Abstract
Dosage compensation is a chromosome-wide regulatory process that balances X-chromosome gene expression between males and females in species whose sex-determining mechanisms require each sex to have a different complement of X chromosomes. Recent advances have clarified the molecular nature of the C. elegans sex-determination signal, which tallies X chromosome number relative to the ploidy and controls both the choice of sexual fate and the process of dosage compensation. Dissecting the sex signal has revealed molecular mechanisms by which small quantitative differences in intracellular signals are translated into dramatically different developmental fates. Recent experiments have also revealed fundamental principles by which C. elegans dosage compensation proteins recognize and bind X chromosomes of XX embryos to reduce gene expression. Dosage compensation proteins function not only in a condensin complex specialized for regulating X-chromosome gene expression, but also in distinct condensin complexes that control other chromosome-wide processes: chromosome segregation and meiotic crossover recombination. The reshuffling of interchangeable molecular parts creates independent machines with similar architecture but distinct biological functions.
Introduction
Organisms that determine sex using chromosome-based mechanisms (e.g. XX female and XY or XO male), have evolved the essential, chromosome-wide regulatory process called dosage compensation to balance sex-chromosome gene expression between the sexes [1]. Strategies for dosage compensation differ, but invariably a regulatory complex is targeted to the sex chromosome of one sex to modulate transcript levels across the entire chromosome. Dosage compensation is exemplary for dissecting the coordinate regulation of gene expression over vast distances and the role of chromosome structure in controlling gene expression.
Mammals randomly inactivate one of the two female X chromosomes using non-coding RNAs that recruit the Polycomb complex [2,3]. Transient pairing of X chromosomes through the X-inactivation center heralds the onset of inactivation and helps specify the X to become inactivated [4,5]. A repressive nuclear compartment reliant on non-coding RNAs recruits the X genes to be silenced [6]. In contrast, flies increase expression of the single male X chromosome through a complex containing non-coding RNAs and MSL (Male Specific Lethal) proteins that binds the male X and acetylates histones [7]. Nematodes reduce expression of both X chromosomes in hermaphrodites by half through a dosage compensation complex (DCC) that binds the two X chromosomes [8]. The DCC resembles condensin, a protein complex conserved from yeast to humans to promote the compaction, resolution, and segregation of chromosomes during mitosis and meiosis [9-12]. In all three examples, selective recruitment of the dosage compensation machinery establishes the epigenetic regulation of X chromosomes that is maintained throughout the lifetime of the animal.
Fundamental issues relevant to all forms of dosage compensation include the regulatory pathway and sex-specific factors that activate dosage compensation in only one sex, the composition of the dosage compensation machinery, the cis-acting sites that selectively target the X chromosome for regulation, and the mechanism of fine tuning gene expression. This review focuses on C. elegans dosage compensation, with brief comparisons to D. melanogaster. Emphasis is placed on recent advances in understanding (1) the C. elegans regulatory hierarchy that controls dosage compensation, including the primary sex-determination signal, (2) fundamental principles by which the DCC recognizes and binds X chromosomes, and (3) the roles of dosage compensation proteins in controlling other chromosome-wide processes through association with distinct condensin complexes: crossover recombination during meiosis and chromosome segregation during mitosis and meiosis.
X:A signal: X and autosomal signal elements oppose each other to determine C. elegans sex
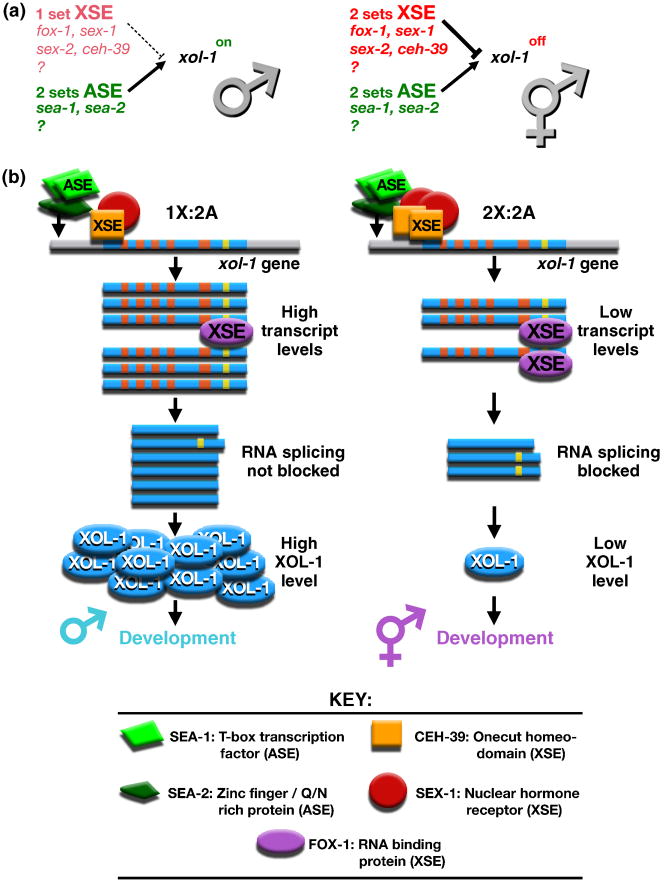
C. elegans determines sex by tallying X-chromosome number relative to the ploidy, the number of sets of autosomes (X:A signal) (Figure 1a) [13,14]. Ratios of 1X:2A and 2X:3A elicit male fate, while 2X:2A and 3X:4A elicit hermaphrodite fate. Not only does the X:A signal dictate sexual fate, it establishes the level of X-linked gene expression by controlling the process of dosage compensation [15,16]. Dissecting the sex determination signal in C. elegans has revealed molecular mechanisms by which small quantitative differences in intracellular signals can be translated into dramatically different developmental fates.
Figure 1. Primary sex determination: the X:A signal model.
(a) In C. elegans, sex is determined by the number of X chromosomes relative to the ploidy, the number of sets of autosomes. This X:A signal is composed of X signal elements (XSEs) that activate the hermaphrodite program of development and Autosomal signal elements (ASEs) that oppose the XSEs to promote male development. The direct target of the X:A signal is the sex determination switch gene xol-1, which controls both the choice of sexual fate and the level of X-linked gene expression achieved through the process of dosage compensation.
(b) X and autosomal signals antagonize each other directly at xol-1 to determine C. elegans sex. Direct rivalry at the xol-1 promoter between XSE transcriptional repressors (the ONECUT homeodomain protein CEH-39 and the nuclear hormone receptor SEX-1) and ASE transcriptional activators (the T-box transcription factor SEA-1 and the zinc finger protein SEA-2) leads to high xol-1 transcript levels in XO embryos and low levels in XX embryos. FOX-1, an RNA binding protein that acts as an XSE, then enhances the fidelity of signaling process by creating an inactive xol-1 mRNA splice variant in a dose-dependent manner. High XOL-1 protein levels in XO animals cause male development, and low XOL-1 levels in XX animals cause hermaphrodite development, including loading of the DCC onto X. Decreasing XSE dose causes XX-specific lethality, while increasing it causes XO-specific lethality. The reciprocal is true for ASEs. Increasing ASE dose causes XX-specific lethality; decreasing it causes XO-specific lethality. (xol-1 gene: promoter, gray; exons, blue; introns, orange and yellow).
X-chromosome number is communicated by a set of trans-acting X-signal elements (XSEs) encoded on the X chromosome (Figure 1a) [17-20]. XSEs act in a cumulative, dose-dependent manner to repress the target gene called xol-1 (XO lethal) in 2X:2A embryos. xol-1 is earliest-acting sex-determining gene and encodes a GHMP kinase family member that specifies the male fate when active and the hermaphrodite fate when inactive [15,19,21,22]. xol-1 also controls dosage compensation and hence viability. Inappropriate xol-1 repression in 1X:2A embryos or inappropriate xol-1 activation in 2X:2A embryos causes lethality due to incorrect levels of X-chromosome gene expression [22]. XSEs were discovered through genetic schemes that identified suppressors of the lethal effects of xol-1 misregulation.
XSE-mediated repression of xol-1 occurs at two distinct levels: transcriptional repression via the nuclear receptor SEX-1 and the ONECUT homeodomain protein CEH-39 and post-transcriptional repression via the RNA binding protein FOX-1 (Figure 1b) [17-20,23]. Both SEX-1 and CEH-39 repress xol-1 directly, by binding to multiple sites in its promoter (B. Farboud, J. Gladden, and B. Meyer (unpublished). Direct RNA splicing control of residual xol-1 transcripts by FOX-1 then enhances the fidelity of the counting process by creating an inactive xol-1 mRNA splice variant in XX embryos (C. Pickle, M. Nicoll, and B. Meyer, unpublished). In summary, multiple repressors act through multiple sites to translate the two-fold difference in X dose between the sexes into the HIGH or LOW activity state of xol-1. Two tiers of xol-1 regulation ensure the correct and stable choice of sexual fate.
Ploidy is communicated by an actual autosomal signal (Figure 1a). The autosomal signal contains discrete trans-acting autosomal signal elements (ASEs) that counter XSEs to coordinately regulate both sex determination and dosage compensation by activating xol-1 [24]. ASEs were identified as suppressors of the XX-specific lethality caused by loss of XSEs. SEA-1, a T-box transcription factor, and SEA-2, a zinc finger protein, act in a cumulative, dose-dependent manner to stimulate xol-1 transcription [24] (P. Nix and B. Meyer, unpublished). Transcriptional activation is direct: ASEs bind to the xol-1 promoter (Figure 1b) (M. Jow, P. Nix, and B. Meyer, unpublished). Thus, SEA-1 and SEA-2 engage in direct molecular warfare with XSEs to overcome their repressive effects, and xol-1 integrates both X and autosomal signals to determine sexual fate. Antagonistic molecular interactions carried out on a single promoter explain how even tiny differences in the X:A ratio can signal different sexual fates: X:A of 0.67 signals male fate, while X:A of 0.75 signals hermaphrodite fate.
The concept of a sex signal composed of zygotic ASEs that oppose zygotic XSEs arose as an hypothesis for fruit flies in 1921 [25] and was soon thereafter presented in textbooks as fact. Ironically, while the worm sex signal fits this simple textbook paradigm, the fly sex signal does not: the effect of ploidy on fly X-chromosome counting is not through ASEs.
The target of the fly X:A signal is Sxl (Sex lethal), the feminizing sex-determination switch gene that induces female development when active and male development, including activation of the MSL dosage compensation complex, when inactive [26,27]. A set of XSEs serve as transcriptional activators of Sxl such that the double dose of XSEs in 2X:2A embryos turns Sxl on [28-32]. Extensive genetic screens to recover mutations in ASEs as suppressors of the XX-specific lethality caused by mutations in XSEs identified only a single ASE that acts as a weak transcriptional repressor of Sxl but cannot account for the effect of ploidy on sex determination [33,34]. Thus, autosomal factors in the form of Sxl repressors do not appear to serve as the monitor of ploidy. Instead, the ploidy affects the timing of cellularization and hence the interval of time during which XSEs can increase in concentration and activate the establishment promoter of Sxl [35]. The lower the ploidy, the longer the XSE promoters remain active and hence the greater the probability of activating Sxl. As a consequence, 1X: 1A embryos become females instead of males, and 2X:3A embryos become mosaic intersexes with less than 100% of cells stably activating Sxl.
The dosage compensation complex
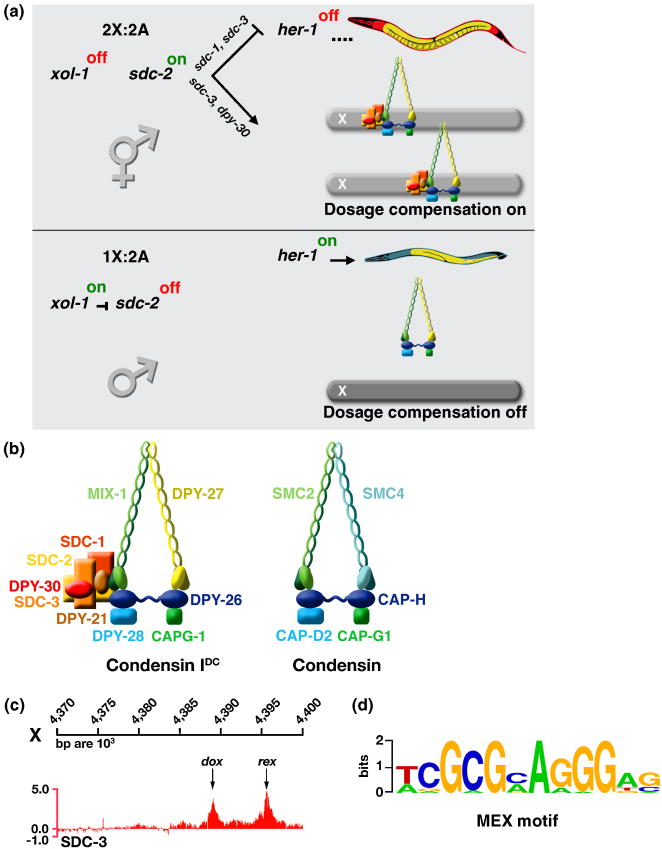
Repression of xol-1 in XX embryos permits the novel, XX-specific protein SDC-2 (Sex Determination and Dosage Compensation) to be active and thereby induce hermaphrodite sexual development and initiate dosage compensation (Figure 2a) [36,37]. SDC-2 acts with the zinc-finger proteins SDC-1 and SDC-3 to induce hermaphrodite development by repressing her-1, a sex-determination gene that elicits male development [37-39]. SDC proteins inactivate her-1 directly by binding to three sites in the gene [38]. SDC-2 also triggers assembly of the DCC onto X through collaboration with SDC-3 and DPY-30 (DumPY) [37,40,41]. DPY-30 is a member of both the DCC and the transcription activation complex called COMPASS, which is responsible for trimethylation of histone H3 at lysine 4 (C. Hassig, R. Auty, W. Kruesi, B. Meyer unpublished) [42]. SDC-2 is the sole dosage compensation protein produced exclusively in XX embryos, and ectopic activation of SDC-2 in XO embryos causes the DCC to assemble onto the male X and the males to die [37]. SDC-2 confers both sex- and X-specificity to DCC binding.
Figure 2. Regulation of Dosage Compensation in C. elegans.
(a) Partial genetic pathway for sex determination and dosage compensation. In XX embryos, xol-1 is repressed by the double dose of XSEs, permitting the XX-specific protein SDC-2 to activate dosage compensation and to repress her-1, a male-specific sex determination gene. SDC-2 acts with SDC-3 and DPY-30 to recruit the DCC condensin subunits to X. SDC-2 acts with SDC-1 and SDC-3 to repress her-1. SDC-2 plays the lead role in recognizing X sequences, while SDC-3 predominates in recognizing the SDC binding sites at her-1. In XO embryos, ASEs overcome XSEs to activate xol-1, resulting in sdc-2 repression and her-1 activation, thereby setting the male sexual fate. The DCC is not loaded onto X.
(b) The DCC consists of five condensin-like components (DPY-27, MIX-1, DPY-26, DPY-28, and CAPG-1) that are homologous to canonical condensin subunits SMC2, SMC4, CAP-H, CAP-D2, and CAP-G1, respectively The DCC also contains at least five additional factors that confer X- and sex-specificity (SDC-2, SDC-3, and DPY-30) or assist in repression (DPY-21 and SDC-1).
(c) DCC binding sites have been mapped by ChIP chip experiments and classified into two categories based on their ability to bind the complex when detached from the X chromosome. rex sites (recruitment elements on X) bind the complex robustly when they are detached from X and present either in multiple copies on extrachromosomal arrays (see Figure 3a) or in low copy number integrated onto an autosome. dox sites (dependent on X) fail to bind the DCC when detached; they depend on the broader X chromosomal context for their ability to associate with the DCC.
(d) Motif searches identified a twelve base pair consensus motif that is enriched at rex sites relative to dox sites and on X chromosomes relative to autosomes. Mutations within the motif disrupt the ability of rex sites to recruit the DCC (see Figure 3a).
The DCC includes ten proteins: the three SDC proteins, DPY-30, DPY-21 (a novel protein), and five proteins that are homologous to the members of condensin (Figures 2a and 3) [9,10,37,38,40,43-46] (W. Kruesi, C. Hassig and B. Meyer, unpublished). The DCC and condensin each contain a pair of SMC proteins (Structural Maintenance of Chromosomes) and three non-SMC proteins [10,11,43,44,47]. The DCC SMC proteins DPY-27 and MIX-1, which are homologs of condensin SMC2 and SMC4, respectively, have nucleotide binding domains (NBDs) at their N- and C-termini that are linked by two long coiled coil domains separated by a hinge domain. Mutation of the NBDs disrupts dosage compensation [43,44]. SMC proteins fold back on themselves to form a central region of two anti-parallel coiled coils flanked by the NBDs and the hinge. The two SMC proteins dimerize through interactions between their hinge domains and use their globular NBDs to bind the non-SMC proteins (DPY-26, DPY-28 and CAPG-1 in the DCC and CAP-H, CAP-D2, and CAP-G1, respectively, in condensin).
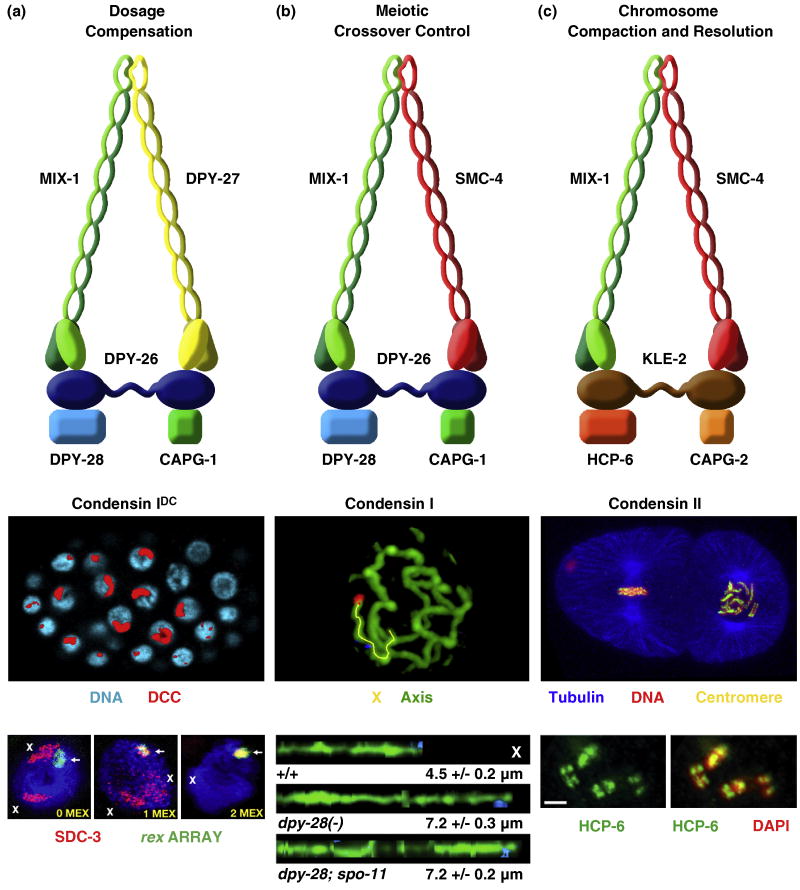
Figure 3. Three condensin complexes carry out distinct functions in C. elegans.
(a) The dosage compensation complex (the DCC, also called condensin IDC) (top) resembles condensin I. It equalizes X-linked expression between the sexes (XX hermaphrodites and XO males) by reducing transcript levels by half in hermaphrodites. Shown is an XX embryo stained with the DCC component SDC-2 (red) and DAPI (blue) (middle). The DCC binds to both X chromosomes. Shown also are images of gut cell nuclei carrying extrachromosomal arrays with multiple copies of rex-1 derivatives stained with DAPI (blue), SDC-3 antibodies (red), and array FISH (green) (bottom). Arrays have rex-1 fragments with different numbers of MEX motifs. Left, a 33 bp rex-1 fragment in which its single MEX motif was mutated, thus abrogating DCC binding. X staining is apparent (red). Middle, a wild-type 33 bp rex-1 fragment with 1 MEX motif. SDC-3 colocalizes with the array and X. Right, a 60 bp rex-1 fragment with 2 MEX motifs. SDC-3 binds robustly to rex-1 and is titrated from X, showing that MEX motifs collaborate to recruit the DCC. An array carrying a rex-1 fragment that titrates the DCC from X can suppress the male lethality caused by mutation of xol-1 in XO embryos.
(b) Condensin I. This complex differs from DCC condensin by a single subunit, SMC-4 (top). This complex controls meiotic DSB distribution through effects on chromosome structure. It also plays minor roles in chromosome segregation in mitosis and meiosis. Shown is a high resolution image of pachytene chromosomes in wild-type animals labeled with the axis protein HTP-3 and FISH probes (blue, red) to X (middle). X chromosomes from wild-type and dpy-28 mutant animals were traced (yellow) in three dimensions and straightened computationally (bottom). Straightened chromosomes are shown horizontally. Genotypes, average total axis length, and standard error of the mean are below each axis. Disruption of dpy-28 dramatically increases the X-chromosome axis length in a manner independent of DSBs made by SPO-11. In response, DSBs are increased in number and redistributed in dpy-28 mutations, causing an increase in crossovers and their redistribution.
(c) Condensin II. This complex shares one subunit (MIX-1) with the DCC (condensin IDC) and two subunits (MIX-1 and SMC-4) with condensin I (top). This complex is the prime condensin complex responsible for mitotic and meiotic chromosome compaction and resolution. Condensin II binds to the centromeres on the holocentric mitotic chromosomes. Shown is a two cell embryo (middle) with one cell in metaphase (left) and one in prometaphase (right). Centromeric proteins (yellow) and condensin II bind along the outer edge of each chromosome, adjacent to where the mitotic spindle (tubulin, blue) attaches. Condensin II also binds to meiotic chromosomes at pachytene exit to create the compact shape (diakinesis bivalents) required for chromosome segregation. Shown are the four sister chromatids of meiotic diakinesis bivalents (bottom) stained with HCP-6 antibodies (green) and DAPI (red). Merge is yellow.
With the exception of MIX-1 and CAPG-1, which were identified through biochemical experiments, the DCC components were discovered through genetic screens that recovered either XX-specific lethal mutations or suppressors of the XO-specific lethality caused by xol-1 null mutations. Dosage compensation proteins (DC proteins) function not only in a condensin complex specialized for dosage compensation, but also in two other biochemically distinct condensin complexes (condensin I and condensin II) that control other chromosome-wide processes (Figure 3) [10,48,49], as will be described subsequently.
Evidence that DC proteins form a complex on X came from multiple lines of experiments. (1) Immunofluorescence experiments using X-specific DNA probes and DC protein antibodies showed co-localization of all DC proteins with X [9,10,37,38,40,43-46,50] (W. Kruesi, C. Hassig and B. Meyer, unpublished). (2) Co-immunoprecipitation experiments using antibodies to each DC protein coupled with protein analysis using Western blots or mass spectrometry showed the association of all DC proteins [9,10,37,38,40,43-46,50] (W. Kruesi, C. Hassig and B. Meyer, unpublished). (3) Functional assays to identify X sites that recruit DC proteins in vivo when detached from X showed all components bind to recruitment sites [51-53]. (4) Chromatin immunoprecipitation experiments using antibodies to six different DC proteins followed by hybridization to genome-wide tiling arrays (ChIP-chip analysis) showed all subunits have similar binding peaks [53,54].
Targeting the DCC to X chromosomes: cis-acting sites
The combination of the genome-wide approach to identify DCC binding sites without regard to recruitment ability and the functional approach in vivo to assess DCC recruitment to sites detached from X showed the DCC binds to discrete, dispersed sites on X that partition into two classes (Figures 2a and 3c) [53]. rex sites (recruitment elements on X) recruit the DCC in an autonomous, DNA sequence-dependent manner using a 12 base pair MEX consensus motif that is enriched on X compared to autosomes (Figure 2d). MEX variants enriched on X by 3.8- to 25-fold are highly predictive (95%) for rex sites, and mutation of MEX motifs disrupts DCC binding (Figure 3a). MEX motifs are clustered in rex sites and collaborate to recruit the DCC[52]. rex sites confer X-chromosome specificity to dosage compensation and occur in only about 200 distinct locations on X.
dox sites (dependent on X) bind the DCC in their native context on X but fail to recruit the DCC when detached from X [53]. dox sites are ∼7-fold more prevalent than rex sites. dox sites lack the MEX variants enriched on X, and motif searches have not identified a compelling motif that distinguishes dox sites from random X or autosomal DNA. dox sites per se are not the catalyst for DCC disbursement along X; rather cis-linkage to rex sites allows dox sites to become fully occupied by the DCC, as will be described subsequently. dox sites do not necessarily need sequences distinct from autosomal sequences, since rex sites mark X chromosomes for DCC binding and repression.
dox sites occur preferentially in highly transcribed promoters, whereas rex sites reside predominantly in intergenic regions [53]. Both the level of DCC binding at dox sites and the probability of a promoter having a dox site correlate directly with the expression level of the gene. Moreover, promoters dynamically regulated during development bind the DCC at higher levels during periods of transcriptional activity, further implicating transcription in binding to dox sites [55] (W. Kruesi and B. Meyer, unpublished). In contrast, rex-site binding remains relatively constant throughout somatic development.
Although the MEX motif is central to the mechanism by which the DCC distinguishes X chromosomes from autosomes, it cannot be the sole source of X specificity [53]. Some rex sites have only MEX variants with consensus matches and distributions similar to those in dox sites, random autosomal sequences, and X sequences not bound by the DCC. A feature other than MEX must designate those sites for recruitment. Nonetheless, mutation of the weak motifs in such rex sites disrupts recruitment, showing that the MEX variants not enriched on X are important for DCC binding in the context of a rex site.
rex and dox sites are interspersed and separated by considerable distances (2-90 kb), implying that long-range communication might facilitate DCC loading onto X. The similarity of the DCC to condensin suggests models involving changes in chromosome structure, such as chromatin looping. Consistent with this view, in yeast, condensin complexes co-localize with widely dispersed tRNA genes to facilitate their clustering in the nucleolus [56]. In C. elegans, tRNA genes on X are highly correlated with DCC binding sites [53], suggesting the association of binding sites might occur.
DCC Loading onto X
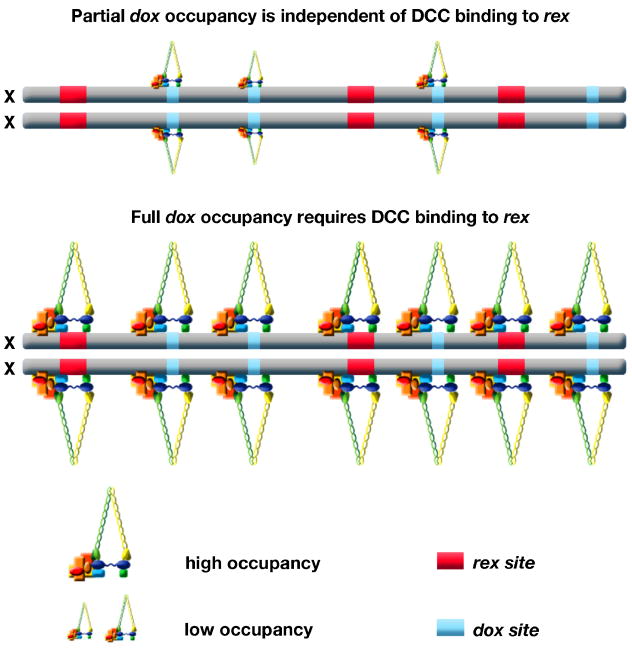
Discovery of autonomous (rex) and dependent (dox) DCC binding sites suggests a model by which the DCC loads onto X chromosomes. The DCC binds to rex sites, which recruit additional complexes to bind along X [53]. This model is supported and refined by two additional sets of experiments that address the relationship between DCC binding at rex and dox sites. First, ChIP-chip analysis of DCC binding in mutants lacking any one of the recruitment proteins SDC-2, SDC-3 or DPY-30 showed that binding to rex sites was essentially abolished, binding to 65% of dox sites was eliminated, and binding to 35% of dox sites was reduced (R. Pferdehirt and B. Meyer, unpublished). These results show that DCC binding to rex sites requires all recruitment proteins and binding to dox sites is both dependent and independent of these proteins. That is, dox sites have an inherent, limited capacity to bind some DCC components, but full occupancy requires the recruitment proteins and presumably their binding to rex sites.
The dual roles of DPY-30 in recruiting the DCC to X and in activating gene expression by acting in COMPASS led to previous speculation that DPY-30 was the primary protein required for “spreading” of the DCC from rex sites to dox sites in active promoters [57]. However, the loss of binding to rex sites in dpy-30 mutants contradicts a selective role for DPY-30 in “spreading” but instead indicates that DPY-30 acts similarly to SDCs in directing DCC binding to X (R. Pferdehirt and B. Meyer, unpublished). Moreover, the robust binding of SDC-2 to both rex and dox sites [53] contradicts the view that SDC-2 is required selectively for DCC binding to rex sites [55].
Second, if the proximity of rex sites to dox sites on X facilitates DCC binding to dox sites, then attaching rex sites to autosomes might enhance binding to autosomes. The DCC binds to autosomes, but at fewer sites (one-fifth) than on X [53]. Binding is not perturbed by disruption of sdc-2, sdc-3 or dpy-30, consistent with the DCC having low, intrinsic binding capability at specific locations (R. Pferdehirt and B. Meyer, unpublished). The autosomal sites usually reside in promoters of expressed genes [53], and DCC binding correlates directly with the dynamic pattern of gene expression during development, as does binding to dox sites on X (W. Kruesi and B. Meyer, unpublished).
Two independent studies found enhancement of DCC binding to autosomal territories located adjacent to X in X-to-autosome fusion chromosomes. In one study [55], DCC binding to wild-type autosomes was negligible and binding to the autosomal region adjacent to X in the fusion chromosome was interpreted as the establishment of new DCC binding sites. In the other study (R. Pferdehirt and B. Meyer, unpublished), DCC binding to the autosomal region adjacent to X generally represented an increase in occupancy at sites usually bound by the DCC on wild-type autosomes. While the mechanistic interpretations of the studies differ in significant ways, both support the view that dox-site binding on X is strengthened by proximity to rex sites. Thus, DCC binding to rex sites confers X specificity to dosage compensation and catalyzes increased DCC binding along X at dox sites (Figure 4).
Figure 4. Model for loading of the DCC onto X.
This model integrates current data. The DCC binds to rex sites in a sequence-dependent manner requiring SDC-2, SDC-3, and DPY-30. rex sites confer X-specificity to DCC binding and recruit additional complexes to bind along X at dox sites, which are unable to recruit the DCC when detached from X. Many dox sites on X have a small, intrinsic DCC binding capability; this binding is enhanced in response to DCC loading onto rex sites. dox sites are located preferentially in promoters of active genes, and positions of dox sites change as gene expression changes, implying that dox-site binding is transcription dependent.
Comparison of X-chromosome recognition and dosage compensation complex binding in flies and nematodes
Although the dosage compensation complexes of flies and nematodes are evolutionarily unrelated and regulate X chromosomes in opposite ways, similar principles appear to govern the X-chromosome targeting and binding of the two complexes. In flies, dosage compensation is achieved by the MSL (male-specific-lethal) complex, which binds to the single X chromosome of males to increase transcript levels [7]. As in worms, about 150 special chromatin entry sites recruit the MSL complex in a DNA-sequence dependent manner using a motif enriched on X [53,58,59]. For both flies and worms, a second mode of binding correlates with high levels of gene expression and appears sequence independent, suggesting that features common to transcribed genes facilitate additional binding on X [53,60-62]. Full binding to the transcription-associated class of sites requires attachment to the sequence-dependent recruitment sites [55,63] (R. Pferdehirt, W. Kruesi and B. Meyer, unpublished). Sequence-independent MSL binding favors the 3′ ends of active genes [64-66], while sequence-independent nematode DCC binding often favors 5′ ends of active genes [53,55].
The C. elegans DCC acts at a distance to repress genes on X
The mechanism of dosage compensation in C. elegans was explored by correlating the locations of DCC binding sites with the genes responsive to dosage compensation [53]. Several striking conclusions emerged. First, the DCC does not compensate all X genes, nor does it always achieve a precise 2-fold repression when it does. Second, although the DCC binds preferentially to more highly expressed genes on X, it can compensate X genes of all expression levels. Third and most unexpected, DCC binding to the promoter or body of a gene is neither necessary nor sufficient to elicit compensation of that gene. Compensated and non-compensated genes alike can have DCC binding sites or not. Thus, additional factors besides the proximity of DCC binding must help define whether a gene responds to the dosage compensation process. Although the DCC could potentially act locally at the sites it binds to modulate gene expression [54], it must act over long range to achieve the compensation of most genes [53]. The DCC likely alters higher-order chromosome structure to control interactions between dispersed regulatory elements that modulate gene expression.
Autosomal gene expression is affected by the DCC
DCC disruption causes opposite effects on X and autosomal genes: X genes have increased expression, and one-fourth of autosomal genes have reduced expression [53]. The DCC thus affects expression throughout the genome. The DCC binding sites on autosomes correlate infrequently with genes affected by dosage compensation mutations. The effect of DCC disruption on autosomal gene expression cannot simply be explained by an indirect effect on X-linked gene expression, because weak dosage compensation mutations have a similar effect on autosomal gene expression as strong mutations, even through strong mutations have a greater effect on X gene expression. However, an “indirect” effect on autosomal gene expression could result from a possible, albeit speculative model of DCC action in which the DCC repels a rate-limiting activator from X, thus making more activators available to autosomes. In dosage compensation mutants, more activators would bind to X and activate X genes, leaving less activator available for autosomal genes.
DC proteins achieve diverse chromosome-wide functions through participation in distinct condensin complexes
DC proteins function not only in a condensin complex specialized for dosage compensation (condensin IDC) but also in two other biochemically distinct condensin complexes in C. elegans (condensin I and condensin II) to carry out independent roles in chromosome segregation and meiotic crossover regulation (Figure 3) [10,12,48,49]. Condensin I differs from condensin IDC by only one subunit: SMC-4 from condensin II replaces the SMC protein DPY-27 from condensin IDC [10,48]. Condensin I controls the number and distribution of double strand breaks (DSBs) in meiosis and thereby the number and position of crossovers (COs) between homologous chromosomes by controlling the higher order structure of meiotic chromosomes (Figure 3b) [48]. Disrupting any condensin I subunit causes a dominant change in the distribution of DSBs, and hence COs, and dramatically lengthens the chromosome axis. Control of CO distribution was thought to be imposed only after DSB formation, affecting the decision of a DSB to become a crossover or noncrossover. Condensin I analysis showed instead that CO number and distribution can also be controlled on a chromosome-wide basis at the level of DSB formation by controlling chromosome structure.
Condensin I also affects mitotic and meiotic chromosome segregation, but in a manner distinct from condensin II, the primary condensin complex in C. elegans to control the compaction and resolution of mitotic and meiotic chromosomes in preparation for their separation (Figure 3c) [9,10,49]. Condensin II shares the SMC protein MIX-1 with condensin IDC and both SMC proteins with condensin I [12,49]. Condensin II has non-SMC proteins that are distinct from, but homologous to, those shared by condensin I and condensin IDC [10,12]. Condensin II binds to the centromeres of mitotic chromosomes and to meiotic chromosomes at pachytene exit to create the compact chromosome shape required for chromosome segregation [12,49]. The participation of dosage compensation proteins in diverse condensin complexes illustrates that reshuffling of homologous, interchangeable molecular parts can create independent machines with similar architecture but distinct biological functions.
Conclusions
Fundamental principles have emerged regarding the X-chromosome-specific targeting and loading of a condensin-like DCC to reduce gene expression from both X chromosomes of C. elegans hermaphrodites. Two distinct mechanisms govern DCC binding: sequence-dependent recruitment to autonomous binding sites that confer X-chromosome specificity (rex) and non-autonomous, sequence-independent binding to sites (dox) correlated with high levels of gene expression. The dosage compensation process is controlled by the primary signal that determines sexual fate, the X:A signal. Dissection of this signal has revealed molecular mechanisms by which small quantitative differences in intracellular signals are translated into alternative developmental fates.
The mechanism of C. elegans dosage compensation has not yet been determined, but given the homology of DCC components to condensin subunits, the long-range communication between rex and dox sites to facilitate DCC loading, the action of the DCC at a distance to regulate X gene expression, and the impact of the DCC on autosomal gene expression, the DCC likely functions by inducing changes in chromosome configuration. Such reconfiguration of chromosome architecture could bring regulatory DNA elements into proximity with genes to be regulated, could create sub-chromosomal territories within X for gene repression, could create a specific sub-nuclear compartment for X to modulate its gene expression, or could even repel a rate-limiting, genome-wide activator from X to reduce gene expression. Precedent exists for such action of condensin. Condensin binds yeast tRNA genes and facilitates the clustering of these widely dispersed loci into the nucleolus [56,67,68]. Indeed, tRNA genes on X are highly correlated with DCC sites, raising the possibility that DCC changes X architecture through association with tRNA genes [53]. In general, condensin can also restrict interactions between chromosomes, thereby causing their isolation. For example, condensin is required for the programmed restructuring and disassembly of Drosophila polytene chromosomes, giant DNA structures with multiple paired chromatids formed by numerous rounds of DNA replication without intervening cell divisions [69]. The compartmentalization of chromatids may be exemplary for how the DC might restrict X chromosomes to domains of gene repression.
Future experiments will reveal the dosage compensation mechanism, a more precise understanding of how the DCC binds to X chromosomes, and the means by which condensin complexes that differ by only a single, homologous subunit can have distinct functions.
Acknowledgments
B.J.M. thanks T. Cline and the Meyer lab for numerous discussions and J. Gunther for figure design. This work was supported by N.I.H. grant GM30702. B.J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of a manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyedin uneditedting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 2.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;(8):293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 5.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 6.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes and Development. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer BJ. WormBook. The C elegans Research Community; X-Chromosome dosage compensation. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22:194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Current Biology. 2009;19:9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Biochemical, cytological, and genetic experiments establish the diverse roles of three separate but inter-related condensin complexes in X-chromosome dosage compensation and mitotic and meiotic chromosome segregation.
- 11.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 12.Chan RC, Severson AF, Meyer BJ. Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol. 2004;167:613–625. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nigon V. Effets de la polyploidie chez un nématode libre. CR Seances Acad Sci Ser D Sci Nat. 1949;228:1161–1162. [PubMed] [Google Scholar]
- 14.Madl JE, Herman RK. Polyploids and sex determination in Caenorhabditis elegans. Genetics. 1979;93:393–402. doi: 10.1093/genetics/93.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller LM, Plenefisch JD, Casson LP, Meyer BJ. xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell. 1988;55:167–183. doi: 10.1016/0092-8674(88)90019-0. [DOI] [PubMed] [Google Scholar]
- 16.Akerib CC, Meyer BJ. Identification of X chromosome regions in Caenorhabditis elegans that contain sex-determination signal elements. Genetics. 1994;138:1105–1125. doi: 10.1093/genetics/138.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- 18.Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–204. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- *19.Gladden JM, Meyer BJ. A ONECUT homeodomain protein communicates X chromosome dose to specify Caenorhabditis elegans sexual fate by repressing a sex switch gene. Genetics. 2007;177:1621–1637. doi: 10.1534/genetics.106.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides the most recent genetic and molecular dissection of X-signal elements that comprise the primary sex-determination signal of C elegans.
- 20.Skipper M, Milne CA, Hodgkin J. Genetic and molecular analysis of fox-1, a numerator element involved in Caenorhabditis elegans primary sex determination. Genetics. 1999;151:617–631. doi: 10.1093/genetics/151.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luz JG, Hassig CA, Pickle C, Godzik A, Meyer BJ, Wilson IA. XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators. Genes Dev. 2003;17:977–990. doi: 10.1101/gad.1082303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhind NR, Miller LM, Kopczynski JB, Meyer BJ. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell. 1995;80:71–82. doi: 10.1016/0092-8674(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 23.Gladden JM, Farboud B, Meyer BJ. Revisiting the X:A signal that specifies Caenorhabditis elegans sexual fate. Genetics. 2007;177:1639–1654. doi: 10.1534/genetics.107.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Powell JR, Jow MM, Meyer BJ. The T-box transcription factor SEA-1 is an autosomal element of the X:A signal that determines C. elegans sex. Dev Cell. 2005;9:339–349. doi: 10.1016/j.devcel.2005.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that ploidy is communicated by an actual autosomal signal. The autosome signal contains discrete autosomal signal elements (ASEs) that counter XSEs to regulate both sex determination and dosages compensation.
- 25.Bridges CB. Triploid intersexes in Drosophila melanogaster. Science. 1921;54:252–254. doi: 10.1126/science.54.1394.252. [DOI] [PubMed] [Google Scholar]
- 26.Cline TW, Meyer BJ. Vive la difference: males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 27.Cline TW. Reflections on a path to sexual commitment. Genetics. 2005;169:1179–1185. doi: 10.1093/genetics/169.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cline TW. Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics. 1988;119:829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;(68):933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- 30.Erickson JW, Cline TW. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993;7:1688–1702. doi: 10.1101/gad.7.9.1688. [DOI] [PubMed] [Google Scholar]
- 31.Erickson JW, Cline TW. Molecular nature of the Drosophila sex determination signal and its link to neurogenesis. Science. 1991;251:1071–1074. doi: 10.1126/science.1900130. [DOI] [PubMed] [Google Scholar]
- 32.Sefton L, Timmer JR, Zhang Y, Beranger F, Cline TW. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405:970–973. doi: 10.1038/35016119. [DOI] [PubMed] [Google Scholar]
- 33.Barbash DA, Cline TW. Genetic and molecular analysis of the autosomal component of the primary sex determination signal of Drosophila melanogaster. Genetics. 1995;141:1451–1471. doi: 10.1093/genetics/141.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrischnik LA, Timmer JR, Megna LA, Cline TW. Recruitment of the proneural gene scute to the Drosophila sex-determination pathway. Genetics. 2003;165:2007–2027. doi: 10.1093/genetics/165.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5:e332. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that ploidy in Drosophila affects the timing of cellularization and thus the interval of time during which XSEs can increase in concentration and activate Sxl.
- 36.Nusbaum C, Meyer BJ. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics. 1989;122:579–593. doi: 10.1093/genetics/122.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- 38.Chu DS, Dawes HE, Lieb JD, Chan RC, Kuo AF, Meyer BJ. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 2002;16:796–805. doi: 10.1101/gad.972702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLong L, Plenefisch JD, Klein RD, Meyer BJ. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics. 1993;133:875–896. doi: 10.1093/genetics/133.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–1031. doi: 10.1242/dev.124.5.1019. [DOI] [PubMed] [Google Scholar]
- 41.Hsu DR, Meyer BJ. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics. 1994;137:999–1018. doi: 10.1093/genetics/137.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonet T, Dulermo R, Schott S, Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol. 2001;(312):367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 43.Chuang PT, Albertson DG, Meyer BJ. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 44.Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 45.Lieb JD, Capowski EE, Meneely P, Meyer BJ. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science. 1996;274:1732–1736. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- 46.Yonker SA, Meyer BJ. Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development. 2003;130:6519–6532. doi: 10.1242/dev.00886. [DOI] [PubMed] [Google Scholar]
- 47.Hirano T. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 2002;16:399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- **48.Mets DG, Meyer BJ. Condensin Regulate Meiotic DNA Break Distribution, thus Crossover Frequency, by Controlling Chromosome Structure. Cell. 2009 doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that dosage compensation proteins participate in a condensin complex that differs from the DCC condensin by only one subunit but has a fundamentally different biological function. The condensin complex controls the distribution of meiotic double strand breaks and hence crossovers by controlling chromosome structure.
- 49.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang PT, Lieb JD, Meyer BJ. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science. 1996;274:1736–1739. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- 51.Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science. 2004;303:1182–1185. doi: 10.1126/science.1092938. [DOI] [PubMed] [Google Scholar]
- 52.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–618. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **53.Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes and Development. 2009;23:602–618. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that the DCC binds to discrete, dispersed sites on X of two types. rex sites recruit the DCC in an autonomous, sequence-dependent manner using a motif enriched on X. dox sites lack the X-enriched motif, are found preferentially in promoters of highly expressed genes, and bind the DCC only when associated with an intact X. A model is formulated in which DCC bindng to rex sites confers X specificty onto dosage compensation and catalyzes increased DCC binding along X at dox sites. This study also correlates DCC binding with function and shows that DCC binding at a gene is neither necessary nor sufficient for the compensation of the gene, implying the DCC acts at a distance to modulate gene expression.
- 54.Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nature Genetics. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ercan S, Dick LL, Lieb JD. The C. elegans dosage compensation complex propagates dynamically and independently of X chromosome sequence. Current Biology. 2009;19:1777–1787. doi: 10.1016/j.cub.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that condensin binds to tRNA genes dispersed throughout the entire genome and faciliates their clustering at the nucleolus.
- 57.Ercan S, Lieb JD. C. elegans dosage compensation: a window into mechanisms of domain-scale gene regulation. Chromosome Research. 2009;17:215–227. doi: 10.1007/s10577-008-9011-0. [DOI] [PubMed] [Google Scholar]
- 58.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 61.Kind J, Akhtar A. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev. 2007;21:2030–2040. doi: 10.1101/gad.430807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- *63.Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev. 2009;23:2266–2271. doi: 10.1101/gad.1840409. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that following recruitment of the MSL complex to chromatin entry sites, transcription of active genes is the main feature that guides MSL binding.
- 64.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 2006;20:871–883. doi: 10.1101/gad.377506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Molecular Biology of the Cell. 2009;(21):254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *69.Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322:1384–1387. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]; This paper demonstrates that condensins can restrict interactions between homologous chromosomes. Loss of condensin function prevents the dissassembly of polytene chromosomes during ovarian nurse cell development and also inhibits transvection, the ability of a gene on one chromosome to influence the activity of an allele on the paired homologous chromosome.