Abstract
Many tumor cells express globally reduced levels of microRNAs (miRNAs), suggesting that decreased miRNA expression in pre-malignant cells contributes to their tumorigenic phenotype. In support of this, Dicer, an RNAse III-like enzyme that controls maturation of miRNA, was recently shown to function as a haploinsufficient tumor suppressor in non-hematopoietic cells. Since the Myc oncoprotein, a critical inducer of B cell lymphomas, was reported to suppress the expression of multiple miRNAs in lymphoma cells, it was presumed that a deficiency of Dicer and subsequent loss of miRNA maturation would accelerate Myc-induced lymphoma development. We report here that, surprisingly, a haploinsufficiency of Dicer in B cells failed to promote B cell malignancy or accelerate Myc-induced B cell lymphomagenesis in mice. Moreover, deletion of Dicer in B cells of CD19-cre+/Eμ-myc mice significantly inhibited lymphomagenesis, and all lymphomas that did arise in these mice lacked functional Cre expression and retained at least one functional Dicer allele. Uncharacteristically, the lymphomas that frequently developed in the CD19-cre+/Dicerfl/fl/Eμ-myc mice were of very early precursor B cell origin, a stage of B cell development prior to Cre expression. Therefore, loss of Dicer function was not advantageous for lymphomagenesis, but rather, Dicer ablation was strongly selected against during Myc-induced B cell lymphoma development. Moreover, deletion of Dicer in established B cell lymphomas resulted in apoptosis, revealing that Dicer is required for B cell lymphoma survival. Thus, Dicer does not function as a haploinsufficient tumor suppressor in B cells and is required for B cell lymphoma development and survival.
Keywords: dicer, myc, lymphoma, B cell
Introduction
Dicer is an RNAse III-like enzyme that processes pre-miRNAs into mature miRNAs. miRNAs are linked to multiple biological processes, including differentiation, apoptosis, and proliferation, that are important for transformation and tumor development (1) Deletion of Dicer in multiple organisms, including mice, is lethal (2), highlighting the importance of Dicer in embryogenesis. However, it has been reported that a global decrease of miRNAs occurs in multiple tumor types (3), implying that decreased miRNA expression contributes to tumorigenesis. Moreover, Jacks and colleagues reported that suppression of Dicer with shRNA or deletion of Dicer increased the growth potential of carcinoma cell lines and oncogenic Ras-induced transformation of murine lung epithelial cells, respectively (4). However, Dicer was recently shown to be a haploinsufficient tumor suppressor in lung epithelial cells and in the retina (5, 6). Heterozygous germline mutations in DICER were also found in a rare pediatric lung tumor (7). In contrast, deletion of Dicer results in growth arrest and p53-dependent senescence in primary fibroblasts and cell death of lung epithelial cells (8, 9). Moreover, Dicer hypomorphic mice have not been reported to have an increased incidence of cancer (10). Therefore, the role Dicer has in tumorigenesis remains unclear.
The oncogene c-Myc, which is frequently overexpressed in human and murine cancers, including B cell lymphomas, was reported to suppress the expression of multiple miRNAs in B cell lymphomas (11). Yet, Myc induces the expression of the microRNA polycistron miR-17∼92 and the miR-106a cluster (12), and constitutive expression of the miR-17∼92 polycistron accelerates Myc-induced B cell lymphoma development (13). Therefore, the role of miRNAs in Myc-induced B cell lymphoma development is unresolved. Recently, deletion of Dicer in very early progenitor B cells by Mb1-cre was shown to result in precursor B cell apoptosis and B cell developmental defects (14). To determine if global loss of miRNA expression would impact B cells that are further differentiated, and more importantly whether Dicer deletion would contribute to Myc-induced B cell lymphoma development, we utilized conditional Dicer knockout mice and CD19-cre and Eμ-myc transgenic mice, which develop pre-B/B cell lymphomas. We observed a small decrease in the numbers of B cells in CD19-cre/Dicerfl/fl mice regardless of Myc status and a significant delay in B cell lymphoma development in CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice. Interestingly, early precursor B cell lymphomas emerged in 40% of the CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice, and all lymphomas regardless of stage of differentiation retained one allele of Dicer due to loss of functional Cre. Loss of one allele of Dicer did not affect B cell development or Myc-induced lymphomagenesis. Deletion of both alleles of Dicer in established B cell lymphomas induced apoptosis. Therefore, we have demonstrated that Dicer is required for B cell lymphomagenesis and the survival of B cell lymphoma that have developed. In addition our data shows that Dicer is not a haploinsufficient tumor suppressor in B cells.
Materials and Methods
Mice
Eμ-myc transgenic mice (15) and CD19-cre (16) mice were mated and then F1s were bred to Dicerfl/fl mice that have loxP sites flanking exons 15-17 (8). F2s were bred to obtain Dicerfl/fl, Dicer+/fl, and Dicer+/+ CD19-cre positive and negative and Eμ-myc positive and negative offspring. All analyses were performed with littermates. Only CD19-cre positive hemizygous mice were used and evaluated in these studies. All mice were carefully monitored and at signs of disease, tumors/tissues were collected and analyzed. Tissues/cells prior to disease were also collected and evaluated. Statistical significance of the survival between the different genotypes of Eμ-myc transgenic mice was determined by log-rank test. All research with mice complied with federal and state guidelines and was approved by the Vanderbilt IACUC committee.
Western and Southern blotting
B cell lymphomas were lysed and proteins Western blotted as previously described (17). Membranes were probed with antibodies specific for Cre (Novagen), Mdm2 (C18, Santa Cruz), p53 (Ab-7; Calbiochem, La Jolla, CA), p19ARF (GeneTex, San Antonio, TX), and β-actin (Sigma). Genomic DNA from B cell lymphomas was Southern blotted for p53 and ARF, and p53 was sequenced as previously described (18, 19).
Phenotype Analysis
Freshly isolated lymphoma cells from CD19-cre+/Eμ-myc mice with none, one, or two floxed Dicer alleles, whole spleens and bone marrow from CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice prior to the development of lymphoma and non-Eμ-myc and/or non-CD19-cre littermates were analyzed by flow cytometry following staining with fluorescent antibodies as previously described (17, 18). Flow cytometry data were evaluated with CellQuest and/or FlowJo software.
Quantitative real-time RT-PCR
Total RNA was isolated from murine cells and tissue with Trizol (InVitrogen) as per manufacture's protocol. Total RNA was further purified with an RNeasy kit (Qiagen). cDNA was generated as previously reported (20). Sequences for β-actin, Dicer, Cre, and CD19-specific primer pairs were obtained from the Primer Bank (Harvard Medical School) and synthesized by Eurofins MWG Operon. Quantitative real-time PCR was performed with SybrGreen (SABiosciences) in triplicate as previously reported (20). The data are expressed in 2-deltaCt using β-actin as a reference. Taqman RT-PCR for miRNA used TaqMan microRNA Assay (Applied BioSystems) in triplicate and compared to RNU6b small RNA expression.
Dicer Gene Rearrangement Analysis
Genomic DNA was isolated using the REDExtract-N-Amp Tissue PCR Kit (Sigma) from lymphomas frozen immediately after harvesting and lymphomas grown in culture. Lymphomas from mice were greater than 85% pure. PCR analysis of DNA was performed under conditions that allow for 15% contaminating normal cells without detection of the unrearranged floxed allele. Primers for detecting unrearranged Dicer alleles have been previously published (8). Primers for detecting Cre-lox deleted Dicer alleles were CCATTGGTGCCAAGACAATG and CAGGCTCCACTCCCTAAC.
Lymphoma Cell Survival Analysis
Isolated primary Dicerfl/fl/Eμ-myc transgenic lymphoma cells were infected with empty pBabe or CreERT2 encoding pBabe retrovirus (21). Infected cells were selected with puromycin. Dicer was deleted in the cells by activating CreERT2 with 1 μM 4-hydroxytamoxifen. Cell numbers and cell viability following activation of CreERT2 was determined by Trypan Blue Dye exclusion assay. Apoptosis as measured by fragmented (sub-G1) DNA was quantified following propidium iodide (PI) staining and flow cytometry, and further verified by Annexin V-FITC and flow cytometry.
Results
Loss of Dicer inhibits B cell lymphoma development
Dicer has been reported to be a haploinsufficient tumor suppressor in non-hematopoietic cells (5, 6). To determine whether Dicer functions as a haploinsufficient tumor suppressor in B cells, we generated single allele conditional Dicer knockout mice (Dicer+/fl) that were transgenic for cre recombinase, which was placed under the transcriptional control of one allele of the endogenous CD19 locus (8, 15, 16, 22). Over a year of observation, B cell malignancies did not emerge in CD19-cre+/Dicer+/fl mice and their survival was similar to that of CD19-cre-/Dicer+/fl and CD19-cre+/Dicer+/+ littermates (data not shown). CD19-cre+/Dicerfl/fl mice also did not have an increased incidence of B cell cancers and had a survival analogous to that of CD19-cre+/Dicer+/fl and CD19-cre+/Dicer+/+ mice. Therefore, loss of one allele of Dicer alone is insufficient to initiate B cell lymphoma.
To test the idea that Dicer is a haploinsufficient tumor suppressor further and to determine whether loss of one allele of Dicer would cooperate with Myc overexpression to promote tumorigenesis in B cells in a similar manner as it did with Ras overexpression in lung epithelial cells (4, 5), we generated CD19-cre+/Dicerfl/fl mice transgenic for Myc (Eμ-myc). Eμ-myc transgenic mice overexpress c-Myc in B cells starting at the pre-B cell stage of development (22). CD19 expression, and thus Cre expression, in CD19-cre mice occurs at the pro-B cell stage and continues as B cell mature (16, 23). Dicer heterozygous floxed CD19-cre+/Eμ-myc mice developed lymphoma and had a survival similar to that of CD19-cre+/Dicer+/+/Eμ-myc transgenics (Fig. 1A), indicating Dicer does not have a haploinsufficiency effect on lymphoma latency or overall survival. However, evaluation of lymphoma development in CD19-cre+/Dicerfl/fl/Eμ-myc mice revealed a significant delay in lymphomagenesis and an extended survival, compared to CD19-cre-/Dicerfl/fl/Eμ-myc (Fig. 1A), CD19-cre+/Dicer+/+/Eμ-myc (Fig. 1C), and CD19-cre-/Dicer+/fl/Eμ-myc littermates (Fig. 1B) (p=0.0001 log-rank tests). The mean survival of CD19-cre+/Dicerfl/fl/Eμ-myc mice was 351 days compared to 204 days for CD19-cre-/Dicerfl/fl/Eμ-myc littermates and 194 days for CD19-cre+/Dicer+/+/Eμ-myc mice. Therefore, CD19-cre+/Eμ-myc mice with both alleles of Dicer floxed did not have an acceleration of lymphomagenesis; instead, they had a significantly protracted rate of lymphoma development. Our results show that a Dicer haploinsufficiency does not cooperate with Myc overexpression in B cells, and that loss of both alleles of Dicer in B cells inhibits Myc-induced B cell lymphoma development.
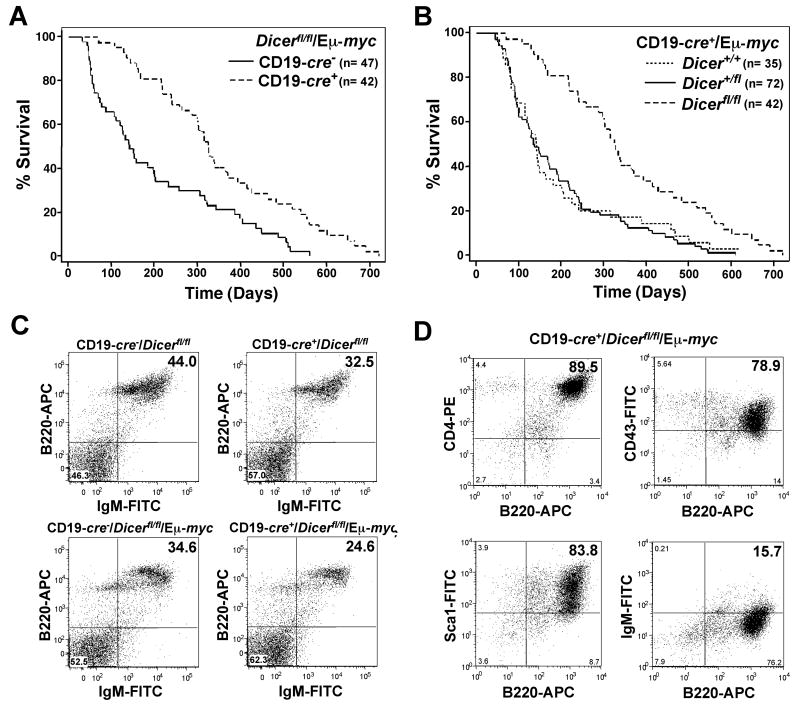
Figure 1. CD19-cre+/Dicerfl/fl/Eμ-myc mice have a protracted rate of lymphomagenesis, decreased precursor B cell numbers, and develop an altered type of lymphoma.
Kaplan-Meier survival curves. (A) CD19-cre+/Dicerfl/fl/Eμ-myc transgenic and CD19-cre-/Dicerfl/fl/Eμ-myc transgenic littermates; p=0.0001 (log-rank test). (B) CD19-cre+/Eμ-myc transgenic littermates with none, one, or two floxed Dicer alleles; p=0.0001 (log-rank test). The number (n) of mice in each group is denoted. (C) Representative dot plots of splenic B cells from the indicated mice (n=6 or 7 littermate matched pairs of each genotype). B220-APC versus IgM-FITC gated on total lymphocytes. (D) Dot plots of lymphoma cells from a CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mouse expressing markers of early precursor B cells. All plots are gated on total lymphocytes. Quadrants were set with fluorochrome-linked isotype controls.
Two floxed alleles of Dicer alter the type of B cell lymphoma that develops
Recently it was shown that deletion of Dicer at a very early stage of B cell development with Mb1-cre led to an almost complete ablation of precursor B cells and subsequently differentiated B cells (14). We evaluated splenic B cells from CD19-cre+/Dicerfl/fl/Eμ-myc mice prior to the development of lymphoma to determine whether mature B cells were present. Wild-type Eμ-myc transgenic mice have a reduced number (approximately 10%) of mature B cells (24), and loss of Dicer further decreased the number of mature B cells present. Specifically, there was a significant decrease in the percentage (and decreased total numbers) of B220+/IgM+ B cells in pre-cancerous CD19-cre+/Dicerfl/fl/Eμ-myc spleens compared to spleens in CD19-cre-/Dicerfl/fl/Eμ-myc littermates (Fig 1C). Comparing seven littermate matched pairs, the mean percentage of B220+/IgM+ B cells in pre-cancerous CD19-cre+/Dicerfl/fl/Eμ-myc spleens was 23.0% ±2.01 and in CD19-cre-/Dicerfl/fl/Eμ-myc littermates was 31.3% ±2.48 (p<0.0001, paired t-test). A comparable decrease in B cells due to Dicer deletion was detected in CD19-cre+/Dicerfl/fl mice in the absence of the Myc transgene (Fig. 1C). Six littermate matched pairs showed a mean of 33.8% ±4.34 B220+/IgM+ B cells in CD19-cre+/Dicerfl/fl mice and 43.3% ±4.64 B220+/IgM+ B cells in CD19-cre-/Dicerfl/fl mice (p=0.0015, paired t-test). No difference in B cell numbers or development was detected in CD19-cre+/Dicer+/fl or CD19-cre+/Dicer+/fl/Eμ-myc mice (data not shown). These results indicate that the CD19-cre+/Dicerfl/fl mice, which express Cre later in B cell development, had only a small reduction (8.3% mean decrease) in their number of B cells, in contrast to the Mb1-cre+/Dicerfl/fl mice.
Since deletion of Dicer in CD19-cre+/Dicerfl/fl and CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice slightly altered B cell development, we sought to determine whether loss of Dicer altered the typical pre-B/B cell lymphomas that arise in Eμ-myc transgenic mice (15, 22). Flow cytometric analysis on freshly isolated tumors was performed. Fourteen of 23 (61%) of the lymphomas analyzed from CD19-cre+/Dicerfl/fl/Eμ-myc mice were typical pre-B and/or B cell lymphomas that expressed B220 and were either IgM- or IgM+, which are characteristic of Eμ-myc lymphomas (22). Surprisingly, 9 of 23 (39%) of the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas analyzed expressed markers characteristic of very early progenitor B cells (B220+, CD4+, CD43+, Sca1+), but lacked markers of differentiated lymphocytes IgM and CD3 (Fig. 1D), which are not observed in wild-type Eμ-myc transgenic mice. However, these early precursor B cell lymphomas were previously detected in Eμ-Bcl2/Eμ-myc double transgenic mice (25). Thus, a significant number of very early precursor B cell lymphomas emerged in CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice, rather than the characteristic pre-B/B cell lymphomas.
Increased frequency of p53 deletions in Dicerfl/fl CD19-Cre+Eμ-myc lymphomas
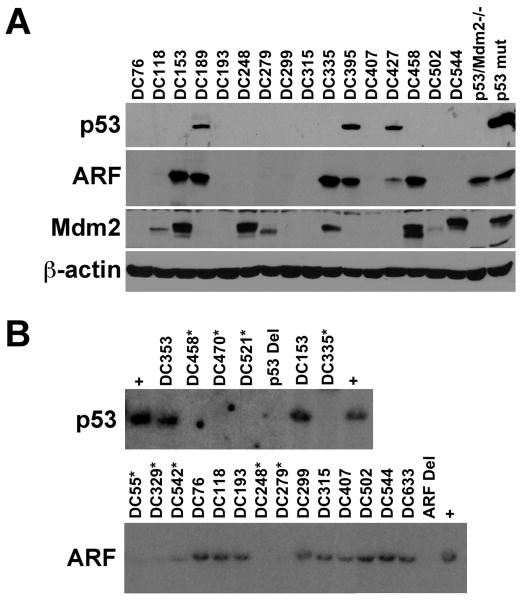
Myc-induced B cell lymphomagenesis proceeds, in part, through inactivation of the ARF-Mdm2-p53 tumor suppressor pathway (17). We analyzed CD19-cre+/Dicerfl/fl/Eμ-myc transgenic B cell lymphomas by Western and Southern blot to determine the frequency of alterations in ARF, Mdm2, and p53 (Fig. 2). Lymphomas that lacked the tumor suppressor ARF protein (Fig. 2A, 16 lymphomas shown out of 25 total analyzed) were subjected to ARF Southern blot analysis. ARF deletions were evident in 20% (5 of 25) of the lymphomas analyzed from CD19-Cre+/Dicerfl/fl/Eμ-myc mice (Fig. 2B), similar to the percentage of ARF deletions in wild-type Eμ-myc lymphomas (17). Mdm2, a negative regulator of p53, was overexpressed in half of the CD19-Cre+/Dicerfl/fl/Eμ-myc lymphomas (Fig. 2A and data not shown), as is typical for wild-type Eμ-myc mice (17). Mutation of the tumor suppressor p53, which is usually evident as increased levels of p53 protein (Fig. 2A), occurs in a quarter of the B cell lymphomas that arise in Eμ-myc mice (17). Similarly, point mutations in p53 were detected in 24% (6 of 25) of the lymphomas from CD19-cre+/Dicerfl/fl/Eμ-myc transgenics. However, p53 deletions were detected in 16% (4 of 25) of the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas (Fig. 2B), whereas deletion of p53 is a rare event, occurring in 3-4% of lymphomas in Eμ-myc mice (17, 18). Mutation or deletion of p53 was detected in early precursor B cell lymphomas and in more mature B cell lymphomas, suggesting that inactivation of p53 does not appear to correlate to the stage of B cell differentiation of the lymphoma in these mice. Together the p53 mutations and deletions were present in 40% of the lymphomas in CD19-cre+/Dicerfl/fl/Eμ-myc mice, indicating there was an increased selection for p53 inactivation in these lymphomas. An evaluation of p53 alterations in lymphomas from CD19-cre+/Eμ-myc mice that were Dicer+/fl revealed a normal frequency of p53 mutations (3 of 11, 27%) and deletions (0 of 11) (data not shown), suggesting that there was no increase in selective pressure for p53 inactivation in CD19-cre+Eμ-myc lymphomas with only one floxed Dicer allele. Therefore, p53 deletion occurred at an increased frequency in CD19-cre+/Eμ-myc lymphomas arising in mice with two floxed Dicer alleles, suggesting that there is increased stress in and/or around the developing B cells in these mice.
Figure 2. Increased frequency of p53 deletions in CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas.
(A) Protein lysates of lymphomas from CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice were subjected to Western blot analysis for p53, ARF, Mdm2, and β-actin. Protein lysates from p53/Mdm2-double null MEFs and a B cell lymphoma containing mutant p53 were used as controls. (B) Southern blots for p53 and ARF of lymphomas from CD19-cre+/Dicerfl/fl/Eμ-myc transgenic mice. Asterisks denote lymphomas that have deleted p53 and ARF. Lymphomas that contain p53 or ARF (plus signs) or that have deleted p53 (p53 Del) or ARF (ARF Del) are denoted.
Loss of Dicer is selected against during B cell lymphomagenesis
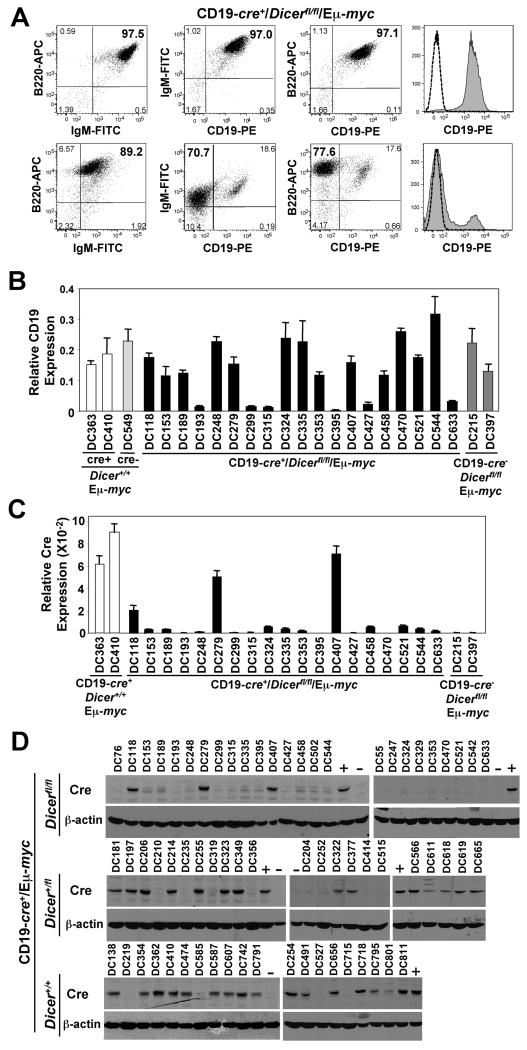
Unexpectedly, 65% (15 of 23) of the lymphomas analyzed that arose in CD19-cre+/Dicerfl/fl/Eμ-myc mice lacked or had reduced expression of CD19, a B cell marker, at their cell surface, as determined by flow cytometry. For example, typical IgM+/B220+/CD19+ (or IgM-/B220+/CD19+) B cell lymphomas were detected in 35% of CD19-cre+/Dicerfl/fl/Eμ-myc mice (Fig. 3A). In contrast, there were CD19-cre+/Dicerfl/fl/Eμ-myc mice that had an IgM+ (or IgM-)/B220+/CD19- (or CD19low) lymphoma (Fig. 3A), which is uncharacteristic for lymphomas in Eμ-myc transgenic mice. Quantitative real-time PCR (qRT-PCR) showed that CD19 mRNA levels were significantly reduced in the lymphomas where cell surface CD19 was absent or very low (Fig. 3B). Since cre is knocked into the CD19 locus and cell surface CD19 was expressed at reduced levels in the majority of the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas, we questioned whether cre expression was also being affected in these lymphomas. Evaluation of cre by qRT-PCR revealed that 89% (17 of 19) of the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas analyzed showed significantly reduced or absent cre mRNA (Fig. 3C). The lymphomas that lacked or had barely detectable cre mRNA did not express Cre protein (Fig. 3D). Only 12% (3 of 25) of the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas analyzed expressed Cre protein. There was a significant difference in Cre protein expression between CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas and lymphomas from Dicer+/fl/CD19-cre+/Eμ-myc and Dicer+/+/CD19-Cre+/Eμ-myc mice (p<0.0005, chi-squared test). Interestingly, the number of floxed Dicer alleles appears to dictate the frequency of loss of Cre protein expression. Specifically, 64% (14 of 22) of Dicer+/fl/CD19-cre+/Eμ-myc lymphomas expressed Cre protein, whereas 80% (16 of 20) of the Dicer+/+/CD19-Cre+/Eμ-myc lymphomas expressed Cre protein (Fig. 3D). Therefore, loss of Cre expression in the lymphomas correlated to the number of floxed Dicer alleles and was a frequent event in CD19-cre+/Dicerfl/fl/Eμ-myc transgenic lymphomas.
Figure 3. Decreased CD19 expression and frequent loss of Cre expression in CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas.
(A) Dot plots of CD19, B220, and IgM expression in two CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas. Quadrants were set with isotype controls for each lymphoma. For histograms, CD19 expression is grey and the isotype control for each is indicted with a dotted line. (B & C) qRT-PCR for CD19 (B) and Cre (C) expression relative to β-actin expression in lymphomas from the indicated genotype. (D) Western blots for Cre and β-actin in protein lysates of CD19-cre+/Eμ-myc lymphomas with none, one, or two floxed Dicer alleles. Protein lysates from lymphomas from CD19-cre+/Dicer+/+/Eμ-myc lymphomas (+) and CD19-cre-/Dicerfl/fl/Eμ-myc lymphomas (-) are indicated.
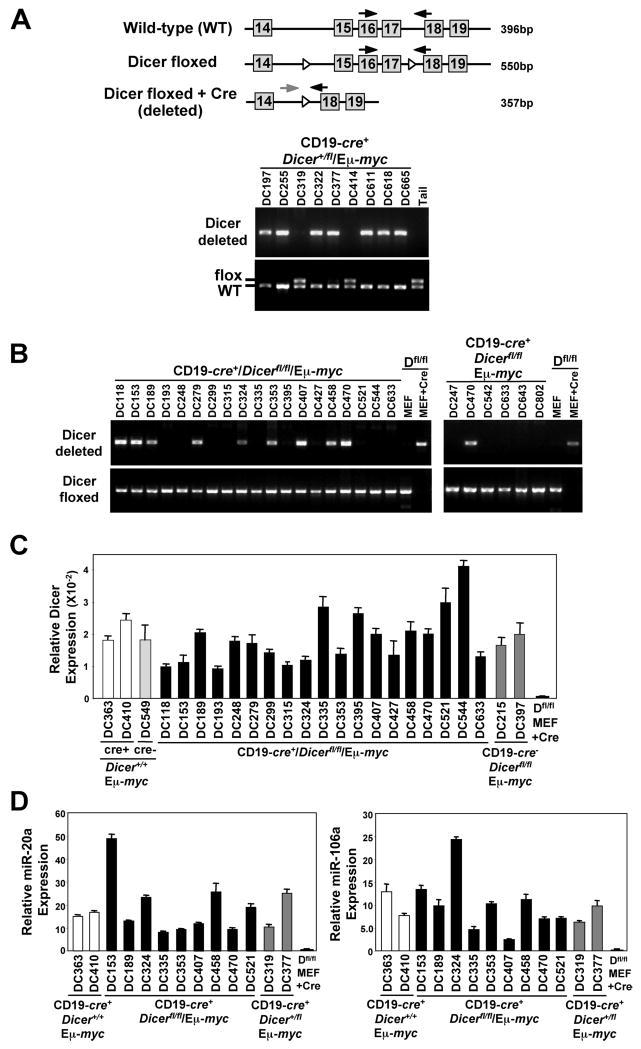
To determine whether functional Cre protein was ever expressed in the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas that lacked Cre protein, and whether the Cre protein expressed in the three lymphomas (DC118, DC279, DC407) was functional, we assessed by PCR genomic DNA for the presence of the region in Dicer that is flanked by loxP sites. First we evaluated lymphomas from CD19-cre+/Dicer+/fl/Eμ-myc lymphomas and observed that 83% (20 of 24) had Cre-lox deletion of their one floxed Dicer allele (Fig. 4A and data not shown), consistent with Hobeika et al. reporting an 80% deletion efficiency in B cells of CD19-cre mice (26). However, not a single lymphoma from a CD19-cre+/Dicerfl/fl/Eμ-myc mouse was found to have deleted both conditional alleles of Dicer (Fig. 4B). Only one conditional allele of Dicer had been deleted in 38% (10 of 26) of the lymphomas from CD19-cre+/Dicerfl/fl/Eμ-myc mice, including the three lymphomas that expressed Cre protein (Figure 4B and data not shown). No deletion of either floxed Dicer allele was detected in 62% (16 of 26) of the lymphomas from CD19-cre+/Dicerfl/fl/Eμ-myc mice. Moreover, all of the CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas analyzed expressed Dicer mRNA (Figure 4C) and miRNAs known to be expressed in B cells (Figure 4D and data not shown), indicating that Dicer was retained and functional in all of the lymphomas. The levels of miRNAs expressed varied between different CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas and the miRNA evaluated, but were similar to controls. These data illustrate that lymphomas do not develop when both alleles of Dicer are deleted, but can emerge when one allele has been deleted. To test this concept further, we evaluated freshly isolated CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas that had been placed into short-term culture and allowed to grow to derive pure populations of lymphoma cells. Again, not a single CD19-cre+/Dicerfl/fl/Eμ-myc lymphoma was detected that had deleted both floxed alleles of Dicer (Figure 4B, right panels). Only 17% (1 of 6) of the cultured lymphomas had deleted one allele of Dicer, whereas the rest had retained both alleles of Dicer. Therefore, loss of Dicer appears to be strongly selected against during B cell lymphoma development in CD19-cre+/Dicerfl/fl/Eμ-myc mice, suggesting that Dicer expression is required for B cell transformation.
Figure 4. CD19-cre+/Dicerfl/fl/Eμ-myc transgenic lymphomas retain at least one Dicer allele.
(A) Schematic of the Dicer locus with loxP sites denoted with open arrowheads. Location of the sites of primer pairs used to detect a region of Dicer by PCR are indicated with arrows. Size in base pairs (bp) of the PCR products from the specific primers is indicated. (A & B) Detection of conditional deleted and floxed Dicer alleles by PCR from genomic DNA from frozen CD19-cre+/Dicer+/fl/Eμ-myc lymphomas (A), frozen CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas (B, left panel), or cultured CD19-cre+/Dicerfl/fl/Eμ-myc lymphomas (B, right panel). Genomic DNA from the tail of a Dicer+/fl mouse with a floxed Dicer allele is a control (A). Genomic DNA from Dicerfl/fl (Dfl/fl) mouse embryo fibroblasts (MEF) with or without Cre recombinase serves as controls for Dicer conditional deletion (B). (C) qRT-PCR for Dicer in individual Eμ-myc lymphomas of the indicated genotype. A Dicerfl/fl (Dfl/fl) MEF expressing Cre recombinase serves as a control for Dicer conditional deletion. (C) Taqman qRT-PCR for miR-20a or miR-106a in the indicated Eμ-myc lymphomas (other miRNA also evaluated with similar results). A Dicerfl/fl (Dfl/fl) MEF expressing Cre recombinase serves as a control for loss of miRNA expression.
Dicer expression is required for B cell lymphoma survival
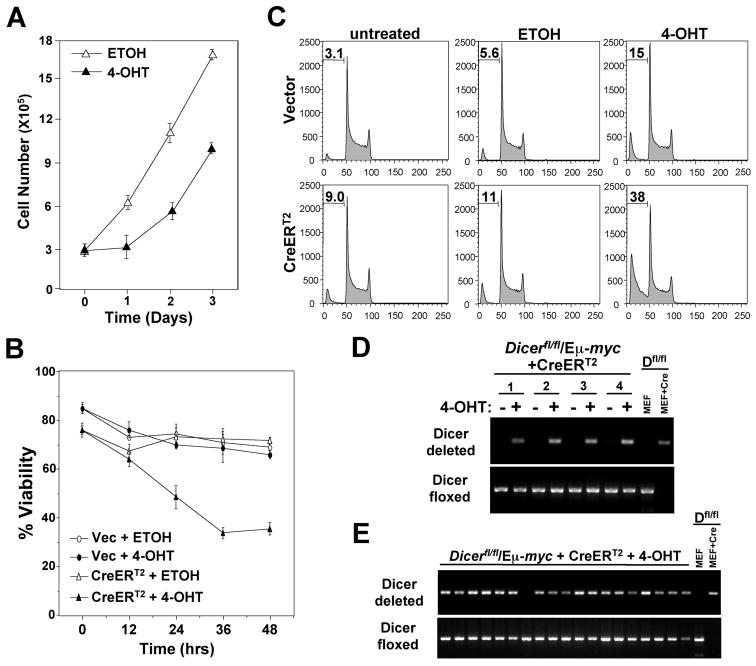
Previously, Jacks and colleagues had reported that suppression of Dicer expression with shRNA resulted in increased proliferation and transformation potential of established epithelial cancer cell lines (4). To determine whether Dicer loss alters the survival or growth of already established B cell lymphomas, lymphomas were isolated from Dicerfl/fl/Eμ-myc mice and infected with a 4-hydroxytamoxifen (4-OHT) inducible Cre retrovirus (CreERT2) (21). Following selection of infected lymphoma cells with puromycin, CreERT2 was activated with 4-OHT. For all cultures of Dicerfl/fl/Eμ-myc lymphomas, the activation of CreERT2 resulted in the induction of apoptosis, whereas addition of vehicle control (ethanol, ETOH) or addition of 4-OHT to empty retroviral infected lymphomas had little effect. The numbers of viable lymphoma cells decreased and the percentage of cells containing fragmented DNA or that were Annexin V positive increased within 24 hours post 4-OHT addition (Fig. 5A-5C and Table 1), indicating that the lymphoma cells were undergoing apoptosis. However, in all cases, 24-48 hours following 4-OHT, surviving lymphoma cells did grow out of the cultures; these cells had similar doubling times and proliferated at a rate analogous to those treated with vehicle control (Fig. 5A and data not shown). Additionally, there were analogous percentages of CreERT2 or empty retroviral expressing lymphoma cells that incorporated BrdU 48 and 72 hours following 4-OHT treatment (data not shown). As expected, PCR analysis showed that the population of lymphoma cells that survived CreERT2 activation had only deleted one allele of Dicer (Fig. 5D).
Figure 5. Dicer loss induces apoptosis of established B cell lymphomas.
(A-E) Lymphomas from Dicerfl/fl/Eμ-myc transgenic mice infected with CreERT2 encoding retrovirus or an empty retrovirus (Vec). 4-OHT or vehicle control (ETOH) was added to lymphoma cell cultures at day or time 0 and cell number (A), viability (B), apoptosis (percentage of cells in subG1 indicated, C), and Dicer gene rearrangement (D) were evaluated at intervals. A, B, and C contain representative data from a minimum of three independent experiments. For D, data for four independent Dicerfl/fl/Eμ-myc CreERT2 expressing lymphomas are shown. (E) Dicer rearrangement PCR of FACS sorted single lymphoma clones that emerged following 4-OHT activation of CreERT2. Nineteen of 43 total clones analyzed are shown. For D and E, a Dicerfl/fl (Dfl/fl) MEF with or without Cre recombinase serves as a control for Dicer deletion.
Table 1. Deletion of Dicer induces apoptosis.
percentage of Annexin V positive primary lymphoma cells
To further test whether a lymphoma cell could survive without Dicer, we placed a single Dicerfl/fl/Eμ-myc lymphoma cell infected with CreERT2 into wells of 96 well plates, added 4-OHT to activate CreERT2, and evaluated the clones that emerged. Only 22% (43 of 192) of the clones survived CreERT2 activation, whereas 98% of the vehicle (ETOH) treated clones survived. Of the 43 Dicerfl/fl/Eμ-myc lymphoma clones that survived 4-OHT treatment, none had deleted both alleles of Dicer (Fig. 5E and data not shown). Forty of the 43 clones had deleted one allele of Dicer, whereas the remaining three clones had retained both alleles of Dicer. These data indicate that deletion of both alleles of Dicer is not advantageous for B cell lymphomas, but instead induces apoptosis. Therefore, one allele of Dicer is required for B cell lymphoma survival. In addition, loss of a single allele of Dicer did not confer a proliferative advantage to B cell lymphomas over those that retained both alleles of Dicer, but did allow cell survival.
Discussion
The role miRNAs and Dicer have in cellular processes necessary for tumorigenesis is incompletely understood. It has been reported that Myc suppresses the expression of many miRNAs, and some tumor cells express globally lower levels of miRNAs or miRNA processing enzymes (3, 4, 11, 27). These studies contributed to the hypothesis that decreased miRNA expression enhances cellular transformation. However, our data here shows that in B cells, deletion of Dicer, which would block the formation of mature miRNAs, did not facilitate Myc-mediated B cell lymphomagenesis. There was no cooperation between loss of Dicer and Myc overexpression in B cell proliferation or tumorigenesis. In fact, in pre-cancerous CD19-cre+/Dicerfl/fl/Eμ-myc mice, there was no detectable increase in the numbers of B cells, which would be indicative of an enhanced proliferative process; instead there were reduced numbers of B cells. Moreover, CD19-cre+/Dicerfl/fl/Eμ-myc mice had a protracted rate of lymphoma development of which the decrease in B cells may have contributed. Importantly, all lymphomas that arose in CD19-cre+/Dicerfl/fl/Eμ-myc mice retained at least one allele of Dicer. The retention of Dicer by a developing B cell is likely to have allowed that B cell to survive (14). The lack or loss of Cre expression would permit a B cell to retain one or both alleles of Dicer, resulting in an increased pool of precursor B cells that do not express Cre and consequently, an increased frequency of lymphomas that lack Cre expression and that have retained Dicer. In support of this idea, 39% of the lymphomas that arose in CD19-cre+/Dicerfl/fl/Eμ-myc mice were early precursor B cell lymphomas that due to their stage of maturation did not yet express CD19, and therefore did not express Cre. Other lymphomas that were more differentiated and should have expressed CD19 appeared to have never expressed Cre or expressed Cre that was only able to delete one allele of Dicer. Consequently, all lymphomas analyzed from CD19-cre+/Dicerfl/fl/Eμ-myc mice, including mature B cell lymphomas, retained at least one allele of Dicer and may or may not have expressed CD19. It is likely the increased stress caused by the altered B cell differentiation in the CD19-cre+/Dicerfl/fl/Eμ-myc mice accounts for the increase in p53 inactivation in the lymphomas that arise. Our results provide strong evidence that loss of Dicer is not advantageous, but instead strongly selected against during Myc-mediated B cell lymphoma development.
Studies with mouse models have shown that Dicer appears to be a haploinsufficient tumor suppressor in lung epithelial, muscle, and retinal cells, leading to an acceleration of cancers in these tissues when Dicer is heterozygous (5, 6). In contrast, Dicer hypomorphic mice do not have an increased incidence of cancers in these or other tissues (10). Moreover, CD19-cre+/Dicer+/fl mice did not have increased numbers of B cells or B cell malignancies. In addition, murine fibroblasts have normal rates of growth when only one allele of Dicer is deleted (8). What distinguishes the studies in non-hematopoietic tissues that show Dicer is a haploinsufficient tumor suppressor is the overexpression of oncogenic Ras and/or deletion of a strong tumor suppressor (p53 or Rb/p107) was required to observe the tumor suppressor phenotype of Dicer heterozygosity (5, 6). However, when the Myc oncogene was overexpressed in B cells that lacked one allele of Dicer this did not result in an acceleration of B cell lymphomagenesis, indicating that Dicer heterozygosity does not cooperate with Myc overexpression in B cells. It will need to be determined whether Dicer heterozygosity can cooperate with Myc overexpression in non-hematopoietic tissues. Therefore in the context of Dicer heterozygosity alone or together with Myc overexpression, Dicer is not a haploinsufficient tumor suppressor in B cells.
Our data indicate certain miRNAs, whose maturation requires Dicer, must be necessary for Myc to induce B cell transformation, since Dicer is retained in all lymphomas arising in CD19-cre+/Dicerfl/fl/Eμ-myc mice. Previously, it was shown that specific miRNAs are induced by Myc and facilitate B cell transformation initiated by Myc (12, 13). For example, the miRNA polycistron miR-17∼92, which is regulated by Myc, is frequently overexpressed in human B cell lymphomas, and when overexpressed in pre-cancerous Eμ-myc fetal liver cells, Myc-mediated lymphomagenesis was accelerated (13). Therefore, Dicer and consequently, miRNAs are required for B cell transformation initiated by Myc. Similarly, one allele of Dicer was retained in tumors arising in a Ras-induced mouse model of lung cancer or soft tissue sarcoma (5), indicating that Dicer is also essential for transformation of non-hematopoietic cells. It will be important in the future to identify the miRNAs required for the transformation of B cells and for non-hematopoietic cells and to determine if there are commonalities.
In addition to Dicer being required for B cell transformation, we also determined that Dicer was necessary for the survival of established B cell lymphomas. Deletion of Dicer led to lymphoma cell apoptosis, whereas retention of at least one allele of Dicer allowed lymphoma cell survival. It is postulated that non-hematopoietic tumor cell lines can survive without Dicer (5). Differences in cell type and/or pre-existing genetic alterations may account for the discrepancy. For example, hematopoietic cells, such as B cells, are poised for apoptosis, whereas fibroblasts and epithelial cells are more prone to undergo senescence. Therefore, it is conceivable that a non-hematopoietic cell could survive long enough to acquire additional genetic alterations permitting it to grow in the absence of Dicer. Consistent with this notion, loss of p53 or the INK4a/ARF locus allowed fibroblasts to delay senescence induced by Dicer deletion (8). Similarly, deletion of Dicer in hepatocytes in mice led to apoptosis, but the few surviving hepatocytes developed into hepatocellular carcinoma late in life, indicting additional genetic events were necessary for hepatocyte survival (28). In contrast, cultured primary B cell lymphomas, all of which have inactivated the p53 pathway, underwent apoptosis upon Dicer deletion. Notably, there was a partial rescue of Dicer deletion-induced apoptosis of B cell precursors by overexpression of Bcl-2 (14), but it is unknown if that resulted in a B cell malignancy later in the life of those mice. Thus, complete loss of Dicer appears to be incompatible with cell viability, unless compensatory mutations occur that allow survival and growth. Therefore it will be important to define the mutations that a cell needs to acquire to live without Dicer and whether hematopoietic cells can ever survive and proliferate without Dicer.
Acknowledgments
The authors would like to thank Dr. William Dupont for Kaplan-Meier plots and log-rank tests, Brandon Metge and Dr. Shiqun Shi for technical assistance, Dr. Jos Jonkers for the CreERT2 retrovirus, Dr. Ursula Lichtenberg and Dr. John Cleveland for CD19-cre mice, Dr. Annette Kim for critically reading of the manuscript, and members of the Eischen lab for helpful discussions. This work was supported by NCI grants R01CA098139, R01CA117935, and P30CA68485 and the Leukemia & Lymphoma Society (C.M.E) and R01CA077735 (S.N.J.).
References
- 1.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 5.Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23(23):2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambertz I, Nittner D, Mestdagh P, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325(5943):965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mudhasani R, Zhu Z, Hutvagner G, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181(7):1055–63. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103(7):2208–13. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita S, Hara A, Kojima I, et al. Dicer is required for maintaining adult pancreas. PLoS One. 2009;4(1):e4212. doi: 10.1371/journal.pone.0004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koralov SB, Muljo SA, Galler GR, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–8. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 16.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–8. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–69. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 2003;22(6):1442–50. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Greiner TC, Lushnikova T, Eischen CM. Decreased Mdm2 expression inhibits tumor development induced by loss of ARF. Oncogene. 2006;25(26):3708–18. doi: 10.1038/sj.onc.1209411. [DOI] [PubMed] [Google Scholar]
- 20.Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27(11):1590–8. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- 21.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237(3):752–7. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 22.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167(2):353–71. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 24.Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47(1):11–8. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 25.Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348(6299):331–3. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- 26.Hobeika E, Thiemann S, Storch B, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103(37):13789–94. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekine S, Ogawa R, Ito R, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136(7):2304–15 e1-4. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]