modulated genes depends strongly on the type and genetic makeup of the cell [17]. Furthermore, amplification and/or overexpression of MYCN oncogene, occurring in poor prognosis tumors, influence the transcriptional response to hypoxia of neuroblastoma cell lines [5].
The identification of molecular markers capable of discriminating the hypoxic status of the tumor may result in the discovery of new risk factors for neuroblastoma patients' stratification and potential targets for tumor therapy. To this end, we were interested in identifying hypoxia signatures that discriminate the hypoxia status of neuroblastoma cell lines. Unsupervised analysis of gene expression profile could not be applied to this system because the overwhelming effect of MYCN amplification on the transcriptome masked the response to hypoxia [5]. The application of a supervised approach represented by regularization with double optimization on microarray data, an embedded feature selection technique proposed by Zou and Hastie [18] and studied by De Mol et al. [19], identified 11 probesets capable of reliably subdividing hypoxic and normoxic cell lines [5]. These results raise the question as to whether this signature is the only possible outcome of the l1-l2 regularization algorithm, and hence the only source of neuroblastoma hypoxia markers, or whether additional signatures, with similar characteristics of performances and robustness can be derived from the experimental data set. Hypoxia induces massive transcriptional changes in the cell [20–22] and it is possible that additional signatures may be found by the l1-l2 algorithm under appropriate conditions.
The l1-l2 regularization algorithm has to deal with heterogeneity of the response of each cell line and with the background noise that is enhanced by the high dimensionality of the system composed by a low number of samples (n = 18 in this work) relative to the large number of the expression values for each sample (p = 54, 613). The n ≪ p scenario is a common issue in signal processing and machine learning [23, 24]. Furthermore, the strong response of each cell line to alteration of the genetic makeup (e.g., MYCN rearrangement) tends to overcame and mask the response to hypoxia. Here, we explore the possibility that l1-l2 feature selection algorithm may generate new hypoxia signatures following prior knowledge-based data filtering techniques as a preprocessing step to feature selection.
Most dimensionality reduction methods, such as PCA and other unsupervised learning methods [25], rely only on the input data and may be driven by strong concurrent signals which are unrelated with, and somehow hide, the problem under study. Alternative strategies of data filtering are based on some form of prior knowledge of the biology of the system. The information collected by Gene Ontology (GO), a project having the aim of classifying gene products in terms of their associated biological processes, cellular and molecular components [26] can help identifying the pathways related to hypoxia and restricts the analysis to smaller sets of data.
In this paper, we demonstrate that l1-l2 regularization applied separately to probesets representing genes belonging to selected GO ontologies has the capability to generate robust hypoxia signatures, equivalent to that generated by the whole data set yielding biologically relevant information in a fraction of computer time.
2. Materials and Methods
2.1. Microarray Experiments
Microarray data were downloaded from the Gene Expression Omnibus public repository at National Center for Biotechnology Information database (accession number GSE15583). These data represent the gene expression profile of nine neuroblastoma cell lines cultured under normoxic (20% O2) or hypoxic (1% O2) conditions for 18 hours as detailed in [5], to obtain a total of 18 samples. Affymetrix HG-U133 Plus 2.0 GeneChip (Affymetrix, SantaClara, CA) were used for this study. Gene expressions were then extracted from CEL files and normalized using the Robust Multichip Average (RMA) method [27] by running an R script using the Bioconductor [28] package affy.
Comparative analysis of hypoxic relative to normoxic expression profiles for each cell line was conducted on GeneSpring 7.3 software (Agilent Technologies). Gene expression data were normalized using “per chip normalization” and “per gene normalization” algorithms implemented in GeneSpring. First, each signal was normalized based upon the median signal in that chip (“per chip normalization”). We then performed a median centering using “per gene normalization” function by which each normalized value is corrected based upon the median of the measurements for that gene in all samples. Finally, only genes that were modulated by at least 2-fold between hypoxic and normoxic cells were considered differentially expressed.
2.2. Gene Ontology
The biological groups were obtained from the literature and they were divided into three main categories depending on the biological characteristics of our experimental system (see Table 2): (i) hypoxia related groups [9, 17, 29]; (ii) MYCN related groups [30–32]; (iii) neuroblastoma related groups [31, 33, 34]. The selected functional groups were then filtered to avoid overlapped or duplicated categories and were defined according to predetermined pathways and functional categories annotated by the Gene Ontology project [26].
Table 2.
Functional gene classes.
| Biological group(1) | Functional class(2) | GO number(3) | no. of probesets(4) | error(%)(5) |
|---|---|---|---|---|
| Hypoxia | Angiogenesis | GO: 1525 | 257 | 39 |
| GO: 6915 | 1366 | |||
| Cell proliferation | GO: 8283 | 1406 | 28 | |
| DNA repair | GO: 6281 | 567 | 44 | |
| Glucose import | GO: 46323 | 10 | 89 | |
| Glucose Transport | GO: 15758 | 48 | 33 | |
| GO: 6096 | 128 | |||
| Iron ion homeostasis | GO: 6879 | 65 | 39 | |
| Notch signaling pathway | GO: 7219 | 134 | 44 | |
| GO: 6119 | 154 | |||
| Oxygen transport | GO: 15671 | 38 | 28 | |
| Regulation of pH | GO: 6885 | 41 | 72 | |
| Response to hypoxia | GO: 1666 | 32 | 28 | |
| MYCN | G1-S transition of mitotic cell cycle | GO: 82 | 62 | 50 |
| Proteasomal ubiquitin-dependent protein catabolism | GO: 43161 | 33 | 50 | |
| Protein folding | GO: 6457 | 601 | 22 | |
| Ribosome biogenesis and assembly | GO: 42254 | 170 | 28 | |
| Structural constituent of ribosome | GO: 3735 | 549 | 44 | |
| Translational elongation | GO: 6414 | 56 | 56 | |
| Translational initiation | GO: 6413 | 173 | 44 | |
| Neuroblastoma | Axon guidance | GO: 7411 | 109 | 50 |
| Axonal fasciculation | GO: 7412 | 4 | 100 | |
| Cell cycle arrest | GO: 7050 | 210 | 39 | |
| Dendrite morphogenesis | GO: 16358 | 12 | 72 | |
| Glial cell migration | GO: 8347 | 2 | 100 | |
| Inactivation of MAPK activity | GO: 188 | 50 | 44 | |
| Nervous system development | GO: 7399 | 1284 | 22 | |
| Neuron migration | GO: 1764 | 7 | 94 | |
| Positive regulation of neuron differentiation | GO: 45666 | 6 | 100 | |
| Regulation of axon extension | GO: 30516 | 14 | 100 | |
| Regulation of G-protein coupled receptor protein signaling pathway | GO: 8277 | 88 | 39 | |
| Regulation of neuronal synaptic plasticity | GO: 48168 | 8 | 100 | |
| Regulation of neurotransmitter secretion | GO: 46928 | 11 | 61 | |
| Synaptic vesicle transporter | GO: 48489 | 36 | 72 | |
| Vesicle organization and biogenesis | GO: 16050 | 6 | 44 | |
(1)Functional classes were clustered into three main biological groups depending on the characteristic of the experimental system and accordingly to the literature. (2)Defined according to the predetermined pathways and functional categories annotated by the Gene Ontology project [26]. (3) Gene Ontology ID [26]. (4)Number of probesets present in Affymetrix HG-U133 Plus 2.0 GeneChip belonging to the selected classes. (5)Leave-one-out error, as calculated by l1-l2 regularization by setting ε = 100 and frequency score = 50. *Functional classes with leave-one-out error <20%.
2.3. Supervised Methods for Gene Selection: l1-l2 Regularization
The core of our approach is the l1-l2 regularization originally presented in [18] and further developed and studied in [35, 36]. To describe such method we first fix some notation in the learning framework. Assume we are given a collection of n examples/subjects, each represented by a p-dimensional vector x of gene expressions. Each sample is associated with a binary label Y, assigning it to a class (e.g., patient or control). The dataset is therefore represented by a n × p matrix X, where p ≫ n and Y is the n-dimensional labels vector. We consider a linear model f(x) = 〈x, β〉. Note that β = β1,…, βp is a vector of weight coefficients and each probeset is associated to one coefficient. A classification rule can be then defined taking sign (f(x)) = sign (〈x, β〉). If β is sparse, that is some of its entries are zero, then some genes will not contribute in building the estimator. The estimator defined by l1-l2 regularization solves the following optimization problem:
| (1) |
where the least square error is penalized with the l1 and l2 norm of the coefficient vector. The least square term ensures fitting of the data whereas adding the two penalties allows avoiding overfitting. The relative weight of the two terms is controlled by the parameter ε. The role of the two penalties is different, the l1 term (sum of absolute values) enforces the solution to be sparse while the l2 term (sum of the squares) preserves correlation among the genes. This approach guarantees consistency of the estimator [19] and enforces the sparsity of the solution by the l1 term, while preserving correlation among input variables with the l2 term. Differently to [18] we follow the approach proposed in [36], where the solution βl1l2, computed through the simple iterative soft thresholding, is followed by regularized least squares (RLSs) to estimate the classifier on the selected features. The parameter ε in the l1-l2 regularization is fixed a priori and governs the amount of correlation. By tuning ε in (0, +∞) we obtain a one-parameter family of solutions which are all equivalent in terms of prediction accuracy, but differ on the degree of correlation among the selected features. In practice, ε has an upper bound, εmax, such that for ε > εmax selection does not change, because all correlated features were already selected with ε = εmax. By setting ε = 100, the maximal value, the maximal gene list, which is correlation aware, is obtained. Conversely, the minimal list is obtained for values of ε equal to or lower than 1.
The training for selection and classification requires the choice of the regularization parameters for both l1-l2 regularization and RLS denoted with λ* and τ*, respectively. Hence, statistical significance and model selection is performed within double-selection bias-free cross-validation loops (see [37] for details). The classification performance of the system is measured by the leave-one-out error that is the percentage of misclassified samples. In other words, leave-one-out error is equal to one minus accuracy. In order to assess a common list of probesets, it is necessary to choose an appropriate criterion [38]. We based ours on the frequency, that is, we decided to promote as relevant variables the most stable probesets across the lists. The complete validation framework comprising the l1-l2 regularization is implemented in MATLAB code.
2.4. Correlation Analysis
The correlation among the probesets selected by the l1-l2 algorithm was performed as previously described in [39]. Briefly, we build blocks of correlated probesets using a variation of well-known agglomerative clustering techniques based on Pearson distance. We first examine the minimal list, which genes are clustered via hierarchical clustering with correlation distance and average linkage. Since no objective algorithm, other than heuristics, is available for establishing the number of clusters, for each GO class the cut of the hierarchical graph determining the number of clusters is chosen following visual examination of the graph. In particular we set the cut at 0.75 of the maximum linkage value in the dendrogram. For each GO class the cut of the hierarchical graph determining the number of clusters is chosen following visual examination of the graph. Each probeset in the maximal list is then assigned to the cluster which average correlation with the given probeset is the highest. In this way we populate the clusters built from the minimal list with all the probesets coming from the maximal list. The correlation analysis was performed using MATLAB Statistic Toolbox.
2.5. HRE Analysis
We mapped the HRE elements in the promoter regions of the genes represented in the Affymetrix HG-U133 Plus 2.0 GeneChip. We downloaded the annotation file for the HG-U133 Plus 2.0 from NetAffx Analysis Center (http://www.affymetrix.com/) and the dataset was restricted to the known mRNA sequences listed in the Ensembl database V56 [40]. The regulatory regions were retrieved from Ensembl database using Ensembl Perl APIs. We operationally defined as ‘‘promoter” the first 2,000 base pairs upstream the transcription initiation site and generated a dataset containing the promoters of the genes coding for the mRNAs spotted on the chip. The HRE matrix has been obtained from 69 experimental validated human HRE sequences [41] with MatDefine tool (Genomatix Software GmbH). HRE consensus elements [(G|C|A|T) (C|G|T|A) (G|C|A|T) (T|G|C|A) (A|G) (CGTG) (C|G|T|A) (G|C|A|T) (G|C|T|A) (C|G|T|A)] were searched in the promoter sequences with MatInspector software (Genomatix) with core similarity = 1 and optimized matrix similarity. About 33% of the promoters contain at least one HRE consensus element. χ2 was used to evaluate the significance of the HRE frequency in the promoter regions of genes belonging to the different signatures. P < .01 was considered significant.
3. Results and Discussion
We studied nine neuroblastoma cell lines [2] heterogeneous with respect to MYCN amplification and morphology (Table 1). The cell lines were cultured under normoxic and hypoxic conditions for 18 hours and the total RNA was tested for gene expression profiling using the Affymetrix HG-U133 Plus 2.0 platform. The response to hypoxia of each individual cell line was first analyzed by measuring the fold change as the ratio of the expression level between hypoxic and normoxic samples. We found that the response of each neuroblastoma cell line to hypoxia is characterized by a high number of modulated genes ranging from 855 to 1609 for the upregulated and from 758 to 1317 for the downregulated probesets (Table 1). However, the modulated genes changed from cell line to cell line (data not shown) and only the application of a strong feature selection technique, represented by the l1-l2 regularization, allowed to identify a single signature of 11 probesets (All-chip signature) discriminating the normoxic and the hypoxic status [5].
Table 1.
Modulation of gene expression by hypoxia in neuroblastoma cell lines.
| cell line | gene expression(1) | |||
|---|---|---|---|---|
| name | Morphology(2) | MYCN amplification(3) | up-regulated | down-regulated |
| ACN | neuroblast (N) | − | 1400 | 1317 |
| SHEP-2 | epithelial (S) | − | 1609 | 1043 |
| GI-ME-N | neuroblast (N) | − | 762 | 881 |
| SK-N-F1 | epithelial (S) | − | 1206 | 1051 |
| SK-N-SH | neuroblast/epithelial (I) | − | 922 | 758 |
| SK-N-BE(2)c | neuroblast/epithelial (I) | + | 855 | 1273 |
| IMR-32 | neuroblast (N) | + | 1000 | 1077 |
| LAN-1 | neuroblast (N) | + | 1061 | 1016 |
| GI-LI-N | neuroblast (N) | + | 1516 | 1002 |
The large amount of hypoxia-modulated genes suggests that additional hypoxia signatures may be identified if we reduce the background noise of the system. To this end, we applied a data filtering strategy based on prior knowledge. We restricted our analysis to the genes known to be involved in the hypoxic response on the bases of our reading of the literature and comprised in the biological processes according to the Gene Ontology (GO) classification [13]. The selection of the GO classes was based on the reports of hypoxia modulated genes without attempting to distinguish the various cell types under investigation. 13 biological processes that are involved in hypoxia response were selected (Table 2). We reasoned that this approach would restrict the analysis to the probesets that have a high impact on the hypoxic response while potentially eliminating the noisy features. To explore the potential interference from MYCN status in the classification process, we selected and tested 7 biological processes involved in MYCN activity (Table 2). Finally, we selected a third group of GO processes related to the neuroblastoma biology as a control. For each of the 38 classes shown in Table 2, the l1-l2 algorithm selected a list of hypoxia discriminating probesets and calculated the corresponding classification leave-one-out error. The output of the l1-l2 regularization algorithm depends on the parameter ε that governs the amount of correlation allowed among the probesets. We set ε = 100, the maximal value, to obtain the most comprehensive signature maximizing the number of correlated probesets to be included in the output [5].
The validation has been performed by leave-one-out cross-validation on the 18 samples. The 18 cross-validation loops produced 18 lists of probesets. Then, a unique list is obtained as the union of the probesets included in the 18 lists, with a frequency score calculated as the frequency of each probeset in the 18 lists generated by the cross validation loops. Stable probesets were defined as those characterized by a frequency score equal to, or higher than, 50% as previously reported in [5]. The use of cross validation allows the selection protocol to generate an unbiased and objective output [42] beyond the theoretical results that guarantee the robustness of the core algorithm [19]. The discriminatory power of the probeset lists is represented by the classification performance. A leave-one-out error of 20% was chosen as the cutoff level for the classification performance. The leave-one-out error of the All-chip signature is 17% [5].
The only classes characterized by a list of selected probesets capable of generating a leave one-out error lower than the 20% cutoff were apoptosis (17%), glycolysis (11%), and oxidative phosphorylation (11%) (Table 2), all of them belonging to the hypoxia biological group. These results demonstrate that, within each of the above classes, there is a list of probesets capable of discriminating the condition of the cell lines thereby defining three new neuroblastoma hypoxia signatures, named apoptosis signature, glycolisis signature, and oxidative phosphorylation signature. As expected, there were no GO classes belonging to the MYCN or neuroblastoma biological groups that generated hypoxia signatures, supporting the validity of our choice of hypoxia-related GO functional classes. Although MYCN represents a strong signal that drives major transcriptome difference in neuroblastoma cell lines [5], our results show that there are no enough discriminatory genes in the MYCN-related processes. These results demonstrate that the feature selection method applied is capable of revealing the differences occurring among hypoxic and normoxic neuroblastoma cell lines by filtering out strong competing signals, such as MYCN amplification status.
The list of the probesets comprising the 11 probesets (All-chip signature) [5] and the newly identified signatures is shown in Table 3 and consists of 10 probesets for apoptosis signature, 3 for glycolysis-signature, and 32 for the oxidative phosphorylation signature. The new signatures highlight 41 probesets that were not previously included in the All-chip signature and contribute to the discrimination of the hypoxic status. Furthermore, the 32 probesets of the oxidative phosphorylation signature does not overlap with the All-chip signature, demonstrating that the increased resolution generated by data filtering allows the identification of previously discarded relevant GO processes. The hypoxia signatures present in the literature show different sizes and gene composition [9, 43–46]. Since different cell types respond heterogeneously to hypoxia by modulating different set of genes, we decided to compare our results with the published hypoxic gene signatures obtained from neuroblastoma cell lines [47] (Table 4). In order to make the comparison feasible, the probesets constituting our signatures have been collapsed to gene symbol. The overlapping genes are underlined in bold in Table 4. While important differences among the signatures exist, the comparison highlights a general consistency. In fact, an overlap is present in All-chip (3/8 genes), apoptosis (2/4 genes), and glycolysis (2/2 genes) signatures. Interestingly, there is no overlap (0/24 genes) among the results published by Jögi et al. [47] and the oxidative phosphorylation signature.
Table 3.
Hypoxia signatures generated after data filtering.
| Signatures | ||||||
|---|---|---|---|---|---|---|
| Probeset(1) | Gene Name | GeneBank(2) | Apo(3) | Gly(3) | OxP(3) | All(3) |
| 201848_s_at | BNIP3 | U15174 | 100 | — | — | 100 |
| 201849_at | BNIP3 | NM_004052 | 83 | — | — | 100 |
| 210512_s_at | VEGF | AF022375 | 78 | — | — | 100 |
| 211527_x_at | VEGF | M27281 | 61 | — | — | — |
| 212171_x_at | VEGF | H95344 | 61 | — | — | — |
| 219232_s_at | EGLN3 | NM_022073 | 61 | — | — | — |
| 210513_s_at | VEGF | AF091352 | 56 | — | — | — |
| 221478_at | BNIP3L | AL132665 | 56 | — | — | — |
| 221479_s_at | BNIP3L | AF060922 | 56 | — | — | — |
| 222847_s_at | EGLN3 | AI378406 | 56 | — | — | — |
| 202022_at | ALDOC | NM_005165 | — | 100 | — | 100 |
| 1558365_at | PGK1 | AK055928 | — | 72 | — | — |
| 228483_s_at | PGK1 | BE856250 | — | 72 | — | — |
| 208972_s_at | ATP5G1 | AF100741 | — | — | 100 | — |
| 222270_at | SMEK2 | BF509069 | — | — | 100 | — |
| 1554847_at | ATP6V1B1 | AY039759 | — | — | 94 | — |
| 218201_at | NDUFB2 | NM_004546 | — | — | 94 | — |
| 203189_s_at | NDUFS8 | NM_005006 | — | — | 89 | — |
| 203371_s_at | NDUFB3 | NM_002496 | — | — | 89 | — |
| 218200_s_at | NDUFB2 | NM_013387 | — | — | 89 | — |
| 203606_at | NDUFS6 | NM_002494 | — | — | 83 | — |
| 204125_at | NDUFAF1 | NM_001687 | — | — | 83 | — |
| 214241_at | NDUFB8 | BE043477 | — | — | 78 | — |
| 230598_at | KIAA1387 | AI742966 | — | — | 78 | — |
| 203190_at | NDUFS8 | NM_002496 | — | — | 72 | — |
| 207335_x_at | ATP5I | NM_006294 | — | — | 72 | — |
| 208745_at | ATP5L | AF092131 | — | — | 72 | — |
| 203039_s_at | NDUFS1 | NM_021074 | — | — | 67 | — |
| 203613_s_at | NDUFB6 | NM_004553 | — | — | 67 | — |
| 208746_x_at | ATP5L | AA917672 | — | — | 67 | — |
| 210453_x_at | ATP5L | U33833 | — | — | 67 | — |
| 211752_s_at | NDUFS7 | AL050277 | — | — | 67 | — |
| 228816_at | ATP6AP1L | AU153583 | — | — | 67 | — |
| 207573_x_at | ATP5L | NM_005176 | — | — | 61 | — |
| 226616_s_at | NDUFV3 | AW241758 | — | — | 61 | — |
| 200096_s_at | ATP6V0E | BC005876 | — | — | 56 | — |
| 214923_at | ATP6V1D | AV717561 | — | — | 56 | — |
| 226209_at | NDUFV3 | BC006215 | — | — | 56 | — |
| 200078_s_at | ATP6V0B | BC035703 | — | — | 50 | — |
| 210206_s_at | DDX11 | AF061735 | — | — | 50 | — |
| 213378_s_at | DDX11 | AV711183 | — | — | 50 | — |
| 214244_s_at | ATP6V0E | AA723057 | — | — | 50 | — |
| 218190_s_at | UCRC | NM_004549 | — | — | 50 | — |
| 241755_at | UQCRC2 | BE467348 | — | — | 50 | — |
| 243498_at | ATP5J | BG010493 | — | — | 50 | — |
| 202887_s_at | DDIT4 | NM_019058 | — | — | — | 94 |
| 223193_x_at | E2IG5 | AF201944 | — | — | — | 94 |
| 224345_x_at | E2IG5 | AF107495 | — | — | — | 89 |
| 225342_at | AK3L1 | AK026966 | — | — | — | 78 |
| 226452_at | PDK1 | AU146532 | — | — | — | 78 |
| 236180_at | — | W57613 | — | — | — | 61 |
| 235850_at | WDR5B | BF434228 | — | — | — | 50 |
(1)Probeset ID according to Affymetrix HG-U133 Plus 2.0 GeneChip. (2)GenBank mRNA accession number. (3)Frequency score as calculated by l1-l2 regularization for the selected probesets in the hypoxia signatures compared to the All-Chip signature. Apo: apoptosis; Gly: glycolisis; OxP: oxidative phosphorylation; All: All-chip.
Table 4.
Hypoxia gene signatures overlap and HRE analysis.
| Signature(1) | Gene Name(2) | HRE(3) |
|---|---|---|
| Apoptosis | BNIP3 | 9 |
| BNIP3L | 5 | |
| EGLN3 | 3 | |
| VEGF | 4 | |
| Glycolysis | ALDOC | 3 |
| PGK1 | 1 | |
| Oxydative Phosphorylation | ATP5G1 | 2 |
| ATP5I | 5 | |
| ATP5L | 0 | |
| ATP6V0B | 6 | |
| ATP6V0E | 2 | |
| ATP6V0E | 2 | |
| ATP6V1B1 | 0 | |
| ATP6V1D | 1 | |
| DDX11 | 1 | |
| LOC92270 | 3 | |
| NDUFAF1 | 1 | |
| NDUFB2 | 1 | |
| NDUFB3 | 1 | |
| NDUFB6 | 1 | |
| NDUFB8 | 2 | |
| NDUFS1 | 3 | |
| NDUFS6 | 9 | |
| NDUFS7 | 2 | |
| NDUFS8 | 3 | |
| NDUFS8 | 3 | |
| NDUFV3 | 7 | |
| SMEK2 | 3 | |
| UCRC | 2 | |
| UQCRC2 | 3 | |
| All-chip | AK3L1 | 4 |
| ALDOC | 3 | |
| BNIP3 | 9 | |
| DDIT4 | 1 | |
| E2IG5 | 5 | |
| PDK1 | 4 | |
| VEGF | 4 | |
| WDR5B | 3 | |
(1)Hypoxia gene signatures. (2)Multiple probesets were collapsed to single genes. The genes overlapping with Jögi et al. hypoxia signature are underlined in bold. (3)Number of HRE sequences found in the promoter region.
About 33% of the genes spotted on the chip present a HRE sequence in the promoter region. We investigated whether there was enrichment in HRE containing promoter in the genes composing our signatures. We found that all the signatures are significantly enriched (P < .01) in genes containing HRE (Table 4). In particular, all the genes included in All-chip, apoptosis, and glycolysis signatures contain at least one HRE, while HRE containing genes constitute 91% of the oxidative phosphorylation signature. These results support the idea that our signatures are associated with the hypoxia status.
The whole signature, rather than individual genes, is important for discriminating the hypoxic status. For example, VEGF is a gene whose expression is strongly related to hypoxia [45] and is part of the apoptosis and angiogenesis classes, both of which are part of the hypoxia biological group. However, the contribution of VEGF probesets is not sufficient to reach the discriminatory power required to generate a significant signature out of the angiogenesis class as opposed to the apoptosis class.
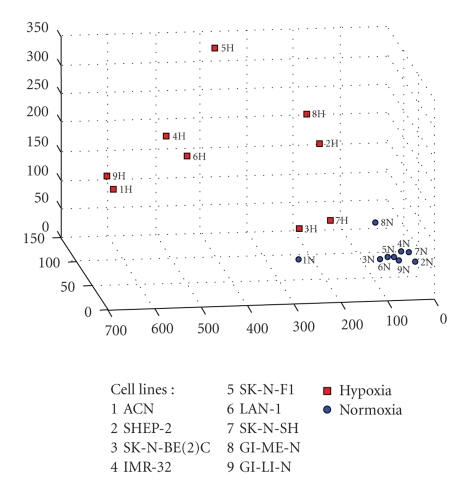
The strong discriminatory power of the signatures can be visualized by a 3-dimensional representation of the probesets projected on their 3 principal components. l1-l2 algorithm produces a multigene model but the multidimensional representation can be well approximated by the tridimensional picture when the number of probesets is not too large. Figure 1 depicts the separation of probesets belonging to the glycolysis and shows that the two classes of normoxic and hypoxic cell lines are clearly separated in the multidimensional space.
Figure 1.
Three-dimensional representation of the principal components analysis of the Glycolysis GO process. Principal components representation of the multivariate analysis performed on the 9 cell lines by l1-l2 algorithm. This figure illustrates a 3-dimensional visualization of the dataset restricted to the selected probesets projected on their 3 principal components. Red squares (H) represent the cell lines in hypoxic status and the blue circles (N) the corresponding cell lines in normoxic status. The numbers indicate the cell lines.
In conclusion, we demonstrate that, upon data reduction the l1-l2 algorithm can identify new hypoxia signatures that have equivalent discriminatory power relative to that obtained by the analysis of the whole transcriptome. From the computational stand point, this process allows to reduce the computer time by approximately 10 times, from days to hours with an average machine, facilitating the analysis of the data. Finally, it is important to highlight the possibility of applying our method to different experimental settings by choosing appropriate selection of GO processes.
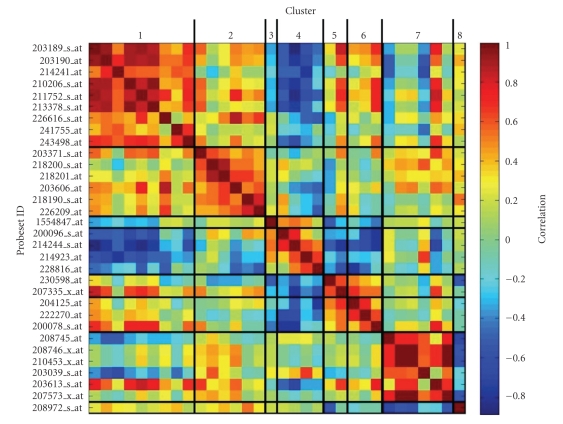
Our prior knowledge-based method produces nested lists of relevant probesets but does not highlight the correlation among them [39], and it should be completed by a postprocessing step depicting the correlation structure. The correlation within the oxidative phosphorylation signature is shown in Figure 2. We computed a distance matrix based on the expression values of the probesets and subdivided it into 8 modules by hierarchical clustering. These modules represent subgroups of correlated probesets that are positively or negatively associated to the hypoxic status. This information is important to pick the correct probesets in order to assess the expression of these markers in the in vivo setting. Furthermore, these data lend themselves to the tuning of the ε parameter that is part of the l1-l2 algorithm.
Figure 2.
Symmetrical heatmap of the correlation analysis for oxidative phosphorylation-signature probesets. The correlation values among the 32 probesets selected by l1-l2 algorithm (ε = 100) for oxidative phosphorylation process are reported. The probesets, named accordingly to Affymetrix HG-U133 Plus 2.0 GeneChip platform, have been subdivided into 8 clusters according to the similarity among their correlation profiles by hierarchical clustering. Cluster ID is indicated at the top of the figure and correlation scale is reported on the right side.
The output of the l1-l2 regularization algorithm depends on the free parameter ε that governs the amount of correlation allowed among the probesets and selects the amount of probesets to be included in the signature. By setting ε = 100, the maximal value, we can obtain a comprehensive signature more descriptive of the biology of the system. By setting ε = 1, we can obtain an equally discriminating signature with fewer genes thereby more effective in identifying critical biomarkers for diagnostic applications [5]. We analyzed the effects of tuning ε on the oxidative phosphorylation signature. The results are shown in Table 5, where the probesets selected by l1-l2 regularization with both ε = 1 and ε = 100 for oxidative phosphorylation are listed. The results demonstrated that the reduction in ε is associated with a smaller signature (from 32 to 16 probesets) as expected by the fact that correlated probesets tend to be discarded.
Table 5.
Tuning the oxidative phosphorylation signature by changing the ε parameter.
| Cluster(1) | Probeset(2) | Gene Name | ε = 1(3) | ε = 100(4) |
|---|---|---|---|---|
| 1 | 203189_s_at | NDUFS8 | 89 | 89 |
| 203190_at | NDUFS8 | 56 | 72 | |
| 214241_at | NDUFB8 | 94 | 78 | |
| 210206_s_at | DDX11 | — | 50 | |
| 211752_s_at | NDUFS7 | — | 67 | |
| 213378_s_at | DDX11 | — | 50 | |
| 226616_s_at | NDUFV3 | — | 61 | |
| 241755_at | UQCRC2 | — | 50 | |
| 243498_at | ATP5J | — | 50 | |
| 2 | 203371_s_at | NDUFB3 | 56 | 89 |
| 218200_s_at | NDUFB2 | 72 | 89 | |
| 218201_at | NDUFB2 | 89 | 94 | |
| 203606_at | NDUFS6 | — | 83 | |
| 218190_s_at | UCRC | — | 50 | |
| 226209_at | NDUFV3 | — | 56 | |
| 3 | 1554847_at | ATP6V1B1 | 100 | 94 |
| 4 | 200096_s_at | ATP6V0E | 50 | 56 |
| 214244_s_at | ATP6V0E | 72 | 50 | |
| 214923_at | ATP6V1D | — | 56 | |
| 228816_at | ATP6AP1L | — | 67 | |
| 5 | 230598_at | SMEK2 | 72 | 78 |
| 207335_x_at | ATP5I | — | 72 | |
| 6 | 204125_at | NDUFAF1 | 67 | 83 |
| 222270_at | SMEK2 | 100 | 100 | |
| 200078_s_at | ATP6V0B | — | 50 | |
| 7 | 208745_at | ATP5L | 89 | 72 |
| 208746_x_at | ATP5L | 72 | 67 | |
| 210453_x_at | ATP5L | 50 | 67 | |
| 203039_s_at | NDUFS1 | — | 67 | |
| 203613_s_at | NDUFB6 | — | 67 | |
| 207573_x_at | ATP5L | — | 61 | |
| 8 | 208972_s_at | ATP5G1 | 100 | 100 |
(1)Cluster number according to Figure 2. (2)Probeset ID according to Affymetrix HG-U133 Plus 2.0 GeneChip. (3)Frequency score (%) as calculated by l1-l2 regularization for the selected probesets by setting ε = 1. (4)Frequency score (%) as calculated by l1-l2 regularization for the selected probesets by setting ε = 100.
4. Conclusions
The identification of signatures discriminating the hypoxic status of the tumor cell may be important for our understanding of the biology of neuroblastoma tumors and for the stratification of the patients in risk groups. One way to generate a robust and reliable hypoxic signature is the application of a supervised approach represented by l1-l2 regularization that generates an 11 probesets signature discriminating the hypoxic status of our panel of nine neuroblastoma cell lines.
Here, we demonstrate that l1-l2 feature selection algorithm generates new and robust hypoxia signatures following prior knowledge-based data filtering techniques as a preprocessing to feature selection. These new signatures have the same discriminatory power as that generated by the whole data set and yield biologically relevant information in a fraction of computer time.
The data filtering is based upon the use of the prior information contained in GO and the literature, and it allows restricting the analysis to smaller data sets. This process filters out not only many noisy probesets but also the probesets selected from the all-chip analysis whose strong relation with hypoxia hid some weaker but important genes. l1-l2 regularization algorithm following data filtering selects probesets that were not the first chosen when all the probesets were considered. The prior knowledge utilized in setting up the filter, comes from the current literature from which we derived the molecular pathways that are important for the response of the cell to the hypoxic environment. These pathways were gathered in the hypoxia biological group. Interestingly, the new signatures were found only in this group and not in other collections of GO pathways like those related to the effects of the MYCN oncogene or to the neuroblastoma biology. In general, the identification of the GO classes related to the phenomenon under investigation may be an empirical, but effective way to target the potential source of signatures to be fed to the l1-l2 regularization. We speculate that this approach could be used to address questions that go beyond the hypoxic status and may find signatures characterizing other pathophysiological situations provided that there is a relevant cellular model and there are sufficient insights in the underlying molecular mechanisms.
The nested structure of the selected gene lists allows the choice of the desired level of complexity, which is the magnitude of signature, maintaining all the information extracted from the data. For example, the minimal list may be preferable when interested in finding biomarkers to be used on large-scale diagnostic tests due to potential constrains on time, cost, and resources.
Finally, working on a limited number of probesets has a major impact on the computational time required for the analysis that changes from days to hours, thereby allowing more leeway to the study of the dataset.
Acknowledgment
This paper was supported by Fondazione Italiana per la Lotta al Neuroblastoma, the Italian Association for Cancer Research (AIRC), the Italian Health Ministry, the EU Integrated Project Health-e-Child IST-2004-027749 and Compagnia di San Paolo Project 4998- ID/CV 2007.0887.
References
- 1.De Preter K, Vandesompele J, Heimann P, et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biology. 2006;7(9, article R84) doi: 10.1186/gb-2006-7-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maris J, Hogarty M, Bagatell R, Cohn S. Neuroblastoma. The Lancet. 2007;369(9579):2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Thiele CJ. Neuroblastoma. In: Master JRW, Palsson B, editors. Human Cell Culture. London, UK: Kluwer Academic; 1999. pp. 21–22. [Google Scholar]
- 4.Weinstein JL, Katzenstein HM, Cohn SL. Advances in the diagnosis and treatment of neuroblastoma. Oncologist. 2003;8(3):278–292. doi: 10.1634/theoncologist.8-3-278. [DOI] [PubMed] [Google Scholar]
- 5.Fardin P, Barla A, Mosci S, Rosasco L, Verri A, Varesio L. The l1 -l2 regularization framework unmasks the hypoxia signature hidden in the transcriptome of a set of heterogeneous neuroblastoma cell lines. BMC Genomics. 2009;10:p. 474. doi: 10.1186/1471-2164-10-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. Targeting HIF-1 for cancer therapy. Nature Reviews Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends in Molecular Medicine. 2002;8(4):S62–S67. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 8.Carmeliet P, Dor Y, Herber J-M, et al. Role of HIF-1α in hypoxiamediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 9.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nature Reviews Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 10.Carta L, Pastorino S, Melillo G, Bosco MC, Massazza S, Varesio L. Engineering of macrophages to produce IFN-γ in response to hypoxia. Journal of Immunology. 2001;166(9):5374–5380. doi: 10.4049/jimmunol.166.9.5374. [DOI] [PubMed] [Google Scholar]
- 11.Talks KL, Turley H, Gatter KC, et al. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. American Journal of Pathology. 2000;157(2):411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. The New England Journal of Medicine. 1999;341(16):1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 13.Jögi A, Øra I, Nilsson H, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmquist-Mengelbier L, Fredlund E, Löfstedt T, et al. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10(5):413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 15.Melillo G, Musso T, Sica A, Taylor LS, Cox GW, Varesio L. A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. Journal of Experimental Medicine. 1995;182(6):1683–1693. doi: 10.1084/jem.182.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melillo G, Sausville EA, Cloud K, Lahusen T, Varesio L, Senderowicz AM. Flavopiridol, a protein kinase inhibitor, down-regulates hypoxic induction of vascular endothelial growth factor expression in human monocytes. Cancer Research. 1999;59(21):5433–5437. [PubMed] [Google Scholar]
- 17.Fredlund E, Ovenberger M, Borg Å, Påhlman S. Transcriptional adaptation of neuroblastoma cells to hypoxia. Biochemical and Biophysical Research Communications. 2008;366(4):1054–1060. doi: 10.1016/j.bbrc.2007.12.074. [DOI] [PubMed] [Google Scholar]
- 18.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society. Series B. 2005;67(2):301–320. [Google Scholar]
- 19.De Mol C, De Vito E, Rosasco L. Elastic-net regularization in learning theory. Journal of Complexity. 2009;25(2):201–230. [Google Scholar]
- 20.Bosco MC, Puppo M, Santangelo C, et al. Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. Journal of Immunology. 2006;177(3):1941–1955. doi: 10.4049/jimmunol.177.3.1941. [DOI] [PubMed] [Google Scholar]
- 21.Ricciardi A, Elia AR, Cappello P, et al. Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Molecular Cancer Research. 2008;6(2):175–185. doi: 10.1158/1541-7786.MCR-07-0391. [DOI] [PubMed] [Google Scholar]
- 22.Chi J-T, Wang Z, Nuyten DSA, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Medicine. 2006;3(3):395–409. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Candès EJ, Romberg J, Tao T. Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Transactions on Information Theory. 2006;52(2):489–509. [Google Scholar]
- 24.Donoho DL. For most large underdetermined systems of equations, the minimal l1-norm near-solution approximates the sparsest near-solution. Communications on Pure and Applied Mathematics. 2006;59(7):907–934. [Google Scholar]
- 25.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York, NY, USA: Springer; 2001. [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31(4, article e15) doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Development Core Team. R: A Language and Environment for Statistical. R Foundation for Statistical Computing; 2004. [Google Scholar]
- 29.Patiar S, Harris AL. Role of hypoxia-inducible factor-1α as a cancer therapy target. Endocrine-Related Cancer. 2006;13(1):S61–S75. doi: 10.1677/erc.1.01290. [DOI] [PubMed] [Google Scholar]
- 30.Boon K, Caron HN, Van Asperen R, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO Journal. 2001;20(6):1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warnat P, Oberthuer A, Fischer M, Westermann F, Eils R, Brors B. Cross-study analysis of gene expression data for intermediate neuroblastoma identifies two biological subtypes. BMC Cancer. 2007;7:p. 89. doi: 10.1186/1471-2407-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell E, Lunec J, Tweddle DA. Cell cycle regulation targets of MYCN identified by gene expression microarrays. Cell Cycle. 2007;6(10):1249–1256. doi: 10.4161/cc.6.10.4222. [DOI] [PubMed] [Google Scholar]
- 33.Asgharzadeh S, Pique-Regi R, Sposto R, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. Journal of the National Cancer Institute. 2006;98(17):1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 34.Fischer M, Oberthuer A, Brors B, et al. Differential expression of neuronal genes defines subtypes of disseminated neuroblastoma with favorable and unfavorable outcome. Clinical Cancer Research. 2006;12(17):5118–5128. doi: 10.1158/1078-0432.CCR-06-0985. [DOI] [PubMed] [Google Scholar]
- 35.Destrero A, Mosci S, De Mol C, Verri A, Odone F. Feature selection for high-dimensional data. Computational Management Science. 2009;6(1):25–40. [Google Scholar]
- 36.De Mol C, Mosci S, Traskine M, Verri A. A regularized method for selecting nested groups of relevant genes from microarray data. Journal of Computational Biology. 2009;16(5):677–690. doi: 10.1089/cmb.2008.0171. [DOI] [PubMed] [Google Scholar]
- 37.Barla A, Mosci S, Rosasco L, Verri A. A method for robust variable selection with significance assessment. In: Proceedings of the European Symposium on Artificial Neural Networks (ESANN ’08); April 2008; Bruges, Belgium. [Google Scholar]
- 38.Jurman G, Merler S, Barla A, Paoli S, Galea A, Furlanello C. Algebraic stability indicators for ranked lists in molecular profiling. Bioinformatics. 2008;24(2):258–264. doi: 10.1093/bioinformatics/btm550. [DOI] [PubMed] [Google Scholar]
- 39.Mosci S, Verri A, Barla A, Rosasco L. Finding structured gene signatures. In: Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW ’08); November 2008; Philadelphia, Pa, USA. pp. 158–165. [Google Scholar]
- 40.Hubbard TJP, Aken BL, Ayling S, et al. Ensembl 2009. Nucleic Acids Research. 2009;37(supplement 1):D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Science’s STKE. 2005;2005(306, article re12) doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 42.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mense SM, Sengupta A, Zhou M, et al. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiological Genomics. 2006;25(3):435–449. doi: 10.1152/physiolgenomics.00315.2005. [DOI] [PubMed] [Google Scholar]
- 44.Kim H, Lee D-K, Choi J-W, Kim J-S, Park SC, Youn H-D. Analysis of the effect of aging on the response to hypoxia by cDNA microarray. Mechanisms of Ageing and Development. 2003;124(8-9):941–949. doi: 10.1016/s0047-6374(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Zhang W, Kondo K, et al. Gene expression profiling in a renal cell carcinoma cell line: dissecting VHL and hypoxia-dependent pathways. Molecular Cancer Research. 2003;1(6):453–462. [PubMed] [Google Scholar]
- 46.Manalo DJ, Rowan A, Lavoie T, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 47.Jögi A, Vallon-Christersson J, Holmquist L, Axelson H, Borg Å, Påhlman S. Human neuroblastoma cells exposed to hypoxia: induction of genes associated with growth, survival, and aggressive behavior. Experimental Cell Research. 2004;295(2):469–487. doi: 10.1016/j.yexcr.2004.01.013. [DOI] [PubMed] [Google Scholar]