Abstract
Previously, we reported modeling synovial sarcomas in mice by conditionally expressing the human t(X;18) translocation-derived SYT-SSX2 fusion protein in Myf5-expressing myoblasts. Using a Tamoxifen-inducible CreER system in mice, we show here that sporadic expression of SYT-SSX2 across multiple tissue types leads to exclusive formation of synovial sarcoma-like tumors while its widespread expression is lethal. Certain clinical and histological features of tumors in this new model suggest additional non-myoblast origin for synovial sarcoma. CreER-based sporadic expression both avoids the severe early developmental phenotypes associated with widespread SYT-SSX2 expression and better models natural pathogenesis of cancers where transformed cells usually arise within an environment of largely normal cells. Furthermore, this strategy may recapitulate multiple potential cellular origins within a single model system.
Keywords: SYT-SSX, Synovial Sarcoma, Mouse Model, Conditional System, CreER
Introduction
Many sarcomas, or mesenchymally-based malignancies, are associated with unique reciprocal chromosomal translocations. These translocations generate chimeric fusion proteins, the presence of which is highly specific to the related sarcoma subtype. Some examples include: PAX3-FKHR in Alveolar Rhabdomyosarcoma, SYT-SSX in Synovial Sarcoma, and EWS-FLI in Ewing's Sarcoma (1). Association of such specific translocation-derived fusion proteins with specific sarcomas leads to the possibility of modeling these diseases in mice by transgenic expression of the fusion product. However, such attempts are often hampered by our lack of knowledge of the cell of origin. The tumorigenic effect of an oncogene is often specific to a cell type and its microenvironment as both intra and extra-cellular factors play important role in tumorigenesis (2). Expression of an oncogene in an inappropriate cell type or microenvironment may lead to no phenotype or tumor-unrelated phenotypes, while ubiquitous expression is often lethal. Therefore, targeting expression of the oncogene in the appropriate cell type within an appropriate microenvironment is crucial to successful tumor modeling and can be technically challenging.
Cre-LoxP based conditional system is widely used in tumor modeling to target oncogene expression to specific cell/tissue types. The most common method of delivering Cre recombinase to the target tissue is by breeding with transgenic mice expressing Cre in a tissue-specific manner. A major limitation of this strategy is that Cre (and hence the oncogene) is expressed in all or most cells in the target tissue leading to significant alteration of the entire tissue and likely affects tumorigenesis by disrupting the required interaction of the oncogene expressing cells with the `normal' cells of the niche. A promising new development has been the establishment of Tamoxifen-inducible CreER systems that allow temporal and even quantitative control over expression in a chosen tissue by controlling nuclear entry of Cre (and hence Cre activity) by measured exposure to exogenous small molecule Tamoxifen (3).
By using a CreER based strategy we demonstrate that random, sporadic expression of SYT-SSX2 in a small subset of cells across multiple tissue types leads to the exclusive generation of synovial sarcoma-like tumors in adult mice.
Materials and methods
Animal study and Genotyping procedures
Studies involving animal subjects were approved by the University of Utah Institutional Animal Care and Use Committee and conducted strictly in accordance with the relevant protocol. The SSM2 conditional mice, R-CreER, and the R-LacZ reporter mouse line were all genotyped with the same set of primers - Forward-WT: GTTATCAGTAAGGGAGCTGCAGTGG, Reverse-targeted: AAGACCGCGAAGAGTTTGTCCTC and Reverse –WT: GGCGGATCACAAGCAATAATAACC. These primers yield a 302 bp band for targeted and a 415 bp band for wild type ROSA locus. Cre-expressing mice were genotyped using a pair of Cre specific primers – Forward: GGATTTCCGTCTCTGGTGTAGC and Reverse: ACCATTGCCCCTGTTCACTATC that yield a 320 bp band.
Tamoxifen was dissolved in warm sterile corn oil at a concentration of 20mg/ml that was used to inject mice intra-peritoneally.
Histology and Immunohistochemistry
For histology, specimens were fixed overnight in 4% paraformaldehyde and embedded in paraffin wax following standard procedure. 4–8 -μm sections were cut and mounted on slides for standard Hematoxylin and Eosin (H & E) staining. Immunohistochemistry on 4 -μm sections of paraffin embedded samples was carried out by the ARUP laboratories at the University of Utah. Counterstaining was done with hematoxylin.
β-galactosidase staining
For whole mount staining; samples were fixed in 1% PFA, 0.2% glutaraldehyde, 0.002M MgCl2, 0.025M EGTA and 0.02% NP-40 in 1× PBS for 2 hrs at 4 degree centigrade followed by overnight staining at RT in 0.005M K3Fe(CN)6, 0.005M K4Fe(CN)6, 0.002M MgCl2, 0.01% Na-doc, 0.02% NP-40 and X-Gal substrate in 1× PBS. Stained tissue were processed for paraffin embedding and sectioned as mentioned above. Counterstaining was carried out with nuclear red following established protocol.
Cell culture
Mouse embryonic fibroblasts were harvested by euthanizing mice and harvesting E13.5 – E14.5 embryos in a sterile environment. Extra-embryonic tissue, head, and internal organs were removed from the embryos in sterile PBS. Embryos were put in sterile tubes containing 50 ml sterile 1× PBS followed by addition of 1ml of cold 0.25% tripsin and incubated at 4 degrees overnight to allow tripsin diffusion within tissue. The next day, all solutions were removed and replaced with 0.25% tripsin solution and incubated at 37 degrees for 10–15 minutes with gentle shaking. This was followed by addition of 10%FBS + DMEM to the tube and vigorous pipetting with medium bore pipet to disrupt the tissue and plated.
Cells were maintained in media containing 10%FBS + DMEM. The production of TAT-Cre is part of a separate manuscript that is being submitted.
Imaging
Computed tomography of sacrificed mice was performed at 93 μm3 voxel resolution using the eXplore RS Small Animal MicroCT Scanner (GE Healthcare). Images were reconstructed and visualized with the manufacturer's MicroView freeware. Fluorescent microscopy was performed using the Axiovert 200M (Carl Zeiss) microscope, image captured using the sensicam (The Cooke Corporation), and the slidebook™ image capture and analysis software (Intelligent Imaging and Innovation Inc). Light microscopy was performed using the same microscope using an Axiocam HRC camera and the Axiovision (4.6) image capture and analysis software.
Microarray Analysis
RNA was extracted from tumors of ROSA-CreER+/−/SSM2+/− mice using TRIzol (invitrogen) and purified using RNeasy kit (Qiagen). Gene expression was measured using Affymetrix oligonucleotide-based GeneChip® microarray technology (Affymetrix, Santa Clara, CA). The microarray data has been uploaded to the GEO website (GEO accession: 14469). Further information on microarray analysis is included in the supplementary document
Results
Targeted expression of SYT-SSX2 in most tissue types is lethal
We previously reported generation of a mouse line (SSM2 mouse line) that expresses the human SYT-SSX2 fusion gene conditionally from the ROSA promoter in the presence of Cre recombinase (4). Expression of SYT-SSX2 is also linked to expression of the Enhanced Green Fluorescent marker protein (EGFP) via an Internal Ribosomal Entry Site (IRES) (Figure 1A). Within the skeletal muscle lineage, the biological effect of SYT-SSX2 expression was found to be dependent upon the differentiation status of the cell such that expression in early progenitors of skeletal muscle led to embryonic lethality, expression in immature but committed myoblasts generated synovial sarcomas, while expression in differentiated skeletal muscle fibers led to severe myopathy. Furthermore, early and ubiquitous expression within developing embryo led to embryonic lethality. (4). This lethality suggested the requirement of more discreet oncogene expression than allowed by many tissue-specific Cre drivers, especially those that are expressed widely during early embryogenesis. To further confirm this, and to investigate alternative tissue sources of synovial sarcoma, we targeted SYT-SSX2 expression to a broad range of tissue types. This was achieved by breeding the SSM2 mice to a broad range of tissue specific Cre expressing mice (Cre drivers) including Ap2-Cre (early ectoderm, neural crest, limb bud mesenchymal lineage) (5), Sox9-Cre (bone, cartilage, and various other tissue) (6), Dermo1-Cre (Mesenchymal lineage) (7), Flk1-Cre (vascular endothelium)(8), Tie2-Cre (endothelial/hematopoetic lineage)(9), and Nestin1-Cre (Neural lineage)(10). No viable SSM2+/−/Cre+/− progenies were obtained (an average of 30 progenies genotyped from each breeding) from any of these breedings, suggesting embryonic lethality, thus further confirming the requirement of more discrete oncogene expression.
Figure 1.
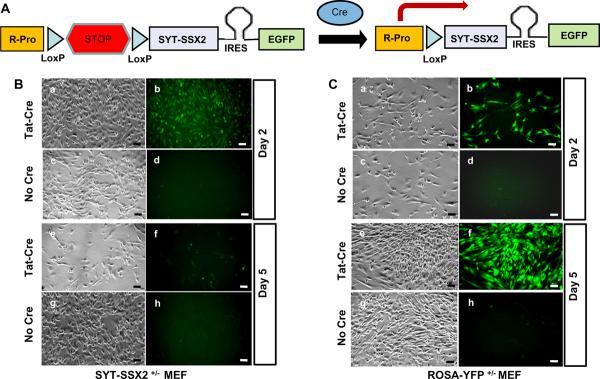
Figure A shows the strategy for conditional expression of SYT-SSX2 and EGFP proteins in the genetically engineered SSM2 mice. Cre mediates recombination between the two LoxP sites in SSM2 mice thereby removing the transcriptional stop signal and allowing transcription (arrow) of the SYT-SSX2 –IRES-EGFP bicistronic mRNA from the endogenous ROSA promoter. The bicistronic mRNA is translated into distinct SYT-SSX2 and EGFP proteins via the EMCV virus derived Internal Ribosomal Entry site (IRES) placed between the coding sequences of SYT-SSX2 and EGFP.
SYT-SSX2+/− fibroblasts (B) from SSM2+/− embryos and control ROSA-YFP+/− fibroblasts (C) from ROSAY-YFP+/− embryos were cultured. SYT-SSX2+/− and ROSA-YFP+/− fibroblasts were divided into two groups; those exposed to TAT-Cre protein at 3 μm concentration for 2 hours and those not exposed to TAT-Cre. By the second day, SYT-SSX2+/− fibroblasts exposed to TAT-CRE (Ba and Bb) showed SYT-SSX2 expression based on EGFP induced fluorescence (Bb) while those not exposed to TAT-CRE (Bc and Bd) showed no fluorescence (Bd). Control ROSA-YFP+/− cells exposed to TAT-Cre (Ca and Cb) showed robust YFP expression (Cb) while those not exposed to TAT-CRE (Cc and Cd) showed no YFP expression (Cd).
Upon serially tracking the same zones, by day five SYT-SSX2+/− fibroblasts exposed to TAT-CRE (Be and Bf) showed significantly less growth (Be) and very few EGFP expressing cells (Bf) while those not exposed to TAT-CRE showed robust growth (Bg) with no EGFP expression (Bh). Control ROSA-YFP+/− cells exposed to TAT-CRE continued to show robust growth (Ce) and YFP expression (Cf) while those not exposed to TAT-CRE showed robust growth (Cg) without YFP expression (Ch). All scale bars = 50 μm.
We previously reported that expression of SYT-SSX2 within Myf5-expressing myoblasts led to apoptosis, except in certain microenvironments where the SYT-SSX2 expressing cells appeared to be protected from apoptosis and thereby allowed progress towards neoplasia. Therefore, it appeared logical to assume that this dominant cytotoxic effect of SYT-SSX2 expression led to lethality in most breeding experiments with tissue specific Cre drivers. To further confirm this in-vitro in a more controlled environment, we harvested mouse embryonic fibroblasts (MEFs) from SSM2+/−mice. As control, we also harvested MEFs from ROSA-YFP+/− (11) mice that expresses the Yellow Fluorescent Protein (YFP) from the ROSA locus in the presence of Cre. These cells were exposed to 3μM of a modified Cre protein that has the protein-transduction-domain of HIV TAT protein fused to the N-terminus of Cre (TAT-CRE) (12). Direct addition of such purified TAT-CRE protein to cultured cells allows for more rapid, stable, and uniform activation of Cre dependent loci than is possible by the more commonly used DNA mediated transfection procedures.
SYT-SSX2+/− MEFs exposed to TAT-CRE (Figure 1Ba and 1Bb) express SYT-SSX2 and turn green due to accumulation of the EGFP protein (Figure 1Bb). Plates were marked for zones showing robust SYT-SSX2 expression (Figure 1Ba and 1Bb) detected via fluorescence and were followed up. Remarkably, after a few days these zones showed few living and very few fluorescent cells (Figure 1Be and 1Bf). This suggests that cells expressing SYT-SSX2 either shut down SYT-SSX2 expression, undergo growth arrest, or cell death. Plates harboring fluorescent SYT-SSX2 expressing MEFs showed significantly higher cellular debris and floating dead cells in the media when compared to control cells. The large increase in the population of dead cells in the media along with an 80%–90% reduction in the number of fluorescent cells in plates harboring SYT-SSX2 expressing cells suggest that SYT-SSX2 expression leads to cell death. Cell death via apoptosis was further suggested in SYT-SSX2 expressing cells by the detection of activated caspase pathway (Supplementary Figure A and B). SYT-SSX2+/− MEFs not exposed to TAT-CRE (Figure 1Bc, 1Bd, 1Bg, and 1Bh) did not show fluorescence (Figure 1Bd and 1Bh) suggesting absence of SYT-SSX2 expression and continue to proliferate robustly (Figure 1Bg and 1Bh).
Control ROSA-YFP+/− MEFs also proliferated robustly with (Figure 1Ca, 1Cb, 1Ce, and 1Cf) and without (Figure 1Cc, 1Cd, 1Cg, and 1Ch) TAT-CRE. It is worth noting here that the fluorescence intensity in TAT-Cre exposed ROSA-YFP+/− MEFs (Figure 1Cb and 1Cf) is higher than in TAT-Cre exposed SYT-SSX2+/− MEFs. This reflects the differences between IRES driven protein expression (EGFP expression in SYT-SSX2 expressing MEFs) vs. direct promoter-driven protein expression (YFP expression in ROSA-YFP MEFs). The TAT-Cre protein used in these experiments has been demonstrated to be about 80% efficient in inducing YFP expression (due to Cre activity) in ROSA-YFP+/− MEFs at 3μM concentration. This and more extensive data on the construction, analysis, use, and in-vivo activity of this modified Cre protein is the subject of an additional manuscript being submitted. The conditional in vitro system we describe here will be a useful tool in the identification of molecular pathways activated by the SYT-SSX2 fusion protein.
Random sporadic expression of SYT-SSX2 in multiple tissue types leads to soft tissue tumors
SYT-SSX2 expression can be confined to a few cells in a given `promoter-defined' tissue by using the tamoxifen- inducible CreER system where the nuclear entry of Cre (and hence SYT-SSX2 expression) can be controlled via exogenous application of Tamoxifen. CreER is a fusion between Cre and a mutant form of ligand binding domain of estrogen receptor (ER™) that keeps the Cre protein sequestered in the cytoplasm via interactions between ER™ and Hsp90.
Exogenous application of Tamoxifen disrupts this interaction and allows nuclear translocation of Cre where it initiates recombination between LoxP sites (3). We obtained the ROSA-CreER mouse line (R-CreER) that expresses CreER ubiquitously (13) from the ROSA locus and bred it to the conditional SSM2 mouse line. The number of cells where Cre is translocated to the nucleus in the R-CreER+/−/SSM2+/− mice can be controlled by the amount and frequency of exogenous application of Tamoxifen. R-CreER+/−/SSM2+/− mice were treated once with low dose Tamoxifen (0.1mg/gm) via intra-peritoneal injection at the age of 3 months. While tumors were generated within all injected (7/7) R-CreER+/−/SSM2+/− mice, control groups comprising R-CreER+/−, R-CreER−/−, SSM2+/−, SSM2−/−, or WT mice did not show any tumors or other discernable mutant phenotypes. Nuclear translocation of Cre has been shown to occur at very low frequency within the R-CreER mice even in the absence of tamoxifen (14). This phenomenon led to development of tumors in R-CreER+/−/SSM2+/− mice (6/6) even without tamoxifen (discussed later).
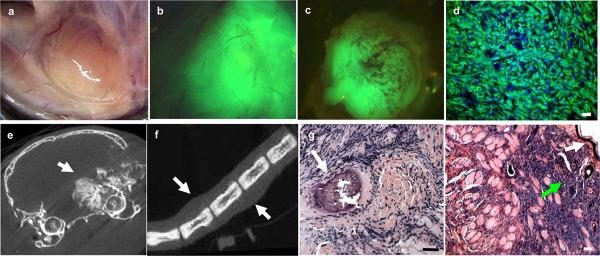
Tumors were first noted in R-CreER+/−/SSM2+/− mice by visual inspection between 5 to 14 months. Tumors in R-CreER+/−/SSM2+/− (Figure 2a to 2h) mice showed intense fluorescence (Figure 2b, 2c, and 2d) demonstrating SYT-SSX2 expression. Multiple tumors (an average of 3 per mouse, n =13) were detected upon necropsy based on fluorescence. CT scanning revealed a variety of tumor characteristics even within the same mouse with some tumors showing evidence of significant calcification (Figure 2e) while others did not (Figure 2f). Tumors were detected in a variety of locations (Figure 2e to 2h) including regions close to bone (Figure 2g) and subcutaneous tissue (Figure 2h).
Figure 2.
Tumors in R-CreER+/−/SSM2+/− mice were detected in a variety of locations such as juxta-articular regions in limbs (a) that is a frequent location in human synovial sarcomas. The tumors showed fluorescence grossly (b and c) and in micrograph (d). Tumors in Tamoxifen exposed (b) and Tamoxifen unexposed (c) mice were fluorescent. CT imaging revealed calcifications (e, arrow) in some tumors such as the one shown in figure e located near the right orbit while others, such as the tail tumor showed in f (arrows show tumor masses), demonstrate exclusive soft tissue involvement.
Histology revealed a variety of tissue involvement such as bone (g, arrow) and subcutaneous tissue (h). White arrow in figure h show keratinized epithelial layer while green arrow show infiltrating tumor cells. All scale bars = 50 μm.
Soft tissue tumors generated are synovial sarcomas
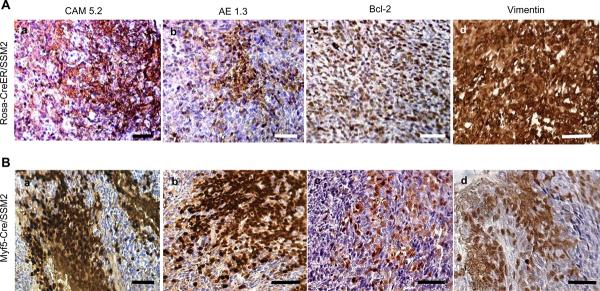
Histology of tumors generated in R-CreER+/−/SSM2+/− was consistent with features of human synovial sarcoma as well as with synovial sarcoma-like tumors previously generated in Myf5-Cre+/−/SSM2+/− mice, which expresses SYT-SSX2 specifically in Myf5-expressing myoblasts (4). Immunohistochemistry revealed expression of synovial sarcoma-associated markers such as cytokeratins, Bcl-2, and Vimentin within tumors of R-CreER+/−/SSM2+/− (Figure 3A) similar to tumors of Myf5-Cre+/−/SSM2+/− mice (Figure 3B), and to human synovial sarcomas. Tamoxifen induces nuclear translocation of CreER, and therefore expression of SYT-SSX2, across multiple tissue types in mice harboring the R-CreER allele. Exclusive generation of synovial sarcoma-like tumors despite oncogene expression in multiple tissue types is remarkable and suggests a dominant role of SYT-SSX2 in directing the tumor phenotype. This is not always the case with translocation products of sarcoma as shown by a recent attempt to generate a mouse model of Ewing's sarcoma by expressing the Ewing's sarcoma-associated EWS-FLI fusion protein that resulted in erythroleukaemia in mice (15).
Figure 3.
Immunohistochemistry revealed similarity between tumors of R-CreER+/−/SSM2+/− (A) and tumors of Myf5-Cre+/−/SSM2+/− tumors (B). Panel includes markers commonly over-expressed in human synovial sarcomas such as Cytokeratins (Aa,Ab,Ba, and Bb), Bcl-2 (Ac and Bc), and Vimentin (Ad and Bd). All scale bars = 50 μm.
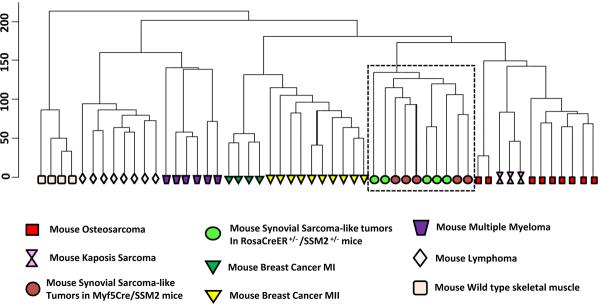
To further confirm the identity of the tumors at the transcriptional level, we extracted RNA from five tumors isolated from five distinct R-CreER+/−/SSM2+/− mice and carried out a microarray hybridization using the Affymetrix mouse whole genome 430 2.0 chip. We have previously generated microarray data from mouse synovial sarcoma-like tumors in Myf5-Cre+/−/SSM2+/− mice and control wild type skeletal muscles using the same Affymetrix platform. We next searched the Gene Expression Omnibus (GEO) database for availability of microarray data generated on Affymetrix mouse 430 2.0 genechip (same platform to reduce cross-platform issues) from other mouse models of tumors and found the following datasets matching our criteria; Kaposi's Sarcoma (16), thymic lymphomas (17), mammary tumors from two distinct models (18) (19), multiple myeloma (20), and osteosarcoma (21). We downloaded these freely available datasets from the GEO web-site. A simple hierarchical clustering using the Genesifter™ microarray analysis suite revealed that our previously reported synovial sarcoma-like tumors from Myf5-Cre+/−/SSM2+/− mice clustered together with our newly generated tumors in R-CreER+/−/SSM2+/− mice, while other tumors clustered separately based on their diagnostic identity (Figure 4). Wild type skeletal muscle samples, used as an internal control, clustered separately from all tumor samples. We previously reported identification of a set of 72 genes as a `synovial sarcoma signature' (4). Clustering based on only this signature gene-set also grouped R-CreER+/−/SSM2+/− and Myf5-Cre+/−/SSM2+/− tumors together, separate from the other mouse tumors (data not shown). These suggest that our newly generated mouse tumors are indeed synovial sarcomas.
Figure 4.
Clustering result of various mouse tumors and normal skeletal muscle samples. The box highlights the similarity between tumors generated in R-CreER+/−/SSM2+/− mice (green circles) and those generated in Myf5-Cre+/−/SSM2+/− mice (red circles). Other tumors cluster in distinct groups based on their diagnostic similarity, while skeletal muscle samples cluster distinct from all tumors.
Low level Spontaneous, background, nuclear translocation of CreER
The R-CreER mouse is known to be `leaky', where some background level of nuclear translocation of CreER is observed even in the absence of Tamoxifen (14). We discovered that R-CreER+/−/SSM2+/− mice develop tumors even without Tamoxifen injection (6/6). The age of onset of tumors (by visual detection) is comparable to those seen in R-CreER+/−/SSM2+/− mice that were injected once with 0.1mg/gm of Tamoxifen. All tumors generated, with or without tamoxifen, were fluoroscent (Figure 2b and 2c). On preliminary examination there was no apparent difference in tumor histology or other characteristics between these tumors. This may suggest that the tumors within R-CreER+/−/SSM2+/− (with or without Tamoxifen) are primarily generated by expression of SYT-SSX2 induced by spontaneous nuclear entry of CreER and that a single pulse of low dose Tamoxifen injection probably did not `induce' the tumors. The fact that none of the SSM2 (>200) mice ever developed tumor in the absence of Cre and the similar latency of tumor induction in single-pulse tamoxifen treated and untreated R-CreER+/−/SSM2+/− mice further supports this hypothesis.
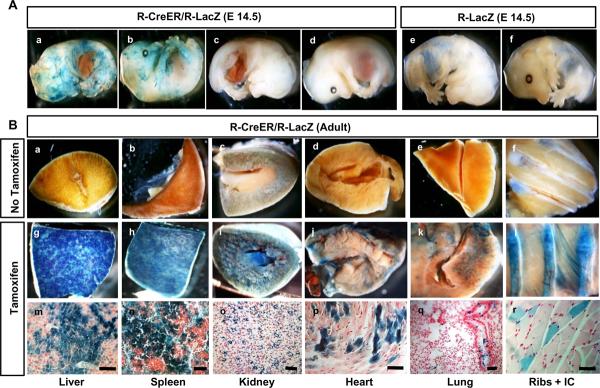
To gain more insight into Tamoxifen induced vs. spontaneous `leaky' nuclear entry of CreER in R-CreER mice, we bred the R-CreER mice to a reporter mouse line conditionally expressing the β-galactosidase marker gene (LacZ) from the ROSA locus (R-LacZ mice) (22). Analysis of R-CreER+/−/R-LacZ+/− embryos revealed that while few a embryos showed significant leakiness in the absence of tamoxifen (Figure 5Aa and 5Ab), most showed no detectable leakiness (Figure 5Ac and 5Ad) compared to control wild type littermate embryos (Figure 5Ae and 5Af). The leakiness was unpredictable, both in terms of number of cells, region, as well as number of individual embryos demonstrating it. CreER in R-CreER+/−/R-LacZ+/− mice showed strong tamoxifen-inducibility (Figure 5Bg to 5Br) compared to controls not exposed to tamoxifen (Figure 5Ba to 5Bf). Surprisingly, skeletal muscles at various locations showed the least nuclear translocation of CreER after intraperitoneal tamoxifen injection (Figure 5Bl and5Br).
Figure 5.
β-galactosidase staining of hemisected R-CreER+/−/R-LacZ+/− E14.5 embryos demonstrated spontaneous nuclear entry (leakiness) of CreER in some embryos (Aa and Ab) while Most R-CreER+/−/R-LacZ+/− embryos did not show detectable leakiness at this stage (Ac and Ad). Control R-LacZ+/− mice were negative for any β-galactosidase staining (Ae and Af).
Various tissues of adult R-CreER+/−/R-LacZ+/− mice did not show β-galactosidase staining in the absence of Tamoxifen (Ba to Bf) while intra-peritoneal injection of 0.1mg/gm Tamoxifen followed by β-galactosidase staining after one week revealed robust β-galactosidase expression in liver (Bg and Bm) and spleen (Bh and Bn), moderated expression in kidney (Bi and Bo) and heart (Bj and Bp), and sparse expression in lungs (Bk and Bq) and Skeletal muscle (Bl and Br).
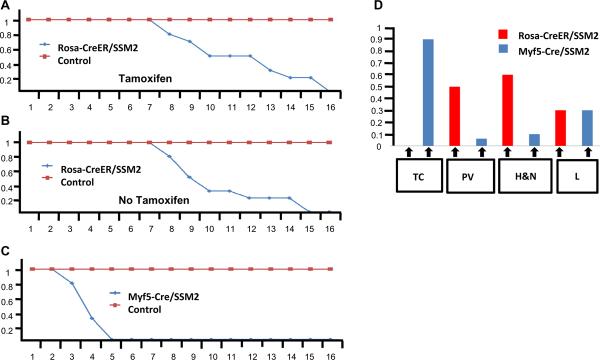
Tumors were generated with complete penetrance in R-CreER+/−/SSM2+/− mice. A single pulse of tamoxifen treatment did not appear to hasten the development of tumors compared to untreated mice (Figure 6A and 6B). Multiple injection of Tamoxifen in R-CreER+/−/SSM2+/− mice led to lethality that was dependent upon both the dosage and frequency of tamoxifen injection (Supplementary Figure C). This again reflects upon the dominant cytotoxic effects of SYT-SSX2 especially when expressed in a large number of cells. We previously reported the generation of the R-DTA mouse line expressing the cytotoxic diptheria toxin from ROSA loucs (23). We next compared the tamoxifen induced lethality between R-CreER+/−/SSM2+/− and R-CreER+/−/R-DTA+/− mice to get an insight into the extent of in-vivo cytotoxic effect of SYT-SSX2 compared to the well known cytotoxic agent diptheria toxin. As expected, tamoxifen injection led to rapid lethality at a lower dosage and frequency within the R-CreER+/−/R-DTA+/− mice when compared to R-CreER+/−/SSM2+/− mice (Supplementary Figure C and D). This suggests that either SYT-SSX2 is not as cytotoxic as the diphtheria toxin or that the cytotoxic effects of SYT-SSX2 are restricted to specific cell types.
Figure 6.
SSM2+/−/R-CreER+/− mice (n=7) and control mice comprising of SSM2+/−, R-CreER+/−, and wild type (n=18) were injected intraperitoneally with one pulse of 0.1 mg/gm of Tamoxifen at 3 months of age. These mice were followed up and the percentage of mice surviving (y axis) was plotted against age (x axis). This reveals that that all SSM2+/−/R-CreER+/− mice injected with Tamoxifen were dead or moribund by 16 months of age while all controls were still alive (6A). A similar comparison was carried out without Tamoxifen injection between SSM2+/−/R-CreER+/− mice (n=6) and control mice comprising of SSM2+/−, R-CreER+/−, and wild type (n=18) that revealed a similar survival differences (6B). In comparison, the survival data from the SSM2+/−/Myf5-Cre+/− mice (n=12) are significantly different where all SSM2+/−/Myf5-Cre+/− mice usually die by 5 months of age (6C).
6D represents the anatomical distribution of tumors detected within SSM2+/−/Myf5-Cre+/− (n=18) and SSM2+/−/R-CreER+/− (n=11) mice. The colored boxes represents the percentage of mice (y axis) harboring tumors in a certain anatomical location (x-axis). TC = Thoracic Cage, PV = Para-vertebral, H&N = Head and Neck, and L = Limbs.
Discussion
Cre-LoxP based conditional systems are considered superior to simple transgenic models due to the ability to target `tumor-inducing' genetic events in specific tissue type. Nonetheless, using standard tissue-specific Cre drivers, a large number of cells within the targeted tissue are affected by the oncogene expression thereby creating a highly un-physiological environment for tumorigenesis. This is a significant limitation of this strategy. Some studies have tried to address this issue such as spontaneous recombination dependent random activation of K-ras leading to lung cancer (24). In this study, we have tried to address this issue by intentionally making Cre activity (and hence SYT-SSX2 oncogene expression) sporadic and randomly distributed within a new mouse model of synovial sarcoma. Although this strategy does not permit tracking of the cell of origin, it does recapitulate the natural pathogenesis more closely by restricting oncogene expression within a small subset of cells, each of which is surrounded by a `normal physiological' microenvironment. In this strategy, random induction of oncogene expression takes place throughout the lifetime of the mouse and across most cell types within a single mouse significantly enhancing the chance of recapitulating the necessary cell and the microenvironment of origin to generate a tumor. Neither a prior knowledge of the cell of origin is required, nor will it be revealed, by this strategy. Although knowledge of the tissue source of a tumor is important from a biological point of view, the ability to rapidly generate mouse models is highly desirable for preclinical applications. Furthermore, the strategy described here is conditional and allows investigators to combine multiple genetic anomalies to attempt at generating a tumor models or investigate the roles of downstream genetic hits within a model. Therefore, with the exception of our inability to trace the origin, the strategy of random induction incorporates the unique advantages of a conditional system minus the disadvantage of `collateral damage' induced by widespread oncogene expression within the target tissue.
Comparing tumors within the new R-CreER+/−/SSM2+/− mouse model with our previous model established by expressing SYT-SSX2 in myoblasts of Myf5-Cre+/−/SSM2+/− mice is instructive. Despite similarity in histology, expression of markers, and transcriptional profile, there are certain differences between these two models. To begin, the age of onset is different in these two models. While Myf5-Cre+/−/SSM2+/− mice died by 3–5 months of age (Figure 6C), R-CreER+/−/SSM2+/− mice live much longer, beyond 1 year, and showed massive tumor load prior to death (Figure 6A and 6B). The tumors within Myf5-Cre+/−/SSM2+/− mice, in most cases, did not appear to be large enough nor numerous enough (based on fluorescence-based visual detection at necropsy) to be the primary cause of death. This suggested either early widespread microscopic metastasis or lethality from a non-tumor related phenotype of widespread SYT-SSX2 expression within Myf5 lineage. In R-CreER+/−/SSM2+/− mice, however, the tumor load was uniformly large enough to be the primary cause of death. This highlights an important limitation of widespread tissue specific oncogene expression. `Collateral damage' induced by oncogene expression in a particular cell lineage may confound phenotype analysis. Of particular concern is pre-clinical application where a candidate drug may have a potent effect on the tumor phenotype, but not on the other `tumor-unrelated phenotype' induced by widespread oncogene expression. In such cases, the mouse model may fail to show a survival benefit despite an effect of the drug on the tumor itself. This is particularly relevant in the case of the synovial sarcoma model since we demonstrate that SYT-SSX2 has two dramatically distinct effects; tumorigenesis and cell death.
Anatomical distribution of tumors in ROSA-CreER+/−/SSM2+/− mice was distinct from Myf5-Cre+/−/SSM2+/− mice, suggesting additional non-myoblast cell of origin in the new model (Figure 6D). Tumors within Myf5-Cre+/−/SSM2+/− mice were observed most frequently in the intercostal region while tumors in the R-CreER+/−/SSM2+/− mice were mostly noted in the para-spinal region (back and tail) and face but not in the intercostal region. Both models, however, often harbored tumors in the limbs. CT scanning and histology revealed significant calcification in some tumors generated in the new model. This fits the range of synovial sarcoma phenotypes in humans where calcification is a relatively synovial sarcoma-specific radiological finding amongst soft tissue sarcomas. Tamoxifen-induced Cre mediated expression of β-galactosidase (LacZ) in R-CreER+/−/R-LacZ+/− mice revealed surprising paucity of LacZ expressing skeletal muscle fibers (Figure 5Bl and 5Br). The differences in age of onset and anatomic location when compared to the myoblast derived synovial sarcomas, makes a non-myogenic origin a possibility in this new model. However, a muscle origin cannot be ruled out in this strategy of random induction. In the light of these findings, the possibility exists that synovial sarcomas have more than one cell-of-origin. Furthermore, the predilection of these tumors to occur in proximity to skeletal elements in both models also raises the issue of whether the cell type or the micro-environment has a more crucial role in the induction of synovial sarcomas. We are currently investigating other potential sources of this lethal disease and address whether origin may have any clinical or prognostic influence.
SYT-SSX2 expression has a prominent cytotoxic effect based on in vivo and in vitro data we show here. It will be very important to investigate the mechanism of cell death and, in particular, how some cells escape this and progress towards neoplasia. From a therapeutic point of view, reactivating these SYT-SSX2 induced pro-apoptotic pathways that were silenced in the tumor cells may be an effective strategy towards designing targeted therapy for this lethal disease. The R-CreER+/−/SSM2+/− random sporadic model in conjunction to the TAT-Cre based in vitro approach we describe here will be very useful in the investigation of cytotoxic pathways activated by SYT-SSX2.
In summary, we describe here a variation on the conditional approach in tumor modeling by taking advantage of the CreER inducible system. We describe a new mouse model of synovial sarcoma utilizing this system and compare it to our previously reported mouse model to highlight the various advantages of this alternate strategy.
Supplementary Material
Acknowledgements
We would like to thank Drs. Lor Randall and Stephen Lessnick for their intellectual input, Drs Cheryl Coffin and Joseph Holden for their expert opinion on pathology, and Dr. Robert B. Weiss, Diane Dunn, and Brett Milash for help with carrying out the microarray and its subsequent analysis. We would also like to thank Sheryl Tripp at the ARUP Laboratories for help with histology and Osama Abdullah at the small animal imaging core for help with the CT scanning. Finally, we would like to acknowledge the efforts by the tissue culture and animal house staff.
Malay Haldar was supported in part by the John E. Goyert Memorial Research Award from the Sarcoma Foundation of America.
References
- 1.Weiss SW, Goldblum JR. Enzinger and Weiss's soft tissue tumors. 4 ed. Mosby, Inc; 2001. [Google Scholar]
- 2.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7(9):645–58. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–18. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 4.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11(4):375–88. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130(25):6361–74. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akiyama H, Kim JE, Nakashima K, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102(41):14665–70. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu K, Xu J, Liu Z, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130(13):3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 8.Licht AH, Raab S, Hofmann U, Breier G. Endothelium-specific Cre recombinase activity in flk-1-Cre transgenic mice. Dev Dyn. 2004;229(2):312–8. doi: 10.1002/dvdy.10416. [DOI] [PubMed] [Google Scholar]
- 9.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–42. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 10.Tronche F, Kellendonk C, Kretz O, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 11.Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10(3):310–5. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 13.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23(6):2314–22. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kemp R, Ireland H, Clayton E, Houghton C, Howard L, Winton DJ. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res. 2004;32(11):e92. doi: 10.1093/nar/gnh090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27(22):7918–34. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutlu AD, Cavallin LE, Vincent L, et al. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi's sarcoma. Cancer Cell. 2007;11(3):245–58. doi: 10.1016/j.ccr.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, Dose M, Kovalovsky D, et al. Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood. 2007;109(12):5463–72. doi: 10.1182/blood-2006-11-059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedlund M, Ng E, Varki A, Varki NM. alpha 2–6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 2008;68(2):388–94. doi: 10.1158/0008-5472.CAN-07-1340. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Tognon CE, Godinho FJ, et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12(6):542–58. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrasco DR, Sukhdeo K, Protopopova M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349–60. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walkley CR, Qudsi R, Sankaran VG, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22(12):1662–76. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 23.Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133(4):581–90. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- 24.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410(6832):1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.