Abstract
Reprogramming of somatic cell nuclei to yield induced pluripotent stem (iPS) cells makes possible derivation of patient-specific stem cells for regenerative medicine. However, iPS cell generation is asynchronous and slow (2–3 weeks), the frequency is low (, 0.1%), and DNA demethylation constitutes a bottleneck. To determine regulatory mechanisms involved in reprogramming, we generated interspecies heterokaryons (fused mouse embryonic stem (ES) cells and human fibroblasts) that induce reprogramming synchronously, frequently and fast. Here we show that reprogramming towards pluripotency in single heterokaryons is initiated without cell division or DNA replication, rapidly (1 day) and efficiently (70%). Short interfering RNA (siRNA)-mediated knockdown showed that activation-induced cytidine deaminase (AID, also known as AICDA) is required for promoter demethylation and induction of OCT4 (also known as POU5F1) and NANOG gene expression. AID protein bound silent methylated OCT4 and NANOG promoters in fibroblasts, but not active demethylated promoters in ES cells. These data provide the first evidence that mammalian AID is required for active DNA demethylation and initiation of nuclear reprogramming towards pluripotency in human somatic cells.
Reprogramming of somatic cell nuclei towards pluripotency has been achieved by nuclear transfer into enucleated oocytes1,2 and the introduction of four defined factors to generate iPS cells3-6. These remarkable advances enable the generation of patient-specific cells for tissue replacement, modelling human diseases in tissue culture, and drug discovery. However, an understanding of the molecular mechanisms underlying nuclear reprogramming has been elusive, largely due to the technical challenges associated with nuclear transfer and the low efficiency of iPS cell generation.
To identify new early regulators essential for nuclear reprogramming towards pluripotency, we capitalized on our previous experience with heterokaryons–cell fusion products that proved useful in determining the principles inherent to the maintenance of the differentiated state of somatic cells7-11. Specifically, these earlier studies, by us and others, showed that the ‘terminally differentiated’ state of human cells was not fixed, but could be altered, and the expression of previously silent genes typical of other differentiated states could be induced11-15. We showed that reprogramming in heterokaryons was influenced by DNA methylation status, tissue of origin, and the relative ratio of nuclei that dictates the balance of regulators11,15-17, consistent with recent experiments in iPS cells18-23. We reasoned that heterokaryons, generated by fusing mouse ES cells and human fibroblasts, could be used to elucidate mechanisms and identify new genes with a role at the onset of reprogramming towards pluripotency because reprogramming takes place in the presence of all ES cell factors, and the onset of reprogramming is synchronously initiated after fusion. The advantages of interspecies heterokaryons over previously used same-species cell fusion hybrids24,25 are that species differences allow a distinction of transcripts derived from the two fused cell types, and reprogramming is assessed immediately after fusion without the need for cell division or DNA replication. Heterokaryons constitute a third complementary approach to nuclear reprogramming that overcomes some of the limitations of nuclear transfer and iPS cell generation by facilitating mechanistic studies.
The use of heterokaryons in this report allowed us to study epigenetic and transcriptional changes critical to the initiation of reprogramming towards pluripotency. We focused on DNA demethylation–a known block to reprogramming that leads to partially reprogrammed iPS cells23, and also a key step for reprogramming by nuclear transfer26. Despite decades of effort, so far no consensus mammalian DNA demethylase has been identified27. Recently, AID has been implicated in DNA demethylation in zebrafish within hours after fertilization, acting in a complex that mediates deamination followed by DNA repair28. In mammals, AID is primarily known for its role in the generation of antibody diversity in Blymphocytes29, but has recently been detected in germ cells30. Our studies demonstrate a new role for AID in active DNA demethylation and reprogramming of mammalian somatic cells towards pluripotency.
Efficient reprogramming towards pluripotency in heterokaryons
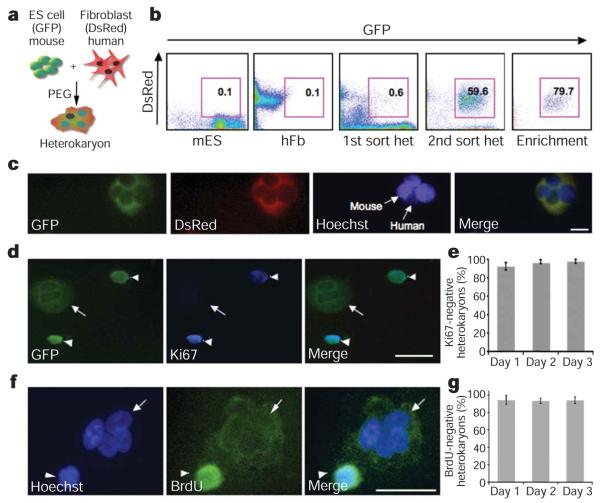
To produce interspecies heterokaryons, mouse ES cells transduced with a green fluorescent protein (GFP) reporter gene were co-cultured with primary human fibroblasts transduced with a DsRed reporter gene, and fused using polyethylene glycol (PEG) (Fig. 1a and scheme in Supplementary Fig. 1). Fused GFP+ DsRed+ heterokaryons, which were readily sorted by fluorescence-activated cell sorting (FACS) (Fig. 1b) and identified using fluorescence microscopy, contained distinctly stained human and mouse nuclei when visualized with Hoechst dye (Fig. 1c, f). Because the efficiency of PEG fusion is low (0.6–1.0%), GFP+ DsRed+ heterokaryons were sorted twice and enriched to 80% purity (Fig. 1b). Using an antibody for Ki67, a nuclear protein present only in proliferating cells, we determined that cell division did not occur in 98±2% (mean±s.e.m.) of heterokaryons over the 3-day time period assayed after fusion (Fig. 1d, e). Furthermore, BrdU labelling was not detected in 94±4% of heterokaryons over the same time period, indicating the absence of DNA replication (Fig. 1f, g and Supplementary Figs 2 and 3). To favour reprogramming towards a pluripotent state, we skewed the ratio of the input cells so that ES cells outnumbered the fibroblasts as gene dosage and the proportion of proteins contributed by each cell type determines the direction of nuclear reprogramming in somatic cells16,31.
Figure 1. Absence of cell division and DNA replication in heterokaryons.
a, Heterokaryon fusion scheme. GFP+ mouse ES (mES) cells were co-cultured with DsRed+ primary human fibroblasts (hFb) and then fused using PEG. b, FACS profiles of GFP+ mES, DsRed+ hFb, and GFP+ DsRed+ day 2 heterokaryons (het). c, Representative image of GFP+ DsRed+ day 2 heterokaryons. Hoechst 33342 (blue) denotes the nuclei. The heterokaryon shown has three distinct, unfused bright mouse nuclei, and one uniformly stained human nucleus. Scale bar, 50 μm. d, GFP+ DsRed+ day 2 heterokaryons were cytospun and stained for Ki67 (blue) to assess cell division. The GFP+ heterokaryon has two distinct nuclei (arrow) that are negative for Ki67 (blue) in contrast to the mononuclear cells (arrowheads) that stain positive for Ki67. Scale bar, 50 μm. e, Heterokaryons on days 1, 2 and 3 after fusion were scored on the basis of Ki67 staining, and 98±2% heterokaryons were non-dividing (mean±s.e.m., P, 0.05). f, Heterokaryons, generated using GFP2 ES cells, were enriched using a human fibroblast marker THY1.1 (see Methods) on day 1 post fusion, and stained for BrdU (green) and nuclei (blue) using Hoechst 33258. The heterokaryon (arrow) has three uniformly stained human nuclei and one bright, punctate mouse nucleus, and is negative for BrdU staining. In contrast, the indicated human mononuclear cell (arrowhead) stains positive for BrdU. Scale bar, 50 μm. g, Day 1, 2 and 3 heterokaryons were scored on the basis of nuclear and BrdU staining. DNA replication did not occur in 94±3% heterokaryons (mean±s.e.m., P, 0.05).
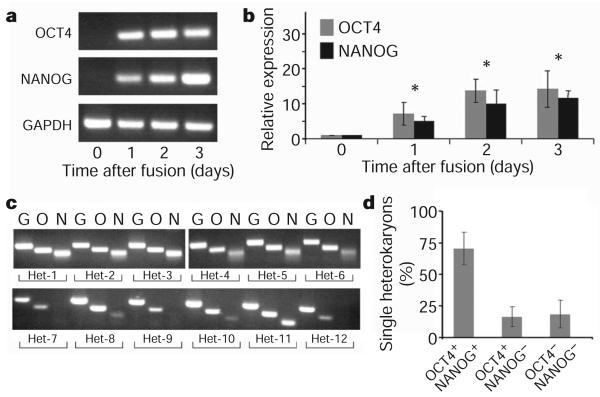
To determine whether ES-cell-specific genes were induced in the human fibroblast nuclei, the induction of human OCT4 and NANOG was assayed relative to ubiquitous GAPDH using species-specific primers (Supplementary Fig. 4). Messenger RNA isolated from sorted heterokaryons 1, 2 and 3 days after fusion was assessed by reverse transcription followed by semiquantitative PCR (RT–PCR) and real-time PCR (Fig. 2a,b). The day 0 controls tested initially were (1) human fibroblasts alone, (2) pre-PEG, unfused co-cultures of mouse ES cells and human fibroblasts, or (3) human fibroblasts treated with PEG to control for the effects of PEG and fusion. All of these day 0 controls gave similar results (data not shown) and the unfused co-culture control was used for all subsequent experiments. Notably, the induction of both human OCT4 and NANOG transcripts was evident as early as day 1 after fusion in heterokaryons but not in controls (Fig. 2a, b and Supplementary Fig. 5), indicating that the onset of expression of two key human pluripotency genes is rapid in heterokaryons. By day 1, expression of human OCT4 and NANOG (normalized to GAPDH in the same samples), had increased 5-fold relative to the control (day 0) and persisted at 10-fold higher levels on days 2 and 3 (Fig. 2b). Human-specific primers were used to determine whether other key pluripotency genes in addition to OCT4 and NANOG were induced using real-time PCR, including KLF4, ESRRB, TDGF1 (also known as CRIPTO), SOX2 and c-Myc (also known as MYC) (Supplementary Fig. 6). ESRRB32 and TDGF1 (ref. 33), which have been shown to be essential for maintaining ES cell self-renewal32 and are targets of OCT4 and NANOG34, were found to be upregulated 3-fold and 2.5-fold, respectively, in heterokaryons on day 2 after fusion (Supplementary Fig. 6). SOX2 is already expressed in human fibroblasts and its promoter is extensively demethylated pre-fusion, in agreement with findings in mouse fibroblasts23; its expression did not increase after fusion. Expression of KLF4 (ref. 35), which is functionally interchangeable with ESRRB, did not change in heterokaryons at day 2 after fusion (Supplementary Fig. 6).
Figure 2. Time course of human fibroblast pluripotency gene expression in heterokaryons at the single-cell level.
a, Human-specific primers against OCT4, NANOG and GAPDH were used for transcript analysis by RT–PCR of unfused co-cultures on day 0 and heterokaryons (mES 3 hFb) isolated on days 1, 2 and 3 after fusion. b, Real-time PCR to assess the upregulation of OCT4 (grey) and NANOG (black) in day 1, 2 and 3 heterokaryons using human-specific primers (mean ± s.e.m.; *P, 0.03). Unfused co-cultures served as day 0 controls and the expression of OCT4 and NANOG was normalized to GAPDH expression. Data shown are from three independent fusion experiments. c, Single heterokaryon nested PCR was used to assess the efficiency of reprogramming in the heterokaryon population. Direct reverse transcription and nested PCR were performed simultaneously on day 3 single heterokaryons, using human-specific primers for GAPDH (G), OCT4 (O) and NANOG (N) as indicated. Twelve heterokaryons analysed from a single fusion experiment are shown. Supplementary Fig. 3 shows 41 heterokaryons analysed from two additional fusion experiments. d, The frequency of heterokaryons expressing both OCT4 and NANOG is 70 ± 13%, showing that a high proportion of heterokaryons initiate reprogramming towards pluripotency. Data shown are a summary of three independent fusion experiments (mean ± s.e.m.).
To assess the efficiency of nuclear reprogramming in human fibroblasts after fusion, single FACS-sorted heterokaryons were analysed by nested RT–PCR for the three human transcripts OCT4, NANOG and GAPDH(control), using two sets of human-specific primers in each case (Fig. 2c). As expected, no human gene products were detected in mouse ES cells (control), and only human GAPDH was detected in human fibroblasts (control) (Supplementary Fig. 4). Notably, 70% of single FACS-sorted heterokaryons from three independent fusion experiments isolated on day 3 after fusion expressed both human OCT4 and NANOG (Fig. 2c, d and Supplementary Fig. 7), showing that a high proportion of heterokaryons initiated reprogramming towards pluripotency. This is in marked contrast to the slow and inefficient induction of OCT4 and NANOG expression in iPS cells (, 0.1% of the total population) in 2–3 weeks3-5,36.
Active DNA demethylation in heterokaryons
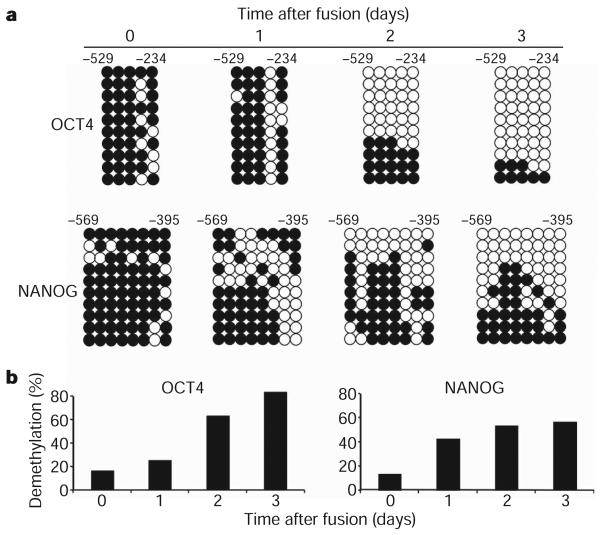
Because DNA demethylation has been shown to be a major limiting step in reprogramming fibroblasts towards iPS cells23, we analysed the time course and extent of demethylation of the human OCT4 and NANOG promoters in heterokaryons. DNA was isolated from heterokaryons on days 1, 2 and 3 after fusion and subjected to bisulphite conversion. Human OCT4 and NANOG promoter regions were amplified by PCR using human- and bisulphite-specific primers (Supplementary Fig. 4) and the products were cloned and sequenced. DNA demethylation was evident at the human OCT4 and NANOG promoters and progressively increased through day 3 (Fig. 3a). In contrast, the β-globin (HBB) HS2 locus remained methylated throughout, indicating that the DNA demethylation was specific (data not shown). The time course and progressive accumulation of demethylated CpG sites in the human OCT4 and NANOG promoters (Fig. 3b) parallel the progressive increase in transcript accumulation observed over the same 3-day time period using real-time PCR (Fig. 2b). Notably, promoter demethylation and activation of pluripotency genes in human somatic cells take place in the absence of Ki67 or BrdU labelling (Fig. 1e, g); thus, demethylation is active and independent of cell division and DNA replication.
Figure 3. Time course of DNA demethylation at pluripotency gene promoters in heterokaryons.
a, Bisulphite sequencing analysis of methylation status of the human OCT4 and NANOG promoter in heterokaryons. Both human OCT4 and NANOG promoters in heterokaryons show rapid and progressive DNA demethylation on days 1, 2 and 3 after fusion compared to the co-culture control. White circles indicate unmethylated, black circles indicate methylated CpG dinucleotides. b, The percentage of demethylation at the human OCT4 and NANOG promoters in heterokaryons after fusion shows a progressive increase in demethylation. Thirty clones were analysed at each time point in two to three independent experiments; ten representative clones are shown.
AID requirement for DNA demethylation and pluripotency
We focused on AID as a candidate factor with a role in mammalian DNA demethylation as it has been detected in mammalian pluripotent germ cells30 and implicated in DNA demethylation in zebrafish post fertilization28. We assayed mouse ES cells and human fibroblasts for AID expression using real-time PCR. Although AID expression in somatic cells is generally thought to be restricted to B lymphocytes, we detected AID mRNA in human fibroblasts as well as mouse ES cells, albeit at greatly reduced levels (5% and 15%, respectively) compared to Ramos, a B-lymphocyte cell line (Supplementary Fig. 8). To investigate the role of AID in these cells, we performed a transient knockdown of mouse and human AID mRNAs by transfection of three distinct, non-overlapping siRNAs to different sequences within the AID coding region, and a fourth siRNA specific to the non-coding 3′ untranslated region (UTR) of AID, to rule out off-target effects and ensure that the results were specific to AID (Supplementary Fig. 9). A fifth siRNA with 50% identity to the AID coding region was used as a control. The extent and timing of knockdown was first confirmed in control mouse ES cells in which siRNA-1, -2, -3 and -4 reduced AID transcripts at day3 post-transfection as compared to the control siRNA (Supplementary Fig. 10, top). AID protein was detected in ES cell lysates. Knockdown of AID protein by 88% correlated well with the mRNA reduction by 81% (Supplementary Fig. 11). In human fibroblasts, AID transcripts were reduced by all of the siRNAs (Supplementary Fig. 10, bottom). These data show that AID is present and can be efficiently reduced by four distinct siRNAs in both ES cells and fibroblasts.
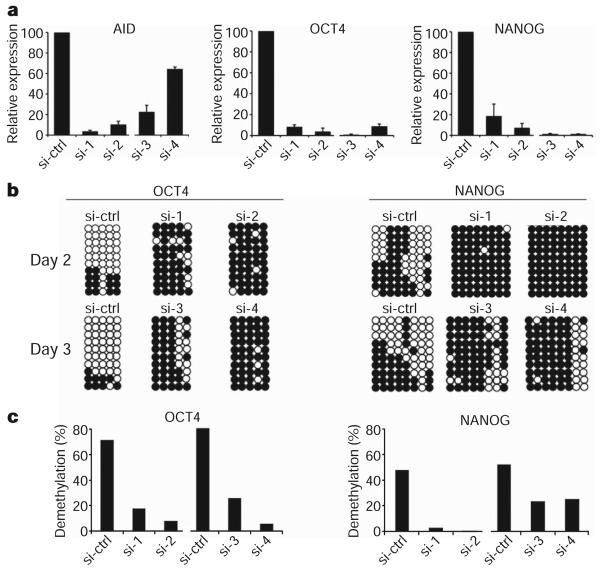
To assess the initiation of reprogramming in heterokaryons subjected to AID knockdown, the expression of human OCT4 and NANOG relative to GAPDH was assessed by real-time PCR. For heterokaryon experiments, siRNAs were transfected into both the mouse ES cells and the human fibroblasts 24 h before fusion (see Supplementary Fig. 1 for scheme). A persistent knockdown of AID was detected by real-time PCR in heterokaryons using each of the four siRNAs on days 2 and 3 after fusion relative to heterokaryons transfected with the control siRNA (Fig. 4a). Notably, both OCT4 and NANOG gene induction was greatly inhibited using siRNA-1 and siRNA-2 on day 2 after fusion, and siRNA-3 and siRNA-4 on day 3 after fusion, as compared to using the control siRNA (Fig. 4a). All siRNAs resulted in a similar block of OCT4 and NANOG activation. These observations indicate that the effect of AID is extremely dosage-sensitive, as a 35% knockdown led to a similar inhibition of pluripotency gene induction as a 96% knockdown. Together, these data show that all four siRNAs to AID used here efficiently inhibited OCT4 and NANOG activation by at least 80%.
Figure 4. Requirement of AID-dependent DNA demethylation for initiation of reprogramming towards pluripotency in heterokaryons.
a, AID and human pluripotency gene expression in heterokaryons subjected to siRNA treatment, as assessed by real time PCR. Day 2 heterokaryons were treated with siRNA-1 and -2 (si-1 and si-2), and day 3 heterokaryons were treated with si-3 and si-4. Total levels of mouse and human AID transcripts were assessed using a set of degenerate primers, whereas human-specific primers were used for human OCT4 and NANOG. Gene expression was normalized internally to GAPDH (degenerate primers) for AID expression, and to human GAPDH for human OCT4 and NANOG expression. The samples were then normalized to the corresponding day 2 or 3 sample treated with the control siRNA (si-ctrl), represented as 100%. b, Human OCT4 and NANOG promoters on days 2 and 3 post fusion after AID knockdown remain methylated. c, The percentage of demethylation at the human OCT4 and NANOG promoters after AID knockdown shows a block in demethylation compared to their respective day 2 and day 3 control samples treated with control siRNA.
To assess the effect of AID knockdown on promoter demethylation, we assayed the CpG methylation status of the human OCT4 and NANOG promoters in heterokaryons. Both day 2 and 3 heterokaryons subjected to AID knockdown using each of the four siRNAs showed a profound inhibition of CpG demethylation in the human OCT4 and NANOG promoters as compared to the control (Fig. 4b, c). A summary of the bisulphite sequencing data for all the siRNA knockdown experiments is shown in Supplementary Fig. 12. In parallel with the reduction in demethylation of the OCT4 and NANOG promoters after AID knockdown, the induction of OCT4 and NANOG transcripts was reduced by at least 80% on days 2 and 3, relative to the control (Fig. 4a). These data show that promoter demethylation is critical to the expression of these two pluripotency genes and that AID is required for mammalian DNA demethylation in somatic cell reprogramming.
To investigate further the requirement and specificity of AID for initiating reprogramming, we tested its ability to accelerate reprogramming, and to rescue the DNA demethylation block caused by the siRNA knockdown in heterokaryons. Therefore, human AID (hAID) was transiently overexpressed in mouse ES cells before siRNA transfection (see scheme in Supplementary Fig. 1). In two separate experiments, when AID was overexpressed twofold or fourfold relative to the control in heterokaryons at day 1 after fusion, there was no acceleration in promoter demethylation or reprogramming (Supplementary Fig. 13). However, after overexpression of hAID in heterokaryons undergoing transient knockdown by siRNA-3, there was a complete rescue of NANOG promoter demethylation and gene expression and a partial rescue of OCT4 promoter demethylation and gene expression (Supplementary Fig. 14). The lack of reprogramming acceleration and partial rescue observed for OCT4 could possibly be due to the kinetics of human OCT4 and NANOG promoter demethylation, which in heterokaryons may require at least 1 day to occur, or by the lack of other factors that work together with AID. These data show that the added hAID is functional and rule out any non-specific effects of the siRNA, further confirming the specific and essential role of AID in DNA demethylation at the onset of reprogramming towards pluripotency.
Binding of AID to methylated pluripotency gene promoters
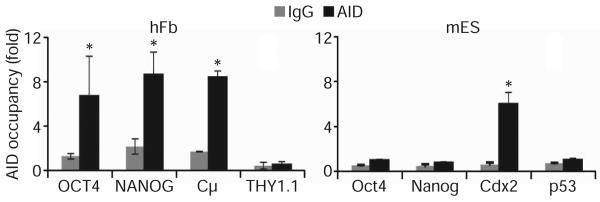
To validate the role of AID in DNA demethylation of human OCT4 and NANOG promoters further, we tested whether AID specifically binds to these promoter regions by performing chromatin immunoprecipitation (ChIP) experiments using an anti-AID antibody, previously validated for ChIP37. The promoter regions assessed in ChIP experiments were designed to be within the OCT4 and NANOG promoter regions that were analysed for CpG demethylation by bisulphite sequencing (Fig. 5 and Supplementary Fig. 15). In human fibroblasts, the ChIP analyses showed significant binding of AID to both human OCT4 (sixfold) and human NANOG (eightfold) promoters (Fig. 5). Thus, AID binds to the heavily methylated promoter regions of human OCT4 and NANOG in fibroblasts that undergo demethylation during reprogramming. As controls, AID binding to the promoter of the IgM constant region (Cμ) was significant, as expected38, whereas no binding was observed for THY1.1 (also known as CD90.1), which is expressed in fibroblasts.
Figure 5. AID binding to pluripotency gene promoters by chromatin immunoprecipitation.
ChIP with anti-AID antibody was performed in human fibroblasts (left) and mouse ES cells (right). AID occupancy is shown relative to background IgG signal (mean±s.e.m.). Significant AID binding was detected in human fibroblasts for the methylated NANOG and OCT4 promoters as well as for the positive control Cμ (P<0.002). In mouse ES cells, AID binding was detected only in the positive control, Cdx2 promoter region (P<0.007), whereas no significant binding was observed for the unmethylated Oct4 and Nanog promoters.
In contrast to fibroblasts, no AID binding was observed at the promoter regions of Oct4 and Nanog in mouse ES cells despite the higher levels of AID protein in ES cells, presumably because these promoters are active and demethylated (Fig. 5). As controls, AID binding was detected at the promoter of Cdx2, a gene not expressed in undifferentiated ES cells39, but was absent from the p53 (also known as Trp53) promoter, as previously reported37. Together, these findings provide strong support for a direct involvement of AID in DNA demethylation and the sustained expression of human NANOG and OCT4 leading to the onset of reprogramming towards pluripotency.
Discussion
DNA demethylation is essential to overcome gene silencing and induce temporally and spatially controlled expression of mammalian genes, yet no consensus mammalian DNA demethylase has been identified despite years of effort27. Evidence of DNA demethylation by 5-methyl-cytosine DNA glycosylases has been shown in plants40,41, but mammalian homologues such as thymine DNA glycosylase (TDG) or the methyl-CpG-binding domain protein 4 (MBD4) have not exhibited comparable functions42,43. As an alternative to direct demethylation, it has been postulated that the deamination of cytosine followed by DNA repair could lead to DNA demethylation44. AID belongs to a family of cytosine deaminases (AID, APOBEC1, 2 and 3 subgroups) that have established roles in generating antibody diversity in B cells, RNA editing and antiviral response45. Both AID and APOBEC1 are expressed in progenitor germ cells, oocytes and early embryos and have a robust 5-methyl-cytosine deaminase activity in vitro30, resulting in a T-G mismatch that is repaired through the base excision DNA repair (BER) pathway, and could theoretically lead to DNA demethylation without replication. Recently in zebrafish embryos, AID was implicated as a member of a tripartite protein complex along with MBD4 and GADD45A, causing cytosine deamination and leading to base excision by MBD4 (ref. 28). The third component GADD45A lacks enzymatic activity and its role in repair-mediated DNA demethylation and gene activation in Xenopus oocytes remains a matter of debate46,47.
Our data provide new evidence implicating AID in active DNA demethylation in mammalian cells, and demonstrate that AID-dependent DNA demethylation is an early epigenetic change necessary for the induction of pluripotency genes in human fibroblasts. Knockdown of AID in heterokaryons prevented DNA demethylation of the human OCT4 and NANOG promoters and the expression of these pluripotency factors by fibroblast nuclei. Indeed, initiation of nuclear reprogramming towards pluripotency was inhibited in human somatic fibroblasts when AID-dependent DNA demethylation was reduced, providing strong evidence that AID is a crucial regulator for the onset of reprogramming. The inhibitory effects of AID reduction were rescued by hAID overexpression with a complete rescue observed for NANOG and a partial rescue observed for OCT4. Moreover, AID binding was observed at silent methylated OCT4 and NANOG promoters in fibroblasts but not in active unmethylated OCT4 and NANOG promoters in ES cells, demonstrating its specific role in DNA demethylation. Previous studies that used cell fusion to analyse reprogramming to pluripotency could not distinguish the products of the two nuclei because the cells were derived from the same species24,25. A recent study used heterokaryons to reprogram lymphocytes towards pluripotency, however, only modest demethylation changes at the OCT4 promoter were observed that did not increase with time48, possibly owing to differences in the cell types and methods used for isolating heterokaryons. The high efficiency of reprogramming in heterokaryons achieved here allowed the discovery of a regulator critical to the induction of five pluripotency genes including OCT4 and NANOG, the first known markers of stable reprogramming leading to the generation of iPS cells. The heterokaryon platform can now be exploited (1) to determine the other components of the mammalian DNA demethylation complex (glycosylase and other DNA repair enzymes) that are likely to work together with AID to mediate active DNA demethylation (Fig. 6), and (2) to perform an unbiased search for other regulators of nuclear reprogramming by screening for human genes that are expressed immediately after cell fusion. Future studies will determine whether expression of AID alone or in conjunction with these other molecules will enhance the generation of iPS cells.
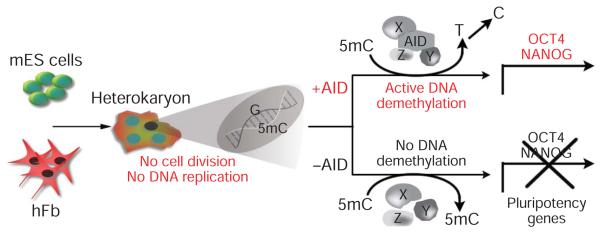
Figure 6. Model for AID-dependent active DNA demethylation in reprogramming.
AID-mediated reprogramming and DNA demethylation in heterokaryons takes place in the absence of cell division or DNA replication, showing that AID is a component of an active DNA demethylation complex in mammals. The other putative components of this mammalian DNA demethylase complex (X, Y and Z) that may act together with the deaminase, AID, remain to be identified. 5mC, 5-methyl-cytosine.
METHODS SUMMARY
Heterokaryon generation and isolation
GFP+ mouse ES cells and DsRed+ human fetal lung primary fibroblasts were generated by transduction with retroviral constructs and fused to form non-dividing, multinucleated heterokaryons as previously described31. GFP+ DsRed+ heterokaryons were sorted twice by flow-cytometry (FACSVantage SE, BD Biosciences) and analysed for gene expression and methylation.
Immunofluorescence
For antibody staining, GFP+ DsRed+ heterokaryons were cytospun and stained. Images were acquired using an epifluorescent microscope (Axioplan2, Carl Zeiss MicroImaging), Fluar ×20/0.75 or ×40/0.90 objective lens, and a digital camera (ORCA-ER C4742-95, Hamamatsu Photonics). The software used for acquisition was OpenLab 4.0.2 (Improvision).
BrdU was added to mouse ES cell and human fibroblast co-cultures 3 h after PEG-induced fusion. Labelling and antibody staining was performed using the BrdU Labelling and Detection Kit I (Roche).
Single-cell RT–PCR
Single cells were directly sorted by FACS. Collection of cells and the subsequent two-step multiplex nested RT–PCR were performed directly from cell lysates using all three human and gene-specific primers (OCT4, NANOG and GAPDH) as described in Methods.
DNA methylation analyses
Bisulphite treatment was performed on DNA isolated from FACS sorted heterokaryons (2,000–10,000 cells) using the Epitect Bisulfite Kit (Qiagen). Nested PCR for regions of the human OCT4 (ref. 18) and NANOG promoters was performed using human- and bisulphite-specific primers. PCR products from second-round bisulphite-specific PCR amplification were cloned and sequenced as described previously17.
siRNA transfection
For siRNA transfection, ES cells and primary fibroblasts were plated at 50–60% confluence the day before transfection. siRNAs (Dharmacon) were transfected using siImporter (Millipore).
ChIP
ChIP was performed as previously described49. ChIP data was presented as normalized to input DNA and as the mean± s.e.m.
Statistical analysis
Data are presented as the mean±s.e.m. Comparisons between groups used the Student's t-test assuming two-tailed distributions.
METHODS
Heterokaryon generation and isolation by flow cytometry
GFP+ mouse ES cells and DsRed+ human fetal lung primary fibroblasts were generated by transduction with retroviral constructs as previously described31, and fused to form non-dividing, multinucleated heterokaryons. Cells were first co-cultured for 12 h in ES cell media and then treated with PEG 1500 (Roche) for 2 min at 37 °C, followed by four successive washes with DMEM. ES cell media was replaced after washing and every 12 h thereafter. GFP+ DsRed+ heterokaryons were sorted twice by flow-cytometry (FACSVantage SE, BD Biosciences) and analysed for gene expression and methylation.
Immunofluorescence
Heterokaryons were sorted twice in PBS with 2.5% (v/v) goat serum and 1 mM EDTA, and cytospun for 5 min. The cytospun GFP+DsRed+ heterokaryons were stained with Hoechst 33342, and imaged. For antibody staining, cytospun cells were fixed, permeabilized and blocked using 20% FBS in PBS. Cells were incubated with the mouse anti-Ki67 antibody (Dako Denmark A/S) at 1:100 dilution in blocking buffer for 1 h, rinsed three times in PBS, and then incubated with a goat anti-mouse Cascade blue secondary antibody (Millipore) at 1:500 dilution for 30 min, rinsed three times, mounted with Fluoromount-G and imaged. Images were acquired using an epifluorescent microscope (Axioplan2, Carl Zeiss MicroImaging), Fluar ×20/0.75 or ×40/0.90 objective lens, and a digital camera (ORCA-ER C4742-95, Hamamatsu Photonics). The software used for acquisition was OpenLab 4.0.2 (Improvision).
BrdU was added to mouse ES cell and human fibroblast co-cultures 3 h after PEG-induced fusion. Labelling and antibody staining was performed using the BrdU Labelling and Detection Kit I (Roche).
Analysis of gene expression
RNA was prepared from ES cells, fibroblasts and twice-sorted heterokaryons at different times post fusion or after siRNA treatment using the RNeasy micro kit (Qiagen). Total RNA for each sample was reverse transcribed using the Superscript First-Strand Synthesis System for RT–PCR (Invitrogen). The reverse-transcribed material was subjected to PCR using Go GreenTaq DNA polymerase (Promega). Human-specific primers were designed for analysing the expression of OCT4, NANOG and GAPDH. Primers used for AID and GAPDH in the siRNA treatment experiments amplify both human and mouse transcripts to assess the total levels of AID and GAPDH in heterokaryons. Human-specific primers used for RT–PCR and quantitative PCR are: OCT4 F 5′-TCGAGAACCGAGTGAGAGGC-3′, R 5′-CACACTCGGACCACATCCTTC-3′; NANOG F 5′-CCAACATCCTGAACCTCAGCTAC-3′, R 5′-GCCTTC TGCGTCACACATT-3′; GAPDH F 5′-TGTCCCCACTGCCAACGTGTCA-3′, R 5′-AGCGTCAAAGGTGGAGGAGTGGGT-3′. Non-species-specific primer sequences for assessing knockdown after siRNA treatment are as follows: GAPDH F 5′-ACCACAGTCCATGCCATCAC-3′, R 5′-TCCACCACCCTGTTGCTGTA-3′; AID F 5′-AAAATGTCCGCTGGGCTAAG-3′, R 5′-AGGTCCCAGTCCGAGATGTAG-3′.
Real-time PCR
Real-time PCR was performed using an ABI 7900HT Real time PCR system using the Sybr Green PCR mix (Applied Biosystems). Samples were cycled at 94 °C for 2 min, 40 cycles of: 94 °C for 20s, 58 °C for 45 s.
DNA methylation analyses
FACS-sorted heterokaryons (2,000–10,000 cells) were collected in 20 μl PBS. DNA was extracted using the DNeasy Tissue Kit (Qiagen). Bisulphite treatment was performed using the Epitect Bisulfite Kit (Qiagen). Nested PCR for regions of the human OCT4 and NANOG promoters was performed using human- and bisulphite-specific primers. Samples were cycled for the first and second nested PCR at 94 °C for 2 min, 30 cycles of 94 °C for 20 s, 61 °C for 30 s, 68 °C for 30 s. PCR products from second-round bisulphite-specific PCR amplification were cloned and sequenced as described before17.
Single heterokaryon nested RT–PCR
Single-cell collection. Single heterokaryons were directly sorted by FACS (FACSVantage SE, BD Biosciences) into PCR tubes containing 9-μl aliquots of RT–PCR lysis buffer. The buffer components included commercial RT–PCR buffer (SuperScript One-Step RT–PCR Kit Reaction Buffer, Invitrogen), RNase inhibitor (Protector RNase Inhibitor, Roche) and 0.15% IGEPAL detergent (Sigma). After a short pulse-spin, the PCR-tubes were immediately shock-frozen and stored at −80 °C for subsequent analysis.
Two-step multiplex nested single-cell RT–PCR. Cell lysates were first reverse-transcribed using the human-specific primer pairs for OCT4, NANOG and GAPDH (Supplementary Table 2, external primers; Supplementary Fig. 1b) using SuperScript One-Step RT–PCR Kit (Invitrogen). In brief, the RT–PCR was performed in the same PCR cell-lysis tubes by addition of an RT–PCR-reaction mix containing the gene-specific primer pairs and RNase inhibitor. Genomic products were excluded by designing and using intron-spanning primer sets for the first and second round PCR and nested RT–PCR to ensure greater specificity. In the first step, the reverse transcription reactions were carried out at 55 °C for 30 min, followed by a 2-min step at 94 °C. Subsequently, 30 cycles of PCR amplification were performed as follows: 94 °C for 30 s; 58 °C for 30 s; 68 °C for 30s. In the final PCR step, the reactions were incubated for 3 min at 68 °C. The completed reactions were stored at 4 °C.
In the second step of the PCR protocol, the completed RT–PCR reaction from the first step was diluted 1:1 with water. One per cent of these reactions were replica transferred into new reaction tubes for the second round of PCR, which was performed for each of the genes separately using nested gene-specific internal primers, for greater specificity, in a total reaction volume of 20 μl (Platinum Taq Super-Mix HF, Invitrogen). Thirty cycles of PCR amplification were performed as follows: 94 °C for 30 s; 58 °C for 30 s; 68 °C for 30 s. In the final PCR step, the reactions were incubated for 3 min at 68 °C. The completed reactions were stored at 4 °C. The second-round PCR products were then subjected to gel electrophoresis using one-fifth of the reaction volumes and 1.4% agarose gels.
THY1.1 enrichment of heterokaryons
GFP− (non-GFP) mES and DsRed+ hFb co-cultures treated with PEG were trypsinized and resuspended in 3 ml FACS buffer. Cells were incubated for 30 min at room temperature with biotin mouse anti-human CD90.1 (BD Pharmingen) at a dilution of 1:5,000. The cells were washed once, resuspended in 3 ml FACS buffer, and incubated for 30 min at room temperature with 10 μl of Dynabeads Biotin Binder (Invitrogen). Beads were collected by magnetic isolation, washed twice and the enriched heterokaryons were cytospun.
Western blots
To visualize AID protein knockdown in mouse ES cells, cell lysates were collected 3 days after transfection with control siRNA or siRNA-3. Detection of AID in these samples was performed from 170 μg of whole cell lysate using anti mouse-AID (L7E7, Cell Signaling, dilution 1:500). The membrane was stripped and probed with anti-mouse α-tubulin (Sigma, dilution 1:20,000) for the loading control.
Supplementary Material
Acknowledgements
We thank O. Alkan for generating retroviral constructs for GFP and DsRed expression; R. Doyonnas for helping to standardize FACS isolation of heterokaryons; Y. Liao for performing the initial experiments showing demethylation in heterokaryons; M. PajÇini for technical assistance with the BrdU experiments; D. Schatz and S. Unniraman for providing the human AID construct; F. Alt for his generous provision of antibody to AID; J. A. Dahl for discussions about chromatin immunoprecipitation assays; H. Chang for use of the Bioruptor sonicator; and M. Wernig, G. Sen, C.-Z. Chen and J. Pomerantz for comments on the manuscript. This work was supported by a National Science Foundation Graduate Research Fellowship awarded to J.J.B., National Institutes of Health (NIH) training grant AI007328 to M.D., and NIH grants AG009521, AG024987 and support from the Baxter Foundation to H.M.B.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Gurdon JB. Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 1962;4:256–273. doi: 10.1016/0012-1606(62)90043-x. [DOI] [PubMed] [Google Scholar]
- 2.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs' eggs. Proc. Natl Acad. Sci. USA. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- 8.Chiu CP, Blau HM. Reprogramming cell differentiation in the absence of DNA synthesis. Cell. 1984;37:879–887. doi: 10.1016/0092-8674(84)90423-9. [DOI] [PubMed] [Google Scholar]
- 9.Chiu CP, Blau HM. 5-Azacytidine permits gene activation in a previously noninducible cell type. Cell. 1985;40:417–424. doi: 10.1016/0092-8674(85)90155-2. [DOI] [PubMed] [Google Scholar]
- 10.Blau HM, et al. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 11.Blau HM, Baltimore D. Differentiation requires continuous regulation. J. Cell Biol. 1991;112:781–783. doi: 10.1083/jcb.112.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron MH, Maniatis T. Rapid reprogramming of globin gene expression in transient heterokaryons. Cell. 1986;46:591–602. doi: 10.1016/0092-8674(86)90885-8. [DOI] [PubMed] [Google Scholar]
- 13.Wright WE. Expression of differentiated functions in heterokaryons between skeletal myocytes, adrenal cells, fibroblasts and glial cells. Exp. Cell Res. 1984;151:55–69. doi: 10.1016/0014-4827(84)90355-0. [DOI] [PubMed] [Google Scholar]
- 14.Spear BT, Tilghman SM. Role of α-fetoprotein regulatory elements in transcriptional activation in transient heterokaryons. Mol. Cell. Biol. 1990;10:5047–5054. doi: 10.1128/mcb.10.10.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blau HM. Differentiation requires continuous active control. Annu. Rev. Biochem. 1992;61:1213–1230. doi: 10.1146/annurev.bi.61.070192.010025. [DOI] [PubMed] [Google Scholar]
- 16.Pavlath GK, Blau HM. Expression of muscle genes in heterokaryons depends on gene dosage. J. Cell Biol. 1986;102:124–130. doi: 10.1083/jcb.102.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Pomerantz JH, Sen G, Palermo AT, Blau HM. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc. Natl Acad. Sci. USA. 2007;104:4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 20.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 21.Eminli S, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nature Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 23.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 25.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 26.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nature Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 27.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 30.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 31.Palermo A, et al. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. FASEB J. 2009;23:1431–1440. doi: 10.1096/fj.08-122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharya B, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 34.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 35.Feng B, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nature Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 36.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nature Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vuong BQ, et al. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nature Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. The AID enzyme induces class switch recombination in fibroblasts. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 39.Deb K, Sivaguru M, Yong HY, Roberts RM. Cdx2 gene expression and trophectoderm lineage specification in mouse embryos. Science. 2006;311:992–996. doi: 10.1126/science.1120925. [DOI] [PubMed] [Google Scholar]
- 40.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 41.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 42.Cortázar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst.) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Millar CB, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 44.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 46.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 47.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira CF, et al. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 2008;4:e1000170. doi: 10.1371/journal.pgen.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature Protocols. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.