Abstract
Data on polyomavirus genomic diversity has greatly expanded in the past few years. The implications of viral DNA sequence variation on the performance of molecular diagnostic assays have not been systematically examined. 716 BK, 1626 JC and 73 SV40 virus sequences available in GenBank were aligned using Clustal-X. Five different published BKV PCR assays currently in use at major medical centers were evaluated for primer and probe mismatches with available GenBank sequences. Coverage of naturally occurring BKV strains varied amongst different assay methods. Targeted viral sequences showed major mismatch with primer or probe sequence in up to 30.7% of known BKV strains. BKV subtypes IVa, IVb, and IVc were more prone to this problem, reflecting common use of Type I Dun sequence for assay design. Despite the known polymorphism of this gene, 484 VP-1 sequences with conserved areas potentially suitable for PCR assay design are available. Assay targets in the Large T-antigen and agnogene are less subject to genetic variation, but sequence information corresponding to the latter two genes is available only for 164 and 174 published strains respectively. Cross reactivity of appropriately selected BKV primers with JCV and SV40 sequences available in current databases was not a significant problem.
INTRODUCTION
Polyomaviruses (PV) belong to the family Polyomaviridae. Virions are 45nm in diameter with a 5 kb circular double stranded genome. The species most relevant to human disease are BK Virus (BKV), JC Virus (JCV) and Simian Virus (SV40). The viral genome is arranged in three general regions: non-coding control region (NCCR), the early coding region (coding for the small and large T antigens), and the late coding region coding for the viral capsid proteins (VP-1, VP-2, VP-3) and agnoprotein (14–19)[Demeter, 1995; Shah, 1995]. The NCCR contains the origin of replication and regulatory regions containing enhancer elements that are important activators of viral transcription. The T antigen promotes viral replication, binds to tumor suppressor proteins Rb and p53, and stimulates host cell entry into the cell cycle [Eckner et al., 1996; Gomez-Lorenzo et al., 2003; Roy et al., 2003; Valls et al., 2003]. VP-1, VP-2, and VP-3 are structural proteins required for the assembly of complete virions.
The viral capsid coding regions display considerable genetic heterogeneity, and this feature has been used to divide BKV into distinct subtypes I, II, III, and IV [Randhawa et al., 2002]. Subtype I is the most prevalent in all major geographic areas with a prevalence range from 46–82%. A possible exception is the Chinese and Mongolian region, where a 54% prevalence for type IV has been reported [Zheng et al., 2007]. Subtype IV is generally the second most prevalent type, and although, subtype IV strains have been reported from Europe and USA [Baksh et al., 2001; Di Taranto et al., 1997; Jin, 1993], these are more frequent in northeast Asia (12–54%). The frequency of subtype IV in Africa is significantly lower than in Europe and Asia. Subtypes II and III are overall quite rare with frequencies of 0–6% and 0–9% respectively. In one African study subtype III was commoner than type IV (9% versus 5%).
There is now enough genetic information available about BKV to suggest the occurrence of subgroups within subtypes I and IV [Ikegaya et al., 2006; Nishimoto et al., 2006; Nishimoto et al., 2007; Takasaka et al., 2004; Zheng et al., 2007; Zhong et al., 2007]. It appears that subgroups of genotype I may have predilection for specific geographic regions, such as subgroup 1a for Africa, 1b-1 for Southeast Asia, 1b-2 for Europe, and 1c for northeast Asia. The proportion of total type 1 subtypes represented by the aforementioned subgroups in the corresponding geographic regions is 75%, 90%, 77.5%, and 64% for 1a, respectively [Zheng et al., 2007]. In one study, differences in prevalence between Europe and northeast Asia are said to be statistically significant [Ikegaya et al., 2006]. There are also differences in geographic distribution for subgroups within subtype IV. Thus, subtype IVa1 comprised 8/15 (53%) of subtype IV strains obtained from southeast Asia (Philippines, Vietnam,and Mynamar). Subtype IVb1 and IVb2 accounted for 40% and 55% respectively of 20 subtype IV strains obtained from Korea and Japan. In contrast, 21/26 (81%) of chinese strains were subtype IVc1, and all 22 subtype IV strains from Europe were subgroup IVc2 [Nishimoto et al., 2007]. It is not yet clear if these geographic variations reflect ethnic background or clinical circumstances of sample collection. Environmental factors involved in person to person transmission may also be important. Japanese-Americans in California tend to carry European subtype 1b-2, and not 1c typical of native Japanese subjects [Yogo et al., 2007].
Currently used PCR assays were developed several years ago when the syndrome of BKVN in kidney transplant patients was first recognized. As noted above, our knowledge of BKV genomic diversity has increased enormously in the last few years [Chen et al., 2006; Chen et al., 2004; Ikegaya et al., 2006; Ikegaya et al., 2005; Krumbholz et al., 2006; Nishimoto et al., 2006; Nishimoto et al., 2007; Nukuzuma et al., 2006; Sharma et al., 2006; Takasaka et al., 2006; Yogo et al., 2007; Zheng et al., 2005a; Zheng et al., 2005b; Zhong et al., 2007]. Some publications report a relatively stable genome in asymptomatic subjects [Takasaka et al., 2006], but there is also evidence that BKV can behave as a rapidly evolving virus in disease states associated with active viral replication[Randhawa et al., 2002]. It has, therefore, become necessary for laboratories to re-evaluate the ability of their assays to capture all the BKV strains currently circulating in humans. PCR monitoring techniques will be less effective without adjustments that take into account continuous appearance of new viral strains. The present study seeks to define the magnitude of this problem by analyzing all BKV sequence information published to date.
MATERIALS AND METHODS
Sequence retrieval
Publicly available whole genome sequences (WGS) and partial sequences were retrieved from GenBank. All data were collected before 1/1/2008. Non coding control region (NCCR) sequences were not considered because of the high rate of genetic recombination in this region.. Redundancies in the data were excluded by observing the following rules: (i) if identical whole genome sequences were submitted by more than one laboratory, only one sequence was retained; (ii) if partial BKV sequences matched a whole genome sequence, only the latter was included in the alignment. Altogether 716 unique BKV sequences were retained, including 161 WGS and 555 partial sequences. For the related JC polyomavirus a total of 1626 unique sequences were retrieved, of which 450 were WGS and 1176 were partial sequences. A much smaller data set of polyomavirus SV40 sequences was available, comprised of 30 unique WGS and 43 unique partial sequences from viral genomic areas outside of the NCCR. A complete listing of the sequences used in this study is provided in supplementary data tables 1–3.
Sequence alignment
DNA sequences were aligned using Clustal X which is a windows version of ClustalW, with default multi alignment parameters [Thompson et al., 1994]. Nucleotides were numbered using the system of Seif et al., in which #1 is assigned to the nucleotide upstream of the start codon of the T antigen [Seif et al., 1979]. BKV MM sequence (V01009) with an alternate numbering system was first such as BKV strain MM (V01109) was first renumbered according to Seif’s schema. The partial BKV sequence D00678 which spans the T-antigen and NCCR regions was truncated at position 1 to facilitate alignment. SV40 sequences AF168993, AF169001, EU268284, AF136002, AF168995, AF136001, AF136000, AF136003, AF168996, AF168997, AF168998, AY148408, AY148409, AY148410, AY148411, AY148412, AY148413, AF169000, AJ276576, BK006135 were reverse translated. Alignments were manually adjusted using BioEdit (Tom Hall, Department of Microbiology, North Carolina State University, North Carolina, USA). http://www.mbio.ncsu.edu/BioEdit/BioEdit.html). Separate alignments were performed for BKV, JCV, and SV40 strains. The BKV Dunlop sequence was included in each alignment as a reference point for evauating primer and probe mismatches.
Analysis of BKV Genomic Variation
Genomic variation analysis was based on 161 unique BKV whole genome sequences. Polymorphic sites were accepted as those (i) where at least two alternate nucleotides were present, and (ii) the frequency of substitutiion was greater than 1%, which means that at least two out of 161 strains possessed variant nucleotides. Areas of deletion were not considered in the evaluation of polymorphism. Assignment of subtype I, II, III or IV to each sequence was based on the designation provided by the submitting investigators, generally based on the schema defined by Jin et al [Jin et al., 1993]. For analysis of Large T gene included only the 2 exons were included; the intron sequence was omitted.
Evaluation of Primer and Probe Sequences Used in Clinical Diagnosis
The purpose of this evaluation was to determine the extent to which diagnostic PCR assays being performed in clinical practice capture BKV genomic diversity. We chose five published assays, four of which are from major transplant centers. Two assays, which we will designate as assay #1 and 2, target the Large T-antigen [Hirsch et al., 2001; Limaye et al., 2001]. Assays #3 and 4 target the VP-1 region [Ding et al., 2002; Randhawa et al., 2004], while assay #5 is based on the agnogene [Leuenberger et al., 2007]. The complete sequence forward (F) and reverse (R) primers and probe (P), and their positions on the genome are provided in Table 1. Assay #3 uses SyBr Green dye instead of a virus specific probe. Primer and probe sequences were systematically evaluated for identity or non-identity with homologous sequences in our BK, JC, and SV40 sequence alignments. Viral strains showing a 100% match were designated as a perfect match (PM) for the primer or probe being evaluated. Any mismatch < 8 nucleotides from the 3’end of a primer was counted as a major mismatch [Rychlik, 1995]. All other mismatches (8 or more nucleotides away) were characterized as a minor mismatch. For DNA probes, mismatches at 5’ and 3’ ends are more likely to cause instability compared to internal mismatches [Suzuki et al., 2007]. Initially, this analysis was carried out on all viral sequences, irrespective of genotype (Tables 2–4). Subsequently a subtype specific subanalysis was performed using 161 BKV WGS that are publicly available (see Table 5).
Table 1.
Primer and Probe Sequences of Assay Evaluated in This Study
| Ass ay |
Target Gene |
Type | Sequence | Genomic position |
Covered sequences |
|---|---|---|---|---|---|
| 1 | Tag | Forward Primer | 5' AGTCTTTAGGGTCTTCTACC 3' | 4392–4411 | 161 |
| ReversePrimer | 5' GGTGCCAACCTATGGAACAG 3' | 4548–4567 | 161 | ||
| Probe | 5' GCAACAGCAGATTCTCAACACTCAACA 3' | 4432–4458 | 161 | ||
| 2 | Tag | Forward Primer | 5' AGCAGGCAAGGGTTCTATTACTAAAT 3' | 4329–4354 | 161 |
| Reverse Primer | 5' GAAGCAACAGCAGATTCTCAACA 3' | 4439–4461 | 161 | ||
| Probe | 5' AAACTGGTGTAGATCAGAGGGAAAGTCTT 3' | 4369–4397 | 161 | ||
| 3* | VP1 | Forward Primer | 5' GCAGCTCCCAAAAAGCCAAA 3' | 1600–1619 | 164 |
| Reverse Primer | 5' CTGGGTTTAGGAAGCATTCTA 3' | 1706–1726 | 704 | ||
| 4 | VP1 | Forward Primer | 5' TGCTGATATTTGTGGCCTGTTTACTA 3' | 2355–2380 | 166 |
| Reverse Primer | 5' CTCAGGCGGATCTTAAAATATCTTG 3' | 2414–2438 | 166 | ||
| Probe | 5' AGCTCTGGAACACAACAGTGGAGAGGCC 3' | 2383–2410 | 166 | ||
| 5 | Agno | Forward Primer | 5’ CCATGGTTCTGCGCCAGCTG 3’ | 386–405 | 161 |
| Reverse Primer | 5' CTAGGAGTCTTTTACAGAGTCT 3' | 567–588 | 161 |
This assay detected the amplified PCR product by the SYBR green method rather than a target specific hydrolysis probe.
Table 2.
Analysis of Primer and Probe Mismatch with All Published BKV Sequences*
| Assay | Primer | Perfect match | Minor Mismatch | Major Mismatch |
|---|---|---|---|---|
| 1 | Forward | 161/161(100%) | 0/161 (0%) | 0/161 (0%) |
| Reverse | 160/161(99.38%) | 1/161 (0.62%) | 0/161 (0%) | |
| Probe | 74/161 (45.96%) | 51/161 (54.04%) | 36/161 (0%) | |
| 2 | Forward | 96/161 (59.63%) | 64/161 (39.77%) | 1/161 (0.62%) |
| Reverse | 105/161(65.22%) | 0/161 (34.78%) | 56/161 (0%) | |
| Probe | 19/161 (11.80%) | 142/161(88.20%) | 0/161 (0%) | |
| 3 | Forward | 163/164(99.39%) | 0/164 (0%) | 1/164 (0.61%) |
| Reverse | 484/704(68.75%) | 283/704(40.91%) | 7/704 (0.28%) | |
| 4 | Forward | 103/166(62.05%) | 63/166 (7.8%) | 0/166 (0%) |
| Reverse | 18/166 (10.84%) | 148/166(89.16%) | 0/166 (0%) | |
| Probe | 107/166(62.65%) | 6/166 (37.35%) | 51/166 (0%) | |
| 5 | Forward | 86/161 (53.42%) | 75/161 (46.58%) | 0/161 (0%) |
| Reverse | 160/161(99.38%) | 1/161 (0.62%) | 0/161 (0%) |
The variable denominators reflect the fact that not viral strains described in the literature have sequence information available corresponding to the area of the viral genome targeted by different assays.
Table 4.
Analysis of Primer and Probe Mismatch with SV40 Virus Sequences*
| Assay | Type | Perfect match | Minor Mismatch | Major Mismatch |
|---|---|---|---|---|
| 1 | Forward | 0/40(0%) | 39/40(97.5%) | 1/40 (2.5%) |
| Reverse | 0/40(0%) | 0/40 (0%) | 40/40(100%) | |
| Probe | 0/40(0%) | 0/40 (0%) | 40/40(100%) | |
| 2 | Forward | 0/36(0%) | 0/36 (0%) | 36/36(100%) |
| Reverse | 0/40(0%) | 0/40 (0%) | 40/40(100%) | |
| Probe | 0/40(0%) | 0/40 (0%) | 40/40(100%) | |
| 3 | Forward | 0/36(0%) | 0/36 (0%) | 36/36(100%) |
| Reverse | 0/35(0%) | 0/35 (0%) | 35/35(100%) | |
| 4 | Forward | 0/43(0%) | 0/43 (0%) | 43/43(100%) |
| Reverse | 0/43(0%) | 0/43 (0%) | 43/43(100%) | |
| Probe | 0/43(0%) | 0/43 (0%) | 43/43(100%) | |
| 5 | Forward | 0/40(0%) | 0/40 (0%) | 40/40(100%) |
| Reverse | 0/39(0%) | 38/39(97.44%) | 1/39 (2.56%) |
Numbers in the first column refer to PCR assays referenced in table 2. The variable denominators in the data cells reflect the fact that not viral strains described in the literature have sequence information available corresponding to the area of the viral genome targeted by different assays.
Table 5.
Subtype specific coverage of different assays.
| As say |
Primer, Probe |
Coverage of different subtype and subgroups | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I(105) | Ia(13) | Ib1(28) | Ib2(42) | Ic(22) | II(3) | III(2) | ||||||||||||||||
| PM | Min or MM |
Major MM |
PM | Min or MM |
Majo r MM |
PM | Min or MM |
Majo r MM |
PM | Min or MM |
Majo r MM |
PM | Min or MM |
Majo r MM |
PM | Min or MM |
Majo r MM |
PM | Min or MM |
Majo r MM |
||
| 1 | Forward | 105 | 0 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 42 | 0 | 0 | 22 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 |
| Reverse | 104 | 1 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 41 | 1 | 0 | 22 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | |
| Probe | 74 | 0 | 31 | 13 | 0 | 0 | 0 | 0 | 28 | 42 | 0 | 0 | 19 | 0 | 3 | 0 | 3 | 0 | 0 | 2 | 0 | |
| 2 | Forward | 96 | 8 | 1 | 13 | 0 | 0 | 27 | 0 | 1 | 34 | 8 | 0 | 22 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 |
| Reverse | 105 | 0 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 42 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | |
| Probe | 14 | 91 | 0 | 12 | 1 | 0 | 2 | 26 | 0 | 0 | 42 | 0 | 0 | 22 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | |
| 3 | Forward | 104 | 0 | 1 | 13 | 0 | 0 | 27 | 0 | 1 | 42 | 0 | 0 | 22 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 |
| Reverse | 105 | 0 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 42 | 0 | 0 | 22 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 0 | |
| 4 | Forward | 105 | 0 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 41 | 1 | 0 | 22 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 |
| Reverse | 105 | 0 | 0 | 13 | 0 | 0 | 3 | 25 | 0 | 42 | 0 | 0 | 22 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | |
| Probe | 105 | 0 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 42 | 0 | 0 | 22 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | |
| 5 | Forward | 83 | 22 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 40 | 2 | 0 | 22 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 |
| Reverse | 105 | 0 | 0 | 13 | 0 | 0 | 28 | 0 | 0 | 42 | 0 | 0 | 22 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | |
| IV(51) | IVa1(4) | IVa2(6) | IVb1(6) | IVb2(5) | IVc1(17) | IVc2(13) | ||||||||||||||||
| 1 | Forward | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | 0 |
| Reverse | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | 0 | |
| Probe | 0 | 46 | 5 | 0 | 0 | 4 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 12 | 1 | |
| 2 | Forward | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 |
| Reverse | 0 | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | |
| Probe | 0 | 50 | 1 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 16 | 1 | 0 | 13 | 0 | |
| 3 | Forward | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | 0 |
| Reverse | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | |
| 4 | Forward | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 |
| Reverse | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | |
| Probe | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | |
| 5 | Forward | 0 | 51 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 |
| Reverse | 50 | 1 | 0 | 3 | 1 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 0 | 13 | 0 | 0 | |
Note: All these subtype specific analysis were based on 161 BKV WGSs. The number of total analyzed strains of different subtypes were marked out behind.
RESULTS
BKV genome diversity
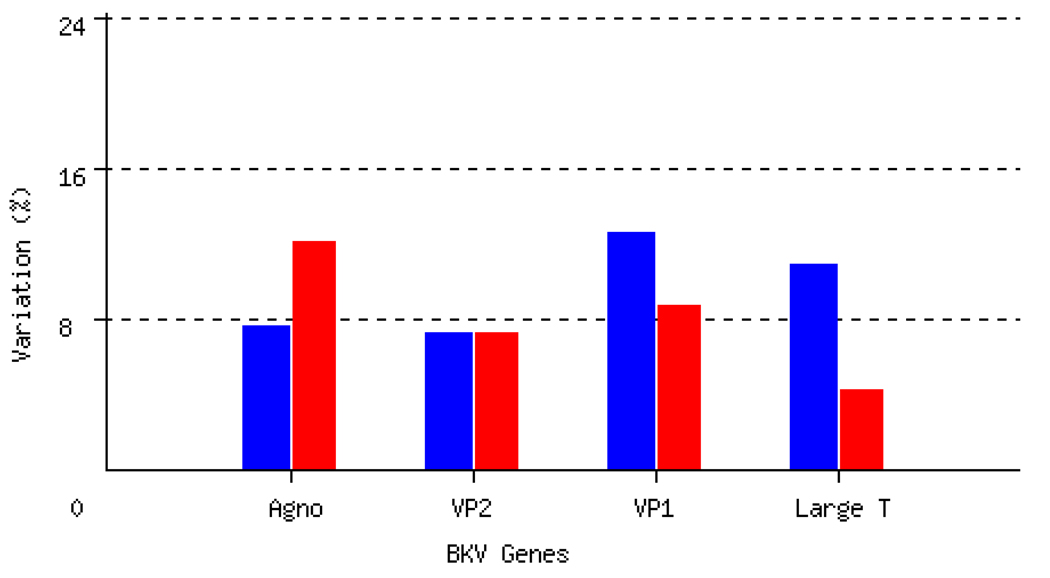
An analysis of the genomic diversity in the NCCR region has been recently published by us, and was not the focus of this study [Sharma et al., 2007] Major rearrangements, deletions and duplications in this region are well known. The coding region of BKV consists of six genes, namely agnogene, VP3, VP2, VP1, small T antigen, and large T antigen (LTA). Rates of inter-strain BKV DNA sequence variation in the Agno, VP2, VP1 and LTA genes were 7.5%, 7.1%, 12.4% and 10.8% respectively (Figure 1). The corresponding rates of amino acid variation were Agno11.9%, VP2 7.1%, VP1 8.6% and LTA 4.2%. The LTA gene was the most conserved at the protein level. There were fewer non-synonymous variants in LTA compared with VP1. Most variants in LTA gene were 3rd position substitutions in codons, and thus synonymous. The most variable coding region was in the VP1 gene spanning amino acids 61 to 83. This region is a part of Jin’s genotyping region and contains 15 variable sites out of 67 possible (22.39%). The amino acid 208–269 region in the VP2 gene was also a highly variable region with substitutions at 19/62 (30.65%) sites.
Fig. 1.
Rates of nucleotide and amino acid variation in the major coding areas of BKV (blue bar: DNA, red bar: amino acids).
Changes other than nucleotide substitutions were relatively uncommon in the coding region of BKV. A duplication of agnogene nucleotides 562–573 was seen in all 161 whole genomes. Several strains had a deletion of nucleotides 553–561 immediately upstream this duplication. The AS Type III whole genome has a unique 37 nucleotide deletion in the NCCR which shifts the agnoprotein start codon closer to the origin of relication. Nucleotides 4590 to 4654 in BKV strain Fin-9 were duplicated, while nucleotides 4590–4855 in the LTA intron were deleted in BKV strain MM. All five type II and III strains deleted nucleotides 2799–2804, and both type III strains lacked nucleotides 2815–2820 in the Large T gene. There were no deletions or duplications in BKV VP1 and VP2 gene. None of these deletions in the LTA or Agnoprotein gene shifted the coding frame or showed any association with a definite disease.
Predicted coverage of known BKV strains
Assay #1: The forward and reverse primers showed a perfect match to all but one of the 161 BKV strains with sequence data available for the targeted LTA region (Table 2). Only the subgroup Ib2 strain LAB-14 demonstrated a mismatch (G->A) 4bp from the 5’ end of the reverse primer. The internal probe matched perfectly 74/161 (45.9%) viral strains, including all 13 Ia, 42 Ib2 and 19 Ic strains (Table 5). All 28 subtype Ib1 strains had a major probe mismatch (A->C) 4 bp from the 3’ end. Three of twenty two (3/22) subtype Ic strains (RYU-2, TW-8, TW-8a) had a major probe mismatch (A->G) at the 3’ terminal nucleotide.
Assay #2: Virtually all subtype I strains perfectly matched the forward primer, an exception being 8 subgroup Ib2_strains (ETH-4, LAB-21, LAB-8, PittNP3,_PittVM1, PittVM3,_PittVR7 and_TUR-5), which showed a mismatch ( A->G) 19 bp from the 3’ end. All subtype II, III, IV strains had a mismatch (G->A/T) 16 bp from the 3’ end; subtypes II and III also demonstrated a mismatch (G->A) 2 bp from the 5’ end. Four subgroup IVb2 strains (JPN-15, JPN-34, KOM-2 and KOM-7) had a mismatch (C->G) with the reverse primer located 11 nucleotides from 3’ end. FNL-17 strain (type IVc) had two mismatches 3 and 4 bp from the 5’ end.
Assay #3: The forward primer was perfectly matched with 163 whole genome sequences (Table 1), but showed a major mismatch (A->G) located 2 bp from the 3’ end of a partial VP-1 sequence of subtype Ib1 from strain CAP-m2 (Table 5). The reverse primer perfectly matched 484 BKV sequences, most of them subtype I strains. This assay based on the VP-1 gene had a potentially higher strain coverage than all other assays. The reverse primer perfectly matched with 484 BKV sequences (Table 2). Incidence of major primer mismatch did not exceed 1/164 (0.62%) for the forward primer. Thus, it is possible to find conserved areas even in the VP1 gene, a somewhat more variable region of the viral genome that is used to define BKV genotypes.
Assay #4: The forward primer matched 103 BKV sequences but the reverse primer matched only 18 (Table 2). The mismatches observed were minor. In genotype and subtype analysis again only minor mismatches were observed for subtype Ib1 and all subtypes of genotype IV.
Assay #5: The reverse primer was a perfect match for 99.38 % of available whole genome sequences. The forward primer showed minor mismatches with 46.58% of available whole genome sequences. No major mismatches were identified (Table 2). There was good predicted coverage of subytpes Ia, 1b1, 1b2, and 1c. Minor mismatches were seen with most type IV sequences (Table V)
Potential for cross amplification of JCV DNA
Assay #1 forward and reverse primers derived from BKV LTA showed perfect match with greater than 98.5% of 447 whole genome JCV sequences available (Table 3). However, the BKV probe showed a major mismatch with all JCV sequences, indicating that cross detection of JCV DNA is unlikely. Cross detection of JCV is also unlikely in Assay #2 where both primers and the detection probe show major mismatches with all 454 JCV Large T-antigen target sequences available. The reverse primer for Assay #3 shows minor mismatches with 95.89% of available JCV sequences. However, this assay is JCV-specific because the forward primer is designed to be complementary to an area of the BKV VP1 that is deleted in all JCV strains. The forward primer of Assay #4 had minor Mismatches with 4 JCV strains, while the reverse primer and probe had major mismatches with all available JCV strains. In the case of assay #5, the reverse primer showed major mismatches with 100% of available JCV sequences.
Table 3.
Analysis of Primer and Probe Mismatch with JC Virus Sequences*
| Primer | Perfect match | Minor Mismatch | Major Mismatch | |
|---|---|---|---|---|
| 1 | Forward | 453/454(99.7%) | 0/454 (0%) | 1/454 (0.22) |
| Reverse | 447/454(98.5%) | 3/454 (0.66%) | 4/454 (0.88) | |
| Probe | 0/454 (0%) | 0/454 (0%) | 454/454 (100%) | |
| 2 | Forward | 0/454 (0%) | 0/454 (0%) | 454/454 (100%) |
| Reverse | 0/454 (0%) | 0/454 (0%) | 454/454 (100%) | |
| Probe | 0/454 (0%) | 0/454 (0%) | 454/454 (100%) | |
| 3 | Forward | 0/487 (0%) | 0/487 (0%) | 487/487 (100%) |
| Reverse | 0/487 (0%) | 467/487(95.89%) | 20/487 (4.11%) | |
| 4 | Forward | 0/1590 (0%) | 2/1590 (0.16%) | 1588/1590(99.84%) |
| Reverse | 0/1590 (0%) | 0/1590 (0%) | 1590/1590(100%) | |
| Probe | 0/1590 (0%) | 0/1590 (0%) | 1590/1590(100%) | |
| 5 | Forward | 0/453 (0%) | 452/453(99.78%) | 1/453 (0.22%) |
| Reverse | 0/453 (0%) | 0/453 (0%) | 453/453 (100%) |
Numbers in the first column refer to PCR assays referenced in table 2. The variable denominators in data cells reflect the fact that not viral strains described in the literature have sequence information available corresponding to the area of the viral genome targeted by different assays.
Possible detection of SV40 DNA
Assay #1 showed minor mismatches with 39 of 40 available sequences (Table 4). However, the non-homologous nature of the reverse primer and probe should prevent amplification of SV40 DNA. Assays #2, #3 and #4 are not expected to have any interference from the (unlikely) presence of SV40 DNA in clinical samples as all show major primer and probe mismatches with simian viral sequences. Likewise, for assay #5, major mismatches with the forward primer provide assurance against unwanted amplification of BKV DNA.
DISCUSSION
Accumulating sequences over the past several years have demonstrated significant variability in regions of the viral genome that are targeted for PCR-based diagnostic assays [Randhawa et al., 2003; Randhawa et al., 2002]. A mismatch between the viral genome and primer or probe sequences could result in either false negative or underquantitated viral load measurements by PCR. Manna et al have described clinical samples which yielded atypical amplification profiles, but the frequency with which this occurs is not clear from their data, which is reported only in abstract form [Manna et al., 2005]. Limaye et al noted an 8% discrepancy rate between two PCR assays, one based on the small t, and the other on the large T-antigen. Moreover, when these 2 assays were compared, a > 1 log difference in viral load was seen for 7/25 (28%) samples [Limaye et al., 2005]. Our data provides a comprehensive assessment of this problem using 716 BKV, 1626 JCV, and 83 SV40 sequences. Available assays differ with regard to primer and probe identity with the targeted viral sequences. Viral genotype and subtype analysis suggests that currently used assays are more likely to have difficultires with subtype IV variants. This is not surprising because many laboratories have designed their primers using a subtype I sequence belonging to the Dun strain. The extent of the problem for subtypes II and III can not be accurately identified at this time due to a paucity of available sequences.
An important caveat of our analysis is that we have limited it to a comparison of primer and probe sequences with viral genomic sequences. It is true that successful PCR requires good binding of forward and reverse primers to the target sequence, as well as probe to the amplicon. However, the efficiency of a PCR reaction also depends on numerous other factors such as primer GC content, melting temperature, and tendency to form hair pin loops and primer dimers. The probability of a successful PCR using partially mismatched primers and probes can not be accurately predicted without actual experimentation. Notwithstanding this limitation, our data demonstrates that available information on BKV genomic diversity has expanded to the point, where it has become important for clinical diagnostic laboratories to re-appraise assays currently being offered by them to transplant physicians. A study on cytomegalovirus demonstrated that 2 mismatches at the 3’ end of a PCR primer can result in 1–2 log differences in calculated viral load [Nye et al., 2005] . Extensive mismatching can result in lack of amplification of the viral target sequence (false negative result) [Whiley and Sloots, 2005]. The challenge faced by laboratory directors today is to develop a PCR test for BKV that covers all known strains, is sensitive, quantitative, and does not amplify JCV or SV40.
Our sequence analysis offers some guidelines to laboratories seeking to develop robust PCR tests for diagnosis of BKV. From the point of view of gene polymorphisms, the agnogene or VP-2 region would be a preferred target site, since it has the lowest rate of genomic variability. However, for the majority of BKV strains studied from clinical samples only VP-1 sequences are known. Notwithstanding its higher variability, relatively conserved sequences can be identified in the VP-1 region: nucleotides 1712–1736 and 1814–1836 are conserved respectively in 484 and 670 BKV strains sequenced to date. In preliminary experiments, we have successfully amplified BKV DNA using the aforementioned genomic regions as the forward and reverse primers. This amplification utilized real time PCR on the TaqMan platform with SyBr Green-based detection of the amplified product. Large T-antigen and agnoprotein are sometimes regarded as more attractive sites than VP-1 for assay development. However, it should be kept in mind that at this time only 164 agnoprotein and 174 large T-antigen sequences are known. It is also likely that additional gene polymorphisms will be discovered in these two areas.
As our knowledge of BKV diversity increases further, it may become necessary to use degenerate primers, or develop multiplex assays interrogating more than one target site. At least one commercial laboratory offers an assay which amplifies both agnoprotein and Large T-antigen sequences in clinical samples. Such multiplex assays will have to be carefully validated qualitatively and quantitatively before being offered to transplant patients. Laboratories may have to keep more than one BKV assay on their diagnostic menu to accurately quantify viral loads of variant BKV strains with problematic gene polymorphisms.
Supplementary Material
Acknowledgments
Supported by grants RO1 AI 51227 and AI 63360 to PR.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the National Institute of Allergy and Infectious Disease, or the National Institute of Diabetes, Digestive and Kidney Diseases.
REFERENCES
- Baksh FK, Finkelstein SD, Swalsky PA, Stoner GL, Ryschkewitsch CF, Randhawa P. Molecular genotyping of BK and JC viruses in human polyomavirus-associated interstitial nephritis after renal transplantation. Am J Kidney Dis. 2001;38(2):354–365. doi: 10.1053/ajkd.2001.26101. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zheng HY, Zhong S, Ikegaya H, He HX, Wei W, He YY, Kobayashi N, Honjo T, Takasaka T, Takahashi S, Kitamura T, Yogo Y. Subtype IV of the BK polyomavirus is prevalent in East Asia. Arch Virol. 2006;151:2419–2429. doi: 10.1007/s00705-006-0814-z. [DOI] [PubMed] [Google Scholar]
- Chen YP, Sharp PM, Fowkes M, Kocher O, Joseph JT, Koralnik IJ. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J Gen Virol. 2004;85:2651–2663. doi: 10.1099/vir.0.79920-0. [DOI] [PubMed] [Google Scholar]
- Demeter LM. JC, BK, and other polyomaviruses; progressive multifocal leukoencephalopathy. In: Mandel GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases. New York: Churchill Livingstone; 1995. pp. 1400–1406. [Google Scholar]
- Di Taranto C, Pietropaolo V, Orsi GB, Jin L, Sinibaldi L, Degener AM. Detection of BK polyomavirus genotypes in healthy and HIV-positive children. Eur J Epidemiol. 1997;13(6):653–657. doi: 10.1023/a:1007371320999. [DOI] [PubMed] [Google Scholar]
- Ding R, Medeiros M, Dadhania D, Muthukumar T, Kracker D, Kong JM, Epstein SR, Sharma VK, Seshan SV, Li B, Suthanthiran M. Noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1 in urine. Transplantation. 2002;74(7):987–994. doi: 10.1097/00007890-200210150-00016. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ludlow JW, Lill NL, Oldread E, Arany Z, Modjtahedi N, DeCaprio JA, Livingston DM, Morgan JA. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16(7):3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Valle M, Frank J, Gruss C, Sorzano COS, Chen XS, Donate LE, Carazo JM. Large T antigen on the simian virus 40 origin of replication: a 3D snapshot prior to DNA replication. EMBO J. 2003;22(23):6205–6213. doi: 10.1093/emboj/cdg612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HH, Mohaupt M, Klimkait T. Prospective monitoring of BK virus load after discontinuing sirolimus treatment in a renal transplant patient with BK virus nephropathy. J Infect Dis. 2001;184(11):1494–1495. doi: 10.1086/324425. [DOI] [PubMed] [Google Scholar]
- Ikegaya H, Saukko PJ, Tertti R, Metsarinne KP, Carr MJ, Crowley B, Sakurada K, Zheng HY, Kitamura T, Yogo Y. Identification of a genomic subgroup of BK polyomavirus spread in European populations. J Gen Virol. 2006;87(Pt 11):3201–3208. doi: 10.1099/vir.0.82266-0. [DOI] [PubMed] [Google Scholar]
- Ikegaya H, Zheng HY, Saukko PJ, Varesmaa-Korhonen L, Hovi T, Vesikari T, Suganami H, Takasaka T, Sugimoto C, Ohasi Y, Kitamura T, Yogo Y. Genetic diversity of JC virus in the Saami and the Finns: Implications for their population history. Am J Phys Anthropol. 2005;128(1):185–193. doi: 10.1002/ajpa.20189. [DOI] [PubMed] [Google Scholar]
- Jin L. Rapid genomic typing of BK virus directly from clinical specimens. Mol Cell Probes. 1993;7(4):331–334. doi: 10.1006/mcpr.1993.1047. [DOI] [PubMed] [Google Scholar]
- Jin L, Gibson PE, Booth JC, Clewley JP. Genomic typing of BK virus in clinical specimens by direct sequencing of polymerase chain reaction products. Journal of medical virology. 1993;41(1):11–17. doi: 10.1002/jmv.1890410104. [DOI] [PubMed] [Google Scholar]
- Krumbholz A, Zell R, Egerer R, Sauerbrei A, Helming A, Gruhn B, Wutzler P. Prevalence of BK virus subtype I in Germany. Journal of medical virology. 2006;78(12):1588–1598. doi: 10.1002/jmv.20743. [DOI] [PubMed] [Google Scholar]
- Leuenberger D, Andresen PA, Gosert R, Binggeli S, Strom EH, Bodaghi S, Rinaldo CH, Hirsch HH. Human polyomavirus type 1 (BK virus) agnoprotein is abundantly expressed but immunologically ignored. Clin Vaccine Immunol. 2007;14(8):959–968. doi: 10.1128/CVI.00123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye AP, Ferrenberg J, Huang ML. Comparison of two separate T antigen region-based PCR assays for quantitation of BKV load in kidney transplant recipients with BKVN. Proceedings from Polyomavirus-Associated Nephropathy Interdisciplinary Conference; May 19–20, 2005; Baltimore, Maryland. p. 1A. [Google Scholar]
- Limaye AP, Jerome KR, Kuhr CS, Ferrenberg J, Huang ML, Davis CL, Corey L, Marsh CL. Quantitation of BK virus load in serum for the diagnosis of BK virus-associated nephropathy in renal transplant recipients. J Infect Dis. 2001;183(11):1669–1672. doi: 10.1086/320711. [DOI] [PubMed] [Google Scholar]
- Manna P, Reddy S, Grantham K. Role of genomic variation on viral load determination for BK virus: potential clinical implications. Am J Transplant. 2005;5:272–273. [Google Scholar]
- Nishimoto Y, Takasaka T, Hasegawa M, Zheng HY, Chen Q, Sugimoto C, Kitamura T, Yogo Y. Evolution of BK virus based on complete genome data. J Mol Evol. 2006;63(3):341–352. doi: 10.1007/s00239-005-0092-5. [DOI] [PubMed] [Google Scholar]
- Nishimoto Y, Zheng HY, Zhong S, Ikegaya H, Chen Q, Sugimoto C, Kitamura T, Yogo Y. An Asian origin for subtype IV BK virus based on phylogenetic analysis. J Mol Evol. 2007;65(1):103–111. doi: 10.1007/s00239-006-0269-6. [DOI] [PubMed] [Google Scholar]
- Nukuzuma S, Takasaka T, Zheng H-Y, Zhong S, Chen Q, Kitamura T, Yogo Y. Subtype I BK polyomavirus strains grow more efficiently in human renal epithelial cells than subtype IV strains. J Gen Virol. 2006;87(7):1893–1901. doi: 10.1099/vir.0.81698-0. [DOI] [PubMed] [Google Scholar]
- Nye MB, Leman AR, Meyer ME, Menegus MA, Rothberg PG. Sequence diversity in glycoprotein B complicates real-time PCR assays for detection and quantification of cytomegalovirus. J Clin Microbiol. 2005;43(10):4968–4971. doi: 10.1128/JCM.43.10.4968-4971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa P, Ho A, Shapiro R, Vats A, Swalsky P, Finkelstein S, Uhrmacher J, Weck K. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J Clin Microbiol. 2004;42(3):1176–1180. doi: 10.1128/JCM.42.3.1176-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa P, Zygmunt D, Shapiro R, Vats A, Weck K, Swalsky P, Finkelstein S. Viral regulatory region sequence variations in kidney tissue obtained from patients with BK virus nephropathy. Kidney Int. 2003;64(2):743–747. doi: 10.1046/j.1523-1755.2003.00132.x. [DOI] [PubMed] [Google Scholar]
- Randhawa PS, Khaleel-Ur-Rehman K, Swalsky PA, Vats A, Scantlebury V, Shapiro R, Finkelstein S. DNA sequencing of viral capsid protein VP-1 region in patients with BK virus interstitial nephritis. Transplantation. 2002;73(7):1090–1094. doi: 10.1097/00007890-200204150-00013. [DOI] [PubMed] [Google Scholar]
- Roy R, Trowbridge P, Yang Z, Champoux JJ, Simmons DT. The cap region of topoisomerase I binds to sites near both ends of simian virus 40 T antigen. J Virol. 2003;77(18):9809–9816. doi: 10.1128/JVI.77.18.9809-9816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik W. Priming efficiency in PCR. Biotechniques. 1995;18(1):84–86. 88–90. [PubMed] [Google Scholar]
- Seif I, Khoury G, Dhar R. The genome of human papovavirus BKV. Cell. 1979;18(4):963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- Shah KV. Polyomaviruses. In: Fields BN, Knipe KM, Howley PM, editors. Virology. Philadlphia: Lippincott-Raven; 1995. pp. 2027–2043. [Google Scholar]
- Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. A phylogenetic analysis of polyomavirus BK sequences. J Virol. 2006;80:8869–8879. doi: 10.1128/JVI.00510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa PS. Polyomavirus BK non-coding control region rearrangements in health and disease. J Med Virol. 2007;79(8):1199–1207. doi: 10.1002/jmv.20909. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ono N, Furusawa C, Kashiwagi A, Yomo T. Experimental optimization of probe length to increase the sequence specificity of high-density oligonucleotide microarrays. BMC Genomics. 2007;8:373. doi: 10.1186/1471-2164-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaka T, Goya N, Ishida H, Tanabe K, Toma H, Fujioka T, Omori S, Zheng HY, Chen Q, Nukuzuma S, Kitamura T, Yogo Y. Stability of the BK polyomavirus genome in renal-transplant patients without nephropathy. J Gen Virol. 2006;87(Pt 2):303–306. doi: 10.1099/vir.0.81368-0. [DOI] [PubMed] [Google Scholar]
- Takasaka T, Goya N, Tokumoto T, Tanabe K, Toma H, Ogawa Y, Hokama S, Momose A, Funyu T, Fujioka T, Omori S, Akiyama H, Chen Q, Zheng HY, Ohta N, Kitamura T, Yogo Y. Subtypes of BK virus prevalent In Japan and variation in their transcriptional control region. J Gen Virol. 2004;85:2821–2827. doi: 10.1099/vir.0.80363-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls E, de la Cruz X, Martinez-Balbas MA. The SV40 T antigen modulates CBP histone acetyltransferase activity. Nucleic Acids Res. 2003;31(12):3114–3122. doi: 10.1093/nar/gkg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley DM, Sloots TP. Sequence variation in primer targets affects the accuracy of viral quantitative PCR. J Clin Virol. 2005;34:104–107. doi: 10.1016/j.jcv.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Yogo Y, Zhong S, Suzuki M, Shibuya A, Kitamura T. Occurrence of the European subgroup of subtype I BK polyomavirus in Japanese-Americans suggests transmission outside the family. J Virol. 2007;81(23):13254–13258. doi: 10.1128/JVI.01018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng HY, Ikegaya H, Takasaka T, Matsushima-Ohno T, Sakurai M, Kanazawa I, Kishida S, Nagashima K, Kitamura T, Yogo Y. Characterization of the VP1 loop mutations widespread among JC polyomavirus isolates associated with progressive multifocal leukoencephalopathy. Biochem Bioph Res Co. 2005a;333(3):996–1002. doi: 10.1016/j.bbrc.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Zheng HY, Nishimoto Y, Chen Q, Hasegawa M, Zhong S, Ikegaya H, Ohno N, Sugimoto C, Takasaka T, Kitamura T, Yogo Y. Relationships between BK virus lineages and human populations. Microbes Infect. 2007;9(2):204–213. doi: 10.1016/j.micinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Zheng HY, Takasaka T, Noda K, Kanazawa A, Mori H, Kabuki T, Joh K, Oh-Ishi T, Ikegaya H, Nagashima K, Hall WW, Kitamura T, Yogo Y. New sequence polymorphisms in the outer loops of the JC polyomavirus major capsid protein (VP1) possibly associated with progressive multifocal leukoencephalopathy. J Gen Virol. 2005b;86:2035–2045. doi: 10.1099/vir.0.80863-0. [DOI] [PubMed] [Google Scholar]
- Zhong S, Yogo Y, Ogawa Y, Oshiro Y, Fujimoto K, Kunitake T, Zheng HY, Shibuya A, Kitamura T. Even distribution of BK polyomavirus subtypes and subgroups in the Japanese Archipelago. Arch Virol. 2007;152(9):1613–1621. doi: 10.1007/s00705-007-0997-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.