Abstract
Within the past few years, microRNAs (miRNAs) and other non-coding RNAs (ncRNAs) have emerged as elements with critically high importance in post-transcriptional control of cellular and, more recently, viral processes. Endogenously produced by a component of the miRNA-guided RNA silencing machinery known as Dicer, miRNAs are known to control messenger RNA (mRNA) translation through recognition of specific binding sites usually located in their 3′ untranslated region. Recent evidences indicate that the host miRNA pathway may represent an adapted antiviral defense mechanism that can act either by direct miRNA-mediated modulation of viral gene expression or through recognition and inactivation of structured viral RNA species by the protein components of the RNA silencing machinery, such as Dicer. This latter process, however, is a double-edge sword, as it may yield viral miRNAs exerting gene regulatory properties on both host and viral mRNAs. Our knowledge of the interaction between viruses and host RNA silencing machineries, and how this influences the course of infection, is becoming increasingly complex. This review article aims to summarize our current knowledge about viral miRNAs/ncRNAs and their targets, as well as cellular miRNAs that are modulated by viruses upon infection.
Keywords: Virus, microRNA, non-coding RNA, RNA interference, post-transcriptional regulation
1. Introduction
1.1. Description of microRNAs
Over the past few years, microRNAs (miRNAs) have established themselves as important post-transcriptional regulators of gene expression. These short ~21 to 24-nucleotides (nt) non-coding RNA (ncRNA) species are expressed in most eukaryotes and by several viruses. They are known to mediate their action through imperfect base-pairing with their target RNA, mainly with the 3′ untranslated region (3′UTR) of messenger RNA (mRNA), to repress translation, thereby inhibiting protein expression. MiRNAs can also bind to mRNA through perfect complementary to induce cleavage of the mRNA (1,2). A recent study has reported that miRNAs could also enhance protein expression during the cell cycle (3); this aspect remains under active investigation. According to the latest release of miRBase (release 12.0, September 2008), the repository of miRNA data on the web, over 695 different miRNAs have been identified in humans as well as more than 130 miRNAs originating from mammalian viruses (4,5). The number of deposited miRNA sequences continues to increase quasi-exponentially since the creation of miRBase in 2004, suggesting that the number of miRNAs may be estimated and reach in the thousands. Moreover, this repository does not include a panoply of recently discovered, small ncRNAs found in living organisms and forming additional classes of gene regulatory RNAs distinct from miRNAs, such as the repeat-associated small interfering RNAs (rasiRNAs) (6), the tiny noncoding RNAs (tncRNAs) (7) and the Piwi-interacting RNAs (piRNAs) (6). Some ncRNAs produced by viruses are hundreds to thousands of nucleotides in length and represent potential regulators of gene expression (8–12).
1.2. MicroRNA biogenesis and function
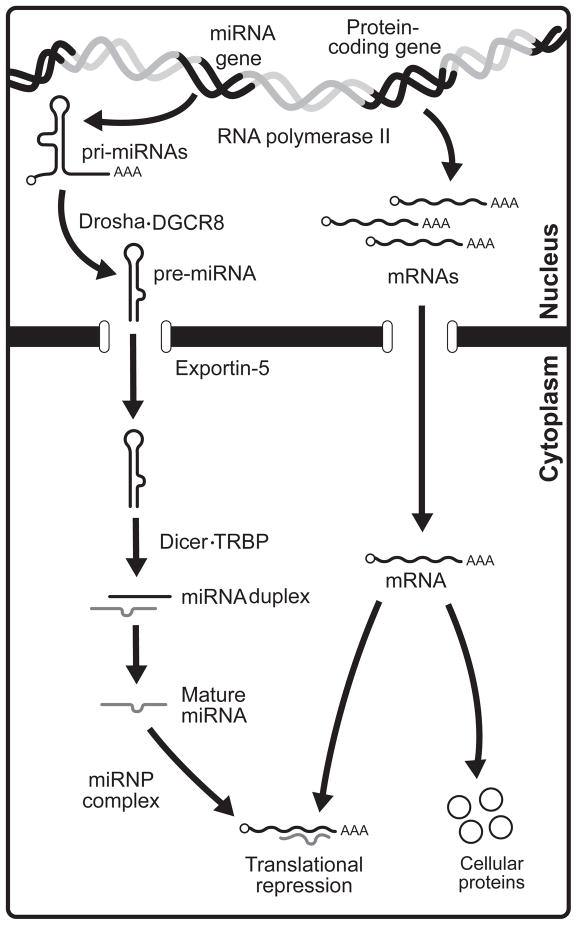
As illustrated in Figure 1, miRNA genes are transcribed in the nucleus mainly by RNA polymerase (pol) II into stem-loop structured primary miRNAs (pri-miRNAs). Harboring a 5′ m7G cap and a 3′ poly(A) tail (13,14), these pri-miRNAs are then trimmed into ~60–70-nt miRNA precursors (pre-miRNAs) by the nuclear ribonuclease (RNase) III Drosha (15), acting in concert with the DiGeorge syndrome critical region 8 (DGCR8) protein within the microprocessor complex (16–19). A non canonical generation of pre-miRNAs, in which certain debranched small introns mimic the structural features of pre-miRNAs to enter the miRNA-processing pathway without Drosha-mediated cleavage, named «mirtrons», has been described initially in Drosophila (20), but are also present in mammals (21). To date, no viral «mirtrons» have been reported. The canonical and non canonical pre-miRNAs are subsequently exported to the cytoplasm via Exportin-5 (22–25) and the base of their stem is recognized by the PAZ domain of Dicer (26). Acting as an intramolecular dimer, Dicer RNase IIIa and IIIb domains cleave the stem at the base of the loop to generate a miRNA:miRNA* duplex (26–29). The transactivating response RNA-binding protein (TRBP) (30) has been shown to operate with Dicer within a pre-miRNA processing complex (31,32), although their precise mechanistic interaction remains elusive. Following a strand selection and separation step, which is based on the thermodynamic stability of the RNA duplex (33), the miRNA strand (~21 to 24-nt) with the least stable 5′ end pairing (called the guide strand) is incorporated into effector miRNA-containing ribonucleoprotein (miRNP) complexes containing Argonaute 2 (Ago2), TRBP and Dicer (32), guiding them towards specific messenger RNAs (mRNAs). The opposite miRNA* strand (also called passenger strand) is encountered much less frequently and is presumably degraded (34). miRNA assembly on specific mRNA sequences may be facilitated by the fragile X mental retardation protein, which can accept and use miRNAs derived from Dicer (35). The targeted mRNA will be primarily subjected to translational repression, although mRNAs containing partial miRNA complementary sites may also be targeted for degradation in vivo (36). These regulatory events may occur at specific cytoplasmic foci referred to as processing bodies (P-bodies) (37,38), or GW182-containing bodies (GW-bodies) (39), which are formed as a consequence of the presence of miRNAs (40). P-bodies are enriched in proteins involved in RNA-mediated gene silencing, such as Ago2 (37), mRNA degradation (41), and nonsense-mediated mRNA decay (42,43).
Figure 1.
Schematic representation of the miRNA-guided RNA silencing pathway in mammalian cells. miRNA genes are transcribed from miRNA genes by RNA polymerase II into primary miRNAs (pri-miRNAs) and then trimmed into miRNA precursors (pre-miRNAs) in the nucleus by the microprocessor complex, which is composed of the ribonuclease Drosha and its cofactor DGCR8. Pre-miRNAs are then exported by Exportin-5 to the cytoplasm where they are cleaved by the pre-miRNA processing complex formed of Dicer and TRBP to generate miRNA:miRNA* duplexes. Following a strand selection and separation step, the mature miRNA is incorporated into an Ago2-containing miRNA ribonucleoprotein complex (miRNP) (or effector complex) to mediate recognition and translational repression of specific cellular mRNAs.
Cellular miRNAs have been shown to control various processes, such as cell proliferation, apoptosis and hematopoietic cell differentiation (44). As for viral miRNAs, which is the subject of this review, they can regulate expression of viral as well as host proteins by directing repression or cleavage of mRNA transcripts, thereby inhibiting key cellular processes involved in the response to viral infection. At the molecular level, recognition of mRNAs by miRNAs is based mainly on imperfect sequence complementarity and the identification of their physiological mRNA targets remains difficult to predict and is rather arduous. Characterization of a few experimentally validated miRNA:mRNA interactions (45), however, allowed to establish a context in which this interaction is favoured. The critical miRNA:mRNA pairing region, referred to as the “miRNA seed”, involves nt 2 to 8 of the miRNA in the 5′ to 3′ orientation.
Because a miRNA can affect a large number of mRNAs, bioinformatic approaches are used in combination with large-scale micro-array-based analyses of regulated mRNAs and/or proteomic analyses of differentially expressed proteins in order to validate miRNA-regulated mRNAs. Whether they originate from the host-infected cells or the viruses themselves, it has become a priority to improve our understanding of the mechanisms by which miRNAs or others ncRNAs mediate their action, to identify the mRNA targets they regulate and to determine how they can modulate the host response to viral infection and the intrinsic replication of the virus. This review article will present and discuss some examples in which miRNAs and ncRNAs can act as key regulators of cellular and viral gene expression in virus-infected cells.
1.3. Techniques for miRNA/ncRNA detection
Over the last several years, many different techniques have been used to predict and identify miRNAs derived from viruses and their host cells. As part of the effort to better understand the interaction between miRNAs and their targets, computational algorithms have been developed based on well defined rules for various molecular features. These in silico approaches provide important tools for miRNA target detection and, together with more elaborated experimental validation strategies, help reveal mRNA targets that are functionally regulated by miRNAs.
Standard Northern blot hybridization, which may be adapted for specific applications, is the most commonly used technique to detect miRNAs and other small ncRNAs, as described recently (46,47). A method of choice to visualize specific miRNAs under various conditions, Northern blotting requires the design and synthesis of as many sequence-specific, complementary DNA probes, which hampers its use in large-scale miRNA detection strategies.
RNase protection assay (RPA) is another indirect technique used to detect unique miRNAs in aqueous solutions. This approach consists in the annealing of our RNA species of interest with a complementary, radiolabeled RNA probe in a test tube, followed by RNAse A/T1 digestion of single-stranded RNA species. Protected upon annealing of the RNA species of interest, the radiolabeled probe is then revealed by denaturing PAGE and autoradiography. Although both methodologies demand that the sequence of the RNA species of interest to be known, which makes them unsuitable for discovering miRNAs of unknown sequences, RPA requires lower amounts of RNA than Northern blot hybridization. Furthermore, whereas both techniques allow discrimination between pre-miRNA and mature miRNA species, RPA may represent a suitable and more sensitive detection method than Northern blot hybridization for miRNAs that are expressed at very low levels, such as those originating from the human immunodeficiency virus type 1 (HIV-1) transactivating responsive (TAR) element (48).
As for the sequence determination of miRNAs of low abundance, primer extension may prove to be more useful and informative than miRNA cloning strategies. It consists in the reverse transcriptase (RT)-driven elongation of a DNA probe designed to be complementary to the 3′ end of the miRNA. Visualized by denaturing PAGE and autoradiography, the length of the elongated product determines the 5′ end of the miRNA under study.
In situ detection approaches of miRNA expression also offers several advantages. Using paraffin-embedded and formalin-fixed tissues, this method may provide access to large repositories of archived biological materials, thereby allowing retrospective studies to be conducted. The Plasterk laboratory, among others, has perform a lot of work in animal embryos by using Locked Nucleic Acid™ (LNA)-modified oligonucleotide probes that contain nucleic acid analogues in which the ribose ring is locked by a methylene bridge connecting the 2′-O atom with the 4′-C atom. This molecular conformation allows higher sensitivity and thermal stability. Specific in situ detection of miRNA is also applicable to whole mounts, thin sections, single cells and frozen samples. LNA™ probes can also find applications in Northern Blot hybridization. As for colorimetric assays using digoxigenin-labelled or biotin-labelled probes, or fluorescence microscopy using fluorescein-labelled probes, it may help visualize the expression profile of specific miRNAs.
The detection and identification of miRNAs and ncRNAs, which may be used as biomarkers, may be clinically relevant and provide critical information for diagnostic purposes. Significant progresses have been made in that area with the development of quantitative bioanalytical methods for the rapid and multiplexed detection of all miRNAs present in a particular cell or tissue sample. Common research laboratory techniques, such as reverse transcription and real-time polymerase chain reaction (RT-PCR), are now being used in combination with microarrays for the identification of pri-miRNAs, pre-miRNAs and mature forms of miRNAs. These techniques, however, often require several cloning steps as well as the need for expensive devices and pieces of equipment to compute and analyze data. High throughput, deep sequencing technologies have been developed recently and have been shown to deliver several orders of magnitude more sequences than is possible with the traditional Sanger method. This advanced technology has already been used to identify conserved and nonconserved miRNAs. Concomitantly, new algorithms have been developed in order to analyze and make sense of the huge amounts of sequencing data that can be generated. Such a bioinformatic tool, miRDeep, uses a probabilistic model of miRNA biogenesis to score compatibility of the position and frequency of sequenced RNA with the predicted secondary structure of the miRNA precursor (49). Sequences obtained by deep-sequencing are aligned to the genome and miRDeep computes their secondary RNA structure. Potential pre-miRNA sequences are then identified and, according to miRDeep algorithm, scored for their likelihood to represent real pre-miRNAs. The output is a list of known and putative pre-miRNAs and mature miRNAs, as well as the probabilities of being false positives.
The increasing importance of endogenous miRNAs and ncRNAs in health and diseases, as well as their potential clinical use, will further encourage the improvement of current methods and stimulate the development of new technologies and strategies that will enhance the chances of successful knowledge transfer from the laboratory to the bedside of patients.
2. Viral miRNAs and ncRNAs in viral-infected cells
2.1. Herpesviridae
The family of Herpesviridae is represented by three virus subgroups (alpha, beta, and gamma) that contain large double-stranded DNA (dsDNA) genomes as long as ~125–230 kilobases (kb) (50). Typically, herpes viruses cause lytic infections or generate latent infections where only a few specialized viral genes are used by the virus to establish lifelong persistence in the human host. Some herpes viruses have been found to express miRNAs (51,52). For instance, a mouse herpesvirus, known as mouse cytomegalovirus (mCMV), contains more than 18 validated miRNAs, all of which are related to the lytic phase of infection (53,54). In this section, we will discuss and provide more details about miRNAs generated by herpes viruses as well as their potential gene targets in infected human cells.
2.1.1. Herpes simplex virus 1 (HSV-1) and 2 (HSV-2)
The first group to study and report the computational predictions and identification of miRNAs derived from viruses was led by Thomas Tuschl (51,52). Using mainly miRNA cloning strategies as their experimental miRNA discovery tool, they were unable either to validate some of their predictions or to confirm the existence of some viral miRNAs discovered later by other groups by means other than miRNA cloning.
For example, Cui et al. (55) have predicted the existence of 13 pre-miRNAs and 24 mature miRNA candidates from 11 genomic loci in HSV-1 (or human herpesvirus type 1, HHV-1), 30% of which are predicted to be conserved in HSV-2 (or human herpesvirus type 2, HHV-2). The authors used Northern blot hybridization to validate the first miRNA in HSV-1, which is encoded upstream of the transcription start site of the latency-associated transcript (LAT); this miRNA was named hsv1-miR-H1. LAT is an ~8.3 kb capped and polyadenylated RNA transcript, not coding for a protein, which is spliced to give an ~2.0 kb stable intron and an unstable exonic RNA of ~6.3 kb (56). Additional small ncRNA species of ~38 and ~110 nt were also identified, although whether they play a role in HSV-1 biology remains unclear.
Although its role and importance in HSV-1 biology remains elusive, hsv1-miR-H1 is expected to act upon and regulate cellular and viral RNA transcripts. Among the predicted pre-miRNAs that could derive from HSV-1, two of them may target UL15, UL15.5 and the intron-containing HSV-1 infected-cell protein 0 (ICP0) transcripts. The latter transcript encodes for an immediate-early polypeptide with transcriptional activation properties found to exert antiproliferative action in infected cells (57) and to favor viral RNA transcription (58). While hsv1-miR-HI targets remain under investigations, two independent groups recently published the identification of novel miRNA candidates and their putative targets for HSV-1 and HSV-2. Umbach et al. (59) used the 454 sequencing technology to analyze 293T cells transfected with a LAT-expressing plasmid as well as the trigeminal ganglia of mice latently infected with HSV-1. They were able to obtain more than 200,000 sequence reads, of which more than half represented cellular miRNAs. The authors found 651 HSV-1-derived miRNAs in 293T cells and more than 815 HSV-1 miRNAs in mouse trigeminal ganglia. Six mature miRNA candidates were identified in both cell types: miR-H2-3p (359 reads), miR-H4-3p (266 reads), miR-H2-5p (10 reads), miR-H4-5p (61 reads), miR-H3 (23 reads), miR-H5 (41 reads). Three of these miRNAs had been computationally predicted (51,55), whereas four derived from exon 2 of the spliced ~6.3 kb LAT transcript, which may explain its characteristic instability and provide a rationale for the existence of spliced LAT transcript, according to Umbach et al. (59). These authors also described that miR-H2-3p and miR-H6 may target ICP0 and ICP4 respectively, which are two transcription factors implicated in HSV-1 replication. If ICP0 and ICP4 proteins can promote exit from latency (60), the regulation of their mRNAs by HSV-1 miRNAs could contribute to maintain the latent state of herpes viral infections.
The existence of viral miRNAs suggest that they may play an important role in viral pathogenesis. For instance, it has been known for some time that the herpesvirus ICP34.5 protein promotes replication of the virus in neuronal cells in vivo (61). Tang et al. (62) recently found a miRNA derived from HSV-1, namely miR-I, which has been shown to reduce the protein expression level of ICP34.5 in transfected or HSV-1-infected cells. These results prompted the authors to hypothesize that the control of ICP34.5 expression in individual infected neurons by these LAT-encoded miRNAs may affect the outcome of viral infection, i.e. productive infection versus latency, leading either to viral spreading to other neurons or establishment of latency, respectively.
2.1.2. Epstein-Barr virus (EBV)
Epstein-Barr virus (EBV), or human herpesvirus type 4 (HHV-4), is a gammaherpesvirus which is maintained in the nucleus of the infected cell as an extra-chromosomal circular episome. Found to be widespread in all human populations, EBV is known to persist in the vast majority of individuals as a lifelong, asymptomatic infection of the B-lymphocyte pool (63,64). In fact, EBV has the capacity to immortalize B cells in culture. This virus has been identified as the causative agent of infectious mononucleosis, Hodgkin’s lymphoma (HL), Burkitt’s lymphoma (BL) and nasopharyngeal carcinoma (NPC) (65). A study of Pfeffer et al. (52) reported the cloning of miRNAs from BL cell lines infected with EBV B95-8 strain, where 4% of the total miRNA content obtained by cloning originated from the EBV genome. They found 5 miRNAs originating from 2 different clusters, i.e. either located within the BHRF1 (Bam HI fragment H rightward open readingframe 1) gene or in the intronic regions of the BART (Bam HI-A region rightward transcript) gene. The existence of miRNAs derived from these clusters in EB has been confirmed by an independent group (66). It is now known that EBV miRNAs originate from 3 clusters and that 2 of the three clusters of miRNAs are made from the BART gene, a set of alternatively spliced transcripts that are highly abundant in NPC but have not been shown to produce detectable levels of proteins (67). Therefore, whereas 3 pre-miRNAs are encoded in the BHRF cluster, more than 20 pre-miRNAs are found within the introns of the BART transcript.
The life cycle of EBV, as others herpesvirus, is biphasic. Upon primary infection, EBV replication is associated with the expression of more than 50 viral proteins that helps the virus establish a latent infection in these cells (68). EBV infection is characterized by 3 different stages of latency (I, II and III), which are each associated with the expression of various subsets of latent genes. Latency I, which is related to BL, is characterized by the expression of latency-associated membrane protein 2a (LMP2A) and EBV-associated nuclear antigen 1 (EBNA1), which is responsible for EBV viral replication. Some lytic events may occur in latency I. Latency II is an intermediate state and is followed by latency III, where all 12 latency genes are expressed, including six nuclear proteins (EBNA-1 to 6), three membrane proteins (LMP-1, LMP-2A and LMP-2B), BART and two small non-translated RNAs (EBER 1 and 2; see Figure 2). This stage of infection is observed in B cells transformed in vitro by EBV or in lymphoproliferative disorders arising in the presence of immunosuppression. Latent EBV gene expression may exhibit cell type-specific patterns in cultured cells, as observed following infection of primary resting human B cells (latency III, the growth program) or in cell lines derived from EBV-associated cancers (latency I or II) (68,69).
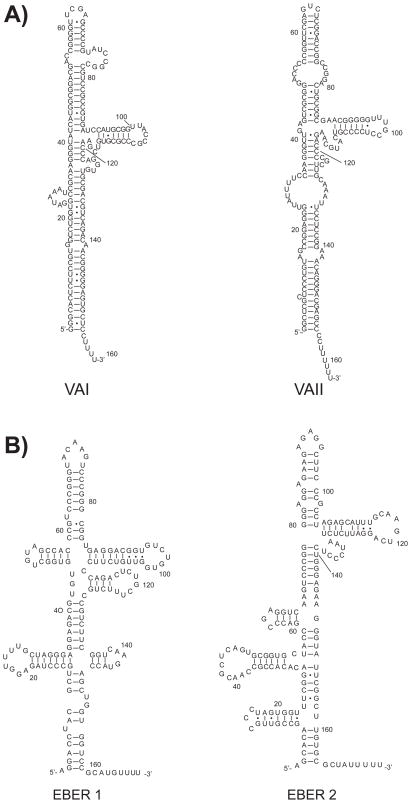
Figure 2.
Representation of non-coding RNAs from Adenovirus and Epstein-Barr virus. A) VAI and VAII RNAs from adenovirus 5 (Ad5). Adapted from Xu et al. (109). B) EBER1 and EBER2 RNAs from Epstein-Barr virus. Adapted from Rosa et al. (123).
The expression pattern of the various EBV miRNAs appears to be fairly complex and depends on both the cell type and on the overall expression pattern of the EBV genes (8). For example, the BART miRNA cluster is well expressed during the lytic process and in cells lines derived from NPC, whereas it is less abundant in B-cells transformed in vitro or derived from BL (70). On the other hand, the BHRF cluster is expressed in BL cells, but not detected in NPC cell lines (70,71). Thus, expression of EBV miRNAs at different stages of infection may underlie their specific role and importance in viral pathogenesis progression.
Several reports have described cellular and viral targets for EBV miRNAs. One of the first targets to be reported was for miR-BART-2. This miRNA was found to target, through perfect complementary, the 3′UTR of the viral polymerase BALF5 mRNA, which is transcribed antisense to miR-BART-2 (52). Evidences for this regulation came from the following observation made after induction of the lytic viral replication cycle in the EBV B95.2 cell line: the decrease in BALF5 3′UTR cleavage was concomitant with that of the level of miR-BART-2 (72). Expressed at very low levels in latently-infected cells, miR-BART-2 could downregulate the levels of aberrant BALF5 mRNA transcripts in order to prevent viral replication during latency.
Other BART miRNAs were recently shown to target an EBV viral gene. MiR-BART-1-5p, miR-BART-16 and miR-BART-17-5p were shown to downregulate EBV latent membrane protein 1 (LMP1) through recognition of the 3′UTR of its mRNA (73). LMP1 have oncogenic properties and is a signaling protein that acts as an active tumor necrosis factor receptor (TNFR) through its resemblance to CD40, thereby activating a number of signalling pathways in a ligand-independent manner (74,75). Another BART miRNA was recently described to target the cellular PUMA mRNA, which encodes for a protein known to up-regulate p53-mediated apoptosis (76). By targeting PUMA mRNA, miR-BART-5 could render NPC and EBV gastric carcinoma cells less sensitive to proapoptotic agents. In that context, apoptosis could be triggered either by depleting miR-BART-5 levels or by enhancing PUMA expression. These findings support a role for miRNAs in the establishment of latent infection through promotion of host cell survival.
Whereas the BART miRNAs may act mainly during the replicative period of EBV infection, BHRF1 miRNAs are produced in latently-infected cells. A cellular target has been validated for one of the 3 BHRF1 miRNAs that are produced by EBV. The BHRF1-3 miRNA has been found to target, through perfect complementary, the 3′UTR of IFN-inducible T-cell attractive chemokine CXCL11/I-TAC (77). Downregulation of CXCL11/I-TAC levels by miR-BHRF1-3 could lead to the immunomodulation of EBV-infected tumor cells by interfering with the IFN-responding pathway. EBV may also interfere with other immune processes of the host cells through the regulation of cellular miRNAs, as discussed in Section 3.
Interestingly, the authors also observed high expression levels of miR-BHRF1-3 in latency type III EBV-infected cells and in primary EBV associated AIDS-related diffuse large B-cell lymphomas (DLBCL) (77). This pattern differs from that of miR-BART-2, which exhibits low expression levels in latency type III EBV-infected cells but high expression, as BHRF1-3, in AIDS-related DLBCL. These observations raise the possibility that EBV miRNA expression may be modulated by HIV-1, or others viruses, and influence the course of EBV infection, which adds to the complexity of coinfection cases.
2.1.3. Human cytomegalovirus (HCMV)
Human cytomegalovirus (HCMV), or human herpesvirus type 5 (HHV-5), was first identified in 1904 as a betaherpesvirus with high prevalence in human adults (50-80%) and mainly asymptomatic, except for immunocompromised individuals, such as very young children, organ transplant recipients, HIV-1-infected persons and those with leukemia. Far from being harmless, this virus has emerged in recent years as the most important cause of congenital infection in the developed world, commonly leading to mental retardation and developmental disability. In healthy people, however, shedding of the virus can occur intermittently, without any detectable signs or symptoms in the long term. The latent form of the virus may persist in T lymphocytes (CD8+) and some hematopoietic cells (78,79).
Pfeffer et al. (51) initially predicted the presence of miRNA precursors in the genome of HCMV, of which 9 of them have been cloned from small RNA libraries isolated from primary infected human fibroblasts. Additional HCMV miRNAs were subsequently reported (80,81). To date, miR-UL112-1 is the only HCMV miRNA for which an mRNA target has been identified. In fact, miR-UL112-1 has been proposed to regulate the immediate-early protein IE72/IE1 of HCMV (82,83) as well as the major histocompatibility complex class I polypeptide-related sequence B, or MICB, of the host (84). HCMV IE72/IE1 is a multifunctional protein involved in many cellular processes including cell cycle regulation, apoptosis, nuclear architecture, and gene expression (85). The observations that miR-UL112-1 is expressed early and is accumulating during HCMV infection led Grey and colleagues to suggest that the regulation of IE72/IE1 protein at later stages of viral replication may attenuate the acute phase of replication (80,86). The biological significance of the IE72/IE1 suppression, however, remains unclear. The other target of miR-UL112-1, MICB, has been postulated to help the HCMV-infected cells evade killing by activated natural killer (NK) cells (84). MICB is a stress-induced ligand of the NK cell activating receptor NKG2D and is critical for the NK cell killing of tumor and virus-infected cells. Suppression of MICB expression by miR-UL112-1 would prevent recognition by NK cells, a function shared by six other HCMV genes encoding proteins UL16, UL18, UL40, UL83, UL141 and UL142 (87). HCMV UL16 protein, like miR-UL112-1, targets MICB (88) may further contribute to HCMV persistence in its host cells.
Although it remains unclear as to how miR-UL112-1 may influence HCMV pathogenesis, through its regulatory effects on a viral and a cellular target, these findings argue for the persistence of HCMV in infected cells through reduction of the viral load and promotion of immune evasion mechanisms.
2.1.4. Kaposi’s sarcoma-associated herpesvirus (KSHV)
This recently known virus, also referred to as human herpesvirus type 8 (HHV-8), is less common among healthy individuals (< 2% of the population of developed countries) and is rather unique in a way that it acquired numerous genes from host cells and has incorporated them into its own genome. Some ot these genes encode for complement-binding protein, IL-6, Bcl-2, cyclin D, a G protein-coupled receptor, interferon regulatory factor, Fas-ligand inhibitory protein (FLIP) and DNA synthesis proteins, such as dihydrofolate reductase, thymidine kinase, thymidylate synthetase, DNA polymerase and several others (89). The gammaherpesvirus KSHV-induced skin lesion manifestations were visualized on KSHV- and HIV-1-infected patients who are immunocompromised. Additional symptoms associated with KSHV infection are primary effusion lymphoma (PEL) and Castleman’s disease (89), which exhibit KSHV-positive tumors hypersecreting IL-6 in response to viral protein vIL-6 (90).
Reported by Pfeffer and collaborators (51), miRNAs are abundant in the KSHV genome. All 10 KSHV miRNA stem–loops identified are located in one short segment in the ~141-kb KSHV genome, and are all oriented in the same direction suggesting that they may all derive from a single pri-miRNA transcript (91). So far, 12 miRNAs derived from the KSHV genome have been identified from 4 independent teams of investigators (see Table 1) (51,66,91,92).
Table 1.
MicroRNAs and non-coding RNAs expressed by human viruses.
| Virus family | Virus name | miRNAs/ncRNAs | Identification of validated miRNAs/ncRNAs | References | |
|---|---|---|---|---|---|
| Predicted | Validated | ||||
| Herpesvirus | Herpes simplex virus type 1 (HSV-1, HHV-1) | (mi) 24 | (mi) 8 | hsv-miR-H1, miR-H2-3p, miR-H4-3p, miR-H2-5p, miR-H3, miR-H5, miR-H6, miR-I | 51,52,61,64 |
| 0 | (nc) 2 | ~38 and ~110 nt from the LAT transcript | 55 | ||
| Herpes simplex virus type 2 (HSV-2, HHV-2) | (mi) 10 | 0 | - | 51,55 | |
| Varicella zoster virus (VZV, HHV-3) | 0 | 0 | - | - | |
| Epstein-Barr virus (EBV, HHV-4) | (mi) 7 | (mi) 23 | miR-BHRF1-1 to BHRF1-3, miR-BART-1 to miR-BART-20 |
51,52,66 70,73 |
|
| (nc) 2 | EBER1, EBER2 | 8,125,126 | |||
| Human cytomegalovirus (HCMV, HHV-5) | (mi) 11 | (mi) 11 | miR-UL22A, miR-UL36, mir-UL70, miR-UL112, miR-UL148D, miR-US4, miR-US5-1,miR-US5-2, miR-US25-1, miR-US25-2, miR-US33 | 50,80,81 | |
| (nc) 1 | β2.7 | 127 | |||
| Roseolos virus (HHV-6) | 0 | 0 | - | - | |
| HHV-7 | 0 | 0 | - | - | |
| Kaposi’s sarcoma-associated herpesvirus (KSHV, HHV-8) | (mi) 8 | (mi) 12 | miR-K12-1 to miR-K12-12 | 51,66,91,92 | |
| Poliomavirus | Simian virus 40 (SV40) | (mi) 1 | (mi) 2 | sv40-miR-S1-5p, sv40-miR-S1-3p | 51,101 |
| Simian virus 12 (SV12) | ND | (mi) 2 | unnamed | 102 | |
| Jamestown Canyon virus (JCK) | (mi) 1 | (mi) 2 | unnamed | 103 | |
| BKV | (mi) 1 | (mi) 2 | unnamed | 103 | |
| Adenovirus | Adenovirus type 2 and 5 | 0 | (nc) 2 | VAI, VAII | 12,104 |
| (mi) 1 | (mi) 3 | mivaRI-137, mivaRI-138 (or 3′svaRNA), mivaRII-138 | 47,104-107 | ||
| Retrovirus | Human immunodeficiency type 1 | 0 | 3 | miR-N367, miR-TAR-5p, miR-TAR-3p | 48,113,116 |
| Human immunodeficiency type 2 | (mi) 2 | 0 | - | 118 | |
| HTLV-1 | 0 | 0 | - | 119 | |
As other herpes viruses, KSHV latency occurs following primary infection through the expression of latency genes, which are restricted to a few viral genes, most of which are located in, or nearby, the region comprising ORF71 to K12 (Kaposin gene), which encodes more than 10 miRNAs (91). This region of the KSHV genome presents polycistronic latency-associated transcripts encoding the latency-associated nuclear antigen (LANA or open reading frame 73; ORF73), v-cyclin (ORF72), and v-Flip (ORF71). MiRNAs and KSHV-associated latency genes are expressed coordinately in latent cells.
Microarray analysis of cell lines stably expressing the KSHV miRNA cluster revealed decreasing levels of several mRNAs, all of which may represent potential cellular targets of KSHV miRNA (93). Using luciferase derepression assays involving cotransfection of 3′UTR-coupled reporter genes and specific 2′OMe antisense RNAs, Samols et al. (93) found that the 2,095 bp-long thrombospondin 1 (THBS1) mRNA 3′UTR is targeted by multiple KSHV miRNAs, in particular miR-K12–1, miR-K12-3-3p, miR-K12-6-3p and miR-K12–11. This cellular protein, which possesses antiproliferative, angiogenic and immunostimulatory functions, has been reported to be downregulated in several types of cancer, including Kaposi sarcoma (KS) (94–96). Cellular proteins exerting a role similar to THBS1 in proliferation, immune modulation, angiogenesis and apoptosis, such as osteopontin (SPP1), the S100 calcium binding protein A2 (S100A2), the plasticity related gene 1 (SRGN or PRG1) and the integral membrane protein 2A (ITM2A) are all downregulated by >4 fold upon expression of the KSHV miRNA cluster. Again, these findings are supportive of a viral miRNA-based escape mechanism from the immune system for KSHV in KS.
Interestingly, miR-K12-11 derived from KSHV contains a seed sequence identical to the oncogenic cellular miR-155, as reported recently in (97,98). In fact, these two independent groups described the downregulation of 20 and 14 cellular genes by both miRK12-11 and miR-155, respectively. A single gene, however, was identified by both teams: BACH-1. This gene is known to encode a transcription factor that negatively regulates transcription of some stress-responding factors in cells (99). Among the other genes identified in these studies are members of cell signaling pathways, cell division, apoptosis, T-cell activation as well as transcription factors. The discrepancies between these studies may be explained by the differences in the cell types and models that were used. Skalsky et al. (98) used PEL-derived cell lines (BC-1, JSC-1, VG-1, BCBL-1, and BCB-1) and overexpression of miR-K12-11 and miR-155 in Hek 293 cells, whereas Gottwein et al. (97) expressed physiological levels of miR-K12-11 and miR-155 from lentivirus in BJAB cells.
2.1.5. Other herpes viruses
To date, no miRNAs have been either predicted or identified for the alphaherpesvirus Varicella zoster, or human herpesvirus type 3 (HHV-3) (51), the causative agent of chickenpox and zona, where symptoms take the form of red papules among the body of the infected host. The same for Roseolovirus, or human herpesvirus type 6 (HHV-6) and HHV-7, which is genetically closely related to HHV-6. In contrast to previously described herpes viruses, the calculated probability to find a miRNA within their genomes is inferior to 50% in HHV-3 and HHV-7. The probability that HHV-6 encodes for a miRNA is high at 84%, although no miRNA has been identified yet. Possibly expressed at very low levels, miRNAs derived from HHV-6 may have escaped detection so far.
2.2. Other DNA viruses (polyomavirus and adenovirus)
Several DNA viruses, in contrast to herpes viruses, contain small genomes of a few thousands bp that encode a small number of proteins. Three polyomavirus have been isolated in humans; Jamestown Canyon virus (JCV), BK virus (BKV) and the simian virus 40 (SV40). Both JCV and BKV can infect humans and cause serious diseases, such as progressive multifocal leukoencephalopathy (PML) upon JCV infection in AIDS-immunocompromised patients and tubulointerstitial nephritis upon BKV infection in kidney transplant patients. Both JCV and BKV may have their importance in cancer (100). As for the monkey SV40 virus, it has contaminated the human population when poliovirus vaccination was used 50 years ago.
Two miRNAs originating from the same pre-miRNA and produced from the SV40 genome, were computationally predicted and experimentally validated by Sullivan et al. (101) in 2005. By using Northern blot and RNase protection assay (RPA), the group mapped the production of sv40-miR-S1-5p and sv40-miR-S1-3p from TC-7/Db cells infected with SV40. They explored the function of these miRNAs by using a mutant that was constructed by mutagenizing selected bases in the region of the predicted pre-miRNA to disrupt the hairpin structure of the pre-miRNA on the late strand, while leaving intact the amino-acid coding potential of the T antigen on the early strand. It came to light that SV40 miRNAs, expressed late in the infection, can direct cleavage of early mRNAs that occur for small- and large-T antigens. Without affecting the infectivity or replication of the virus, downregulation of the T antigens seemed to decrease the susceptibility to cytotoxic T lymphocytes (CTL) and cytokine release, again suggesting a role for viral miRNAs in escape mechanisms from the host immune system. The same year, another group published the identification of miRNAs from the simian virus 12 (SV12) with intriguing sequence complementarities with the SV40 antigen (102).
Three years later, new miRNA sequences from polyomavirus genome were identified using similar experimental approaches. JCV, BKV and SV40 were reported to share homologous pre-miRNA hairpins, both arms of which are processed into mature miRNAs (103). Despite being only ~65% identical (5p miRNAs are ~55% identical, and 3p miRNAs are ~75% identical), all three pre-miRNAs share several atypical properties in terms of processing and abundance (103). Interestingly, the authors have reported that both arms of the precursor hairpin can be active on the same target, in this case viral early RNAs. They reported the same directed cleavage of these mRNAs by JCV and BKV miRNAs, as for those from SV40, by using 5′RACE to map the early mRNA cleavage products. Finally, detection of JCV miRNAs in the brain tissues of PML patients may help create therapies based on interference with existing miRNAs.
Bigger than polyomavirus, adenoviruses are medium size, dsDNA viruses known to infect mammalian cells (104). Most infections with adenovirus result in problems with the upper respiratory tract of the infected host. Other pathologies were also assigned to adenoviruses, such as ear infection, gastroenteritis, cough and, rarely, viral encephalitis. Some miRNAs have been predicted for a few species of adenoviruses (51) but, until recently, no miRNA function has been described. A particular aspect of the adenovirus 2 biology is worth noting and pertains to the production of two non-coding RNAs, named VAI and VAII (See Figure 2A) (105). The ~160-nt long VAI RNA accumulates in large amounts during adenovirus infection. As discussed later, VAI RNA has the ability to interfere with the interferon-related cellular defense mechanism and, thus, with protein synthesis by blocking the activity of protein kinase R (PKR) leading to the inhibition of eukaryotic initiation factor 2a (eIF2a) phosphorylation (10,106).
VAI RNA also exhibits a secondary structure formed of two short imperfectly base-paired stems, referred to as the terminal and apical stems, respectively, and a structurally complex domain, known as the central domain (See Figure 2A) (11). Although the ability of the VAI RNA to interfere with the cellular components of the miRNA pathway of the host will be discussed later (see Section 2.4), its processing by components of the miRNA pathway, such as Dicer, is a recent concept supported by two independent groups (107,108). Sano et al. (108) found that VAI RNA is processed by Dicer in the cytoplasm and identified by Northern blot hybridization one miRNA originating from the right strand of the 3′ terminal stem region. They determined the cleavage sites by S1 nuclease mapping, which consists in the degradation by S1 nuclease of single-stranded RNA (ssRNA) (or ssDNA), while preserving dsRNA hybrids (108). Although the authors have shown that the small RNA derived from VAI is functional and can downregulate a reporter gene in cultured cells, whether it influences expression of any cellular genes remains to be determined.
Other groups have reported the identification of one to three different miRNAs generated from VAI and VAII RNAs of the adenovirus type 5, and originating from the 3′ stem region (107,109,110). VAII RNA represented a more potent substrate for Dicer and ~2-fold more VAII RNA-derived small RNAs were present in the total pool of small RNAs. Approximately 1.5% of VAII RNA is cleaved by Dicer, resulting in the production of ~75,000 copies of mivaRII-138 miRNA in late-infected cells. This huge amount of mivaRII-138 miRNA may exert potent gene regulatory effects and influence the outcome of adenoviral infections. Whether the effects of VAI RNA on host cells relate to the biogenesis of small regulatory RNAs derived from VAI or to the inhibitory interference of VAI RNA with Dicer remains unclear.
These issues warrant further investigations, considering that adenoviral vectors are among the most commonly used vectors for gene therapy, aimed at delivering small interfering RNAs (siRNAs) and short hairpin RNAs (shRNAs) to target cells, second only to retroviruses (111).
2.3. Retrovirus
In contrast to DNA viruses, members of the retrovirus family possess an RNA genome. Replication of retroviruses occurs via a DNA intermediate and involves the viral RT enzyme, which converts RNA into DNA. The viral DNA is then integrated into the host genome for subsequent viral gene transcription and virus production. Retroviral genomes commonly contain three open reading framse that encode for structural proteins (eg, gag), for a RT, for an integrase and protease (pol) and for retroviral coat proteins (env). Among the subfamilies of retroviruses, the group of lentiviruses is represented by many well-known members, including human immunodeficiency viruses 1 and 2 (HIV-1 and HIV-2) and simian immunodeficiency virus (SIV), whereas the deltaretrovirus subfamily is represented by human T cell leukemia/lymphoma virus type 1 (HTLV-1).
After cell entry, RNA decapsidation occurs in the cytoplasm and likely exposes some RNA structures to cellular components of the miRNA pathway, such as Dicer. Transcription of viral mRNAs in the nucleus may also expose the viral RNA structures that may be recognized and processed by the microprocessor complex containing Drosha. However, initial studies have failed to identify miRNAs originating from HIV-1 or other retroviruses in virus-infected cells. Using better adapted and more sensitive approaches, other groups were able to detect miRNAs derived from the RNA of retroviruses, such as HIV-1.
Using a bioinformatic tool designed to uncover well-ordered folding patterns in nucleotide sequences, five candidate pre-miRNAs encoded by different regions of the HIV-1 genome were initially flagged (112). Omoto and colleagues (113) then reported a miRNA, named miR-N367, derived from the nef region, an accessory gene partially overlapping with the 3′ long terminal repeat (LTR). This HIV-1 miRNA have been detected by Northern blot hybridization in MT-4 T cells persistently infected with HIV-1 IIIB strain and cloned from a ~25-nt RNA sub-population. Overexpression of miR-N367, which shows perfect complementarity with nef, seemed to suppress HIV-1 LTR-driven transcription in reporter gene assays (114), suggesting that this nef-derived miRNA could act as a negative regulator of HIV-1 transcription. The biogenesis and function of this viral miRNA requires further investigations.
Another study reported that the HIV-1 RNA genome also encodes an siRNA derived from the env gene (115). The authors observed that an RNA strand forming a perfect 19-bp duplex, joined by an extended 198-nt loop, could be converted into an siRNA upon incubation with recombinant Dicer in vitro. A probe specific to this viral siRNA detected a ~24-nt signal not seen in mock-infected cells by Northern blot analysis (115). Overexpression of this viral siRNA effectively reduced Env mRNA levels and viral replication, whereas its neutralization with complementary 2′O-methyl oligonucleotides led to a dose-dependent increase in HIV-1 replication in human cells (115). These results suggest that an HIV-1-derived siRNA can modulate virus production.
Another HIV-1 RNA structure, the TAR element, was reported to be cleaved by Dicer into miRNAs (48,116). Klase et al. (116) suggested that HIV-1 TAR-derived miRNAs may act by recruiting the histone deacetylase HDAC-1 to the HIV-1 LTR promoter in the nucleus in order to silence transcription by chromatin remodeling, a concept that has been proposed previously (117). The authors hypothesize that this sequence of events may suppress transcription of viral as well as cellular genes, thereby influencing particular steps of HIV-1 pathogenesis, such as latency.
More recently, our group demonstrated that the TAR element of HIV-1 could release two miRNAs, namely miR-TAR-5p and miR-TAR-3p, through an asymmetrical processing reaction involving Dicer (48). This reaction led to the preferential release and accumulation of miR-TAR-3p from the right arm of the TAR element, which may explain, at least in part, the superior potency of miR-TAR-3p in mediating gene silencing in vivo. Although HIV-1 can generate two miRNAs from the same hairpin, i.e. TAR element, like JCV, BK and SV40 viruses, it remains unclear as to whether miR-TAR-5p and miR-TAR-3p could regulate the same transcript(s). Computational analysis of the TAR element of HIV-2, which is larger (~123 bp) than that of HIV-1, suggest that it may also be a source of two miRNAs (118), whose existence remains to be validated experimentally.
A recent study by Lin and Cullen (119) is challenging the existence of miRNAs derived from primate retroviruses, such as HIV-1 and HTLV-1. We cannot exclude the possibility that the identification of some miRNAs may be restricted to specific viral strains or that miRNAs may have escaped detection by standard small RNA cloning strategies, since methylation of the 2′ hydroxyl of the terminal ribose significantly reduces the cloning efficiency of silencing-associated small RNAs (120). This would explain some of the discrepancies observed between laboratories using different techniques to identify viral miRNAs. Related to that issue, the recent report on the presence of highly homologous and/or larger numbers of miRNAs resulting from retroviruses that have been integrated into the human genome, such as human endogenous retrovirus L (HERV-L), simian foamy viruses, human foamy viruses and HTLV-1, is intriguing and suggests that this may be a mechanism by which the retrovirus-infected host is more or less susceptible to subsequent viral infections (121).
2.4. Non-coding RNAs expressed from viruses
Several DNA viruses have been shown to generate ncRNAs. The first ncRNA to be described is VAI RNA from adenovirus 2. As previously mentioned, VAI RNA was found to interfere with the PKR response of virus-infected cells, although it may also take part in the strategy that adenoviruses have evolved to counteract cellular antiviral defense systems based on the miRNA pathway. Trancribed by RNA polymerase III, VAI RNA (see Figure 2A) has been shown to inhibit the nuclear export of pre-miRNAs by competition with Exportin-5 nuclear export factor as well as Dicer activity through direct binding to the enzyme (12). Another group observed the direct binding of VAI and VAII RNAs to Dicer as well as to the RNA-induced silencing complex (RISC) or miRNP (107). These observations are counterbalanced by the fact that, in contrast to two similar non-coding RNAs, i.e. EBER-1 from EBV (122,123) and hY1 from human cells (124), that are refractory to processing by Dicer (108), VAI and VAII RNAs may be processed by Dicer into miRNAs (107–110), which may exert gene regulatory functions in adenovirus-infected cells. Nevertheless, the VAI/VAII RNA-mediated attenuation of the cellular miRNA pathway may confer an advantage to the virus through a decreased level in cellular antiviral miRNAs.
Epstein-Barr virus-encoded small RNAs (EBERs) have also been proposed, like VAI RNA, to bind PKR (9). EBER1 (167 nt) and EBER2 (172 nt) (see Figure 2B) are generally the most abundant RNAs in EBV-infected cells (8,122). Structural similarities were observed between EBERs and VAI/VAII ncRNAs and, interestingly, the generation of adenoviral mutants, in which VA RNAs were replaced by EBERs, showed that EBERs could substitute for VA RNA function (125,126). Several roles have been attributed to EBERs as cell transformation and regulation of gene expression, where nuclear EBER1 and EBER2 ncRNAs may act as transcriptional regulators for cytokines and growth factors expression, and inhibit apoptosis via binding to PKR. More recently, it has been speculated that EBER RNAs could be involved in the rescue of EBV from cytoprotective transcriptional repression under particular stress conditions in vivo (for a review of EBER RNA function, see (8)). VA and EBER ncRNAs may thus counteract the activation of host defenses in virus-infected cells.
Additional viral ncRNAs have been found in the genome of HCMV. One of them, a ~2.7 kb transcript called β2.7 RNA, is particularly abundant in early time of infection (127). Reeves et al. (128) reported the ability of the β2.7 RNA transcript to interact with a subunit of the mitochondrial enzyme complex I (reduced nicotinamide adenine dinucleotide–ubiquinone oxido-reductase), suggesting a role for β2.7 RNA in protecting cells from apoptosis induced via that complex. It is now clear that viruses have developed refined strategies based on ncRNAs in order to circumvent key cellular processes that allow them to persist in their hosts.
Recently, a hepatitis delta virus (HDV) small RNA of ~24–25 nt was reported to be expressed from the bottom strand of the antigenomic pode of the viral RNA genome (129) [pode and antipode refer to both extremities of the HDV RNA hairpin ends, excluded from the coding region of hepatitis delta antigen (HDAg)]. HDV is a subviral satellite that encodes for a single protein, the HDAg, and can propagate solely in the presence of hepatitis B virus (HBV) or in cells previously infected with HBV. After cell entry, HDV circular genomic RNA becomes a template for rolling-circle replication in the nucleus to produce antigenomic RNA multimers, which are further cleaved into monomers by the intrinsic ribozyme activity of the antigenomic strand. Characterizing the extremities of the HDV RNA hairpin, Haussecker et al. (129) found a 5′ capped, 2′-3′-hydroxylated small RNA from HDV. Position of the small RNA within the HDV sequence led the authors to suggest that it could be involved in HDV transcription initiation. They identified and cloned additional small RNAs from both HDV genomic and antigenomic polarities that may function as small priming RNA (spRNAs) to help transcription initiation and, consequently, the success of the HDV infection.
Together, the discovery, role and importance of viral ncRNAs raise several important issues: How many small or large ncRNAs are produced from viral genomes? How do they contribute either to facilitate the infectious process or to assist in immune evasion mechanisms? What is their impact in viral pathogenesis? In cell biology? Answers to these questions will help us better understand viral infections and, perhaps, create new therapeutic opportunities to fight viruses.
3. Cellular miRNAs in virus-infected cells
3.1. Effects of cellular miRNAs on virus biology
In non-infected cells, miRNAs have been proposed to regulate up to 90% of the genes (130). A case of intricate relationship between cellular miRNAs and viruses came from the observation that replication of hepatitis C virus (HCV) was dependent on cellular miR-122 expression (131). They demonstrated that HCV RNA can replicate in miR-122-expressing Huh-7 cells, but not in HepG2 cells lacking miR-122. A binding site for miR-122 was predicted to reside close to the 5′ end of the viral genome, and creation of a loss-of-function mutation led to reduced levels of intracellular viral RNA. MiR-122 is expressed at high levels in human hepatocellular carcinoma (HHC), a well-known consequence of the HCV chronic infection (132). Increased miR-122 expression may lead to the regulation of anti-apoptotic genes (133) and enhance viral replication to promote cell proliferation. The exact mechanism by which miR-122 favors HCV replication in hepatocytes, however, remains the subject of speculations (134).
More recent studies have explored the importance of the cellular miRNA pathway in the control of HIV-1 replication (135,136). Using siRNAs against Drosha and Dicer in peripheral blood mononuclear cells (PBMCs) isolated from HIV-1-infected patients, Triboulet et al. (135) observed a faster virus replication kinetics in Drosha- or Dicer-depleted cells, as compared to cells treated with a control siRNA. The authors also confirmed a role for Drosha and Dicer in the suppression of HIV-1 replication in latently infected U1 cells.
Huang et al. (136) showed that the 3′ UTR of almost all HIV-1 mRNAs produced in resting primary CD4+ T lymphocytes during latency contain a 1.2-kb fragment that can be recognized by cellular miRNAs, with a negative impact on viral protein production. Combined with the relatively inefficient synthesis of Tat and Rev, miRNAs expressed by resting CD4+ T cells may participate in the post-transcriptional regulation of HIV-1 mRNA and contribute to keep the virus in its latent phase, as observed in patients with suppressive highly active antiretroviral therapy (HAART) (136). These new elements contribute to our understanding of the molecular basis of viral latency and may help us design therapeutic strategies aimed at purging HIV-1-infected patients of the quiescent virus.
3.2. Role of cellular miRNAs in host defenses against viruses
As discussed above, ncRNAs derived from some viruses are able to interact with PKR, an effector of the interferon (IFN) pathway, which is activated upon viral infection. It is relevant to note here that IFN may be link to the generation of miRNAs. Pedersen et al. (137) recently reported the modulation of miRNAs upon IFNβ induction. Interestingly, 8 of these IFNβ-induced miRNAs have sequence-predicted targets within the HCV genomic RNA. Individual transfection experiments revealed that miR-196, miR-296, miR-351, miR-431 and miR-448 were able to substantially attenuate HCV replication, whereas miRNAs miR-1, miR-30 and miR-128 had no effect alone. The anti-HCV properties of IFNβ may also be mediated through downregulation of miR-122 expression, which appears to be important for HCV replication (131). These results suggest that the IFNβ-induced expression of endogenous miRNAs may confer and help establish antiviral properties to human cells.
Another example of the complex interaction between viruses and cellular components is the miRNA miR-K12-11 derived from KSHV that functions as an ortholog of miR-155, an oncogenic miRNA upregulated in lymphomas (137). The sequence identity between miR-K12-11 and miR-155, which implies a common set of regulated genes, suggest that KSHV may thwart its host through a miRNA mimicking strategy.
A few years ago, some HIV-1 gene candidates were predicted to be controlled by host miRNAs in view of thermodynamically favorable miRNA:RNA target base pairing (138). In addition, changes in miRNA expression profiles, more specifically the downregulation of a large pool of cellular miRNAs, have been observed in human HeLa cells transfected with the infectious molecular HIV-1 clone pNL4-3 (139). Similar observations were reported recently, in which HIV-1 infection was associated with either up- or down-regulation of specific miRNA clusters located within the host genome. For example, expression of the miR-17/92 cluster, which encodes 7 miRNAs, among which miR-17-5p and miR-20 may target the histone acetyltransferase and HIV-1 Tat cofactor p300/CBP-associated factor (PCAF), was substantially downregulated (135). The authors proposed that this gene regulatory axis may help understand how latent virus reservoirs could be activated.
The presence of miRNAs has been documented in mature neuronal dendrites, suggesting that they may be involved in controlling local protein translation and synaptic function. A rare clinical manifestation of HIV-1 infection, HIV-1 encephalopathy (HIVE), results in neuronal damage and dysfunction. If neurons are rarely infected by HIV-1, they are nevertheless exposed to the viral components of HIV-1, including its transactivating protein Tat. Eletto et al. (140) recently showed that Tat deregulates expression of selected miRNAs in primary cortical neurons, including the neuronal miR-128, which has been shown to normally inhibit expression of the pre-synaptic protein SNAP25. This is yet another mechanism by which HIV-1 and its components can perturb normal cellular activities and compromise health of the infected individuals.
4. Concluding remarks
Whereas some viruses may take advantage of the cellular and viral miRNAs for their own replication, others may be targeted by cellular miRNAs that function as part of the host defenses against viruses. Similarly, while some viral RNAs may be processed by components of the host miRNA pathway, such as Dicer, others have evolved countermeasures in the form of various inhibitors of RNA silencing. Elucidation of the complex relationship between viruses and their hosts involving miRNAs and ncRNAs is mandatory for the development of antiviral gene therapies based on the neutralization and/or promotion of miRNA/ncRNA expression.
Table 2.
The viral and cellular messenger RNA targets of viral microRNAs.
| Origin | miRNA | …target… | mRNA | Target | References |
|---|---|---|---|---|---|
| Viral miRNA | HSV-1 miR-H1 | → | HSV-1 ICP0 | Viral mRNA | 55 |
| HSV-1 miR-H2-3p | → | HSV-1 ICP0 | 59 | ||
| HSV-1 miR-H6 | → | HSV-1 ICP4 | 59 | ||
| HSV-1 miR-I | → | HSV-1 ICP34.5 | 62 | ||
| EBV miR-BART-2 | → | EBV BALF5 | 52 | ||
| EBV miR-BART-1-5p, miR-BART-16, miR-BART-17-5p | → | EBV LMP1 | 73 | ||
| HCMV miR-UL112-1 | → | HCMV IE72/IE1 | 82,83 | ||
| SV40 sv40-miR-S1-5p, sv40-miR-S1-3p | → | SV40 T-antigen | 101 | ||
| HIV-1 miR-N367* | → | HIV-1 Nef | 114 | ||
| HIV-1 TAR miRNA | → | HIV-1 LTR | 116 | ||
| EBV miR-BART-5 | → | PUMA | Cellular mRNA | 76 | |
| EBV BHRF1-3 | → | CXCL11/I-TAC | 77 | ||
| HCMV miR-UL112-1 | → | MICB | 84 | ||
| KSHV miR-K12–1, miR-K12-3-3p, miR-K12-6-3p and miR-K12–11 | → | Thrombospondin 1 | 93 | ||
| KSHV miRK12-11 | → | BACH-1 | 97,98 | ||
Please note that, according to the miRBase registry, this sequence should be considered at risk of deletion from future releases.
Table 3.
The viral and messenger RNA targets of cellular microRNAs upon viral infection.
| Origin | miRNA | …target… | mRNA | Target | References |
|---|---|---|---|---|---|
| Cellular miRNA | hsa-miR-122 | → | HCV 5′NTR (upon HCV infection) | Viral mRNA | 131 |
| hsa-miR-196,miR-296, miR-351, miR-431, miR-448 | → | HCV genome (upon IFN response) | 139 | ||
| hsa-miR-155 (KSHV miRK12-11 ortholog) | → | BACH-1 (upon KSHV infection) | Cellular mRNA | 97,98 | |
| hsa-miR-17-5p and miR-20 | → | PCAF (upon HIV-1 infection) | 134 | ||
| hsa-miR-128 | → | SNAP25 (upon HIV-1 infection) | 138 | ||
Acknowledgments
We are grateful to the Illustration Department of the CHUL Research Center for graphic illustrations. P. P. is a New Investigator of the Canadian Institutes of Health Research (CIHR) and Junior 2 Scholar from the Fonds de la Recherche en Santé du Québec. This work was financially supported by grant HOP-83069 from Health Canada/CIHR to P.P.
References
- 1.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 2.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 3.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 8.Swaminathan S. Noncoding RNAs produced by oncogenic human herpesviruses. J Cell Physiol. 2008;216:321–326. doi: 10.1002/jcp.21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens MJ, Laing KG, Jeffrey IW, Schofield A, Sharp TV, Elia A, Matys V, James MC, Tilleray VJ. Regulation of the interferon-inducible eIF-2 alpha protein kinase by small RNAs. Biochimie. 1994;76:770–778. doi: 10.1016/0300-9084(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 10.Kitajewski J, Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Thimmappaya B, Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 11.Furtado MR, Subramanian S, Bhat RA, Fowlkes DM, Safer B, Thimmappaya B. Functional dissection of adenovirus VAI RNA. J Virol. 1989;63:3423–3434. doi: 10.1128/jvi.63.8.3423-3434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 17.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 25.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 28.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 31.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 34.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-Strand Cleavage Facilitates Assembly of siRNA into Ago2-Containing RNAi Enzyme Complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 35.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. Rna. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. Rna. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14–3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 44.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 45.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 48.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 50.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 52.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 53.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81:13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bloom DC. HSV LAT and neuronal survival. Int Rev Immunol. 2004;23:187–198. doi: 10.1080/08830180490265592. [DOI] [PubMed] [Google Scholar]
- 57.Cuchet D, Ferrera R, Lomonte P, Epstein AL. Characterization of antiproliferative and cytotoxic properties of the HSV-1 immediate-early ICPo protein. J Gene Med. 2005;7:1187–1199. doi: 10.1002/jgm.761. [DOI] [PubMed] [Google Scholar]
- 58.La Frazia S, Amici C, Santoro MG. Antiviral activity of proteasome inhibitors in herpes simplex virus-1 infection: role of nuclear factor-kappaB. Antivir Ther. 2006;11:995–1004. [PubMed] [Google Scholar]
- 59.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J Virol. 2001;75:6143–6153. doi: 10.1128/JVI.75.13.6143-6153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson RL, Stevens JG. Biological characterization of a herpes simplex virus intertypic recombinant which is completely and specifically non-neurovirulent. Virology. 1983;131:171–179. doi: 10.1016/0042-6822(83)90543-3. [DOI] [PubMed] [Google Scholar]
- 62.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brady G, MacArthur GJ, Farrell PJ. Epstein-Barr virus and Burkitt lymphoma. J Clin Pathol. 2007;60:1397–1402. doi: 10.1136/jcp.2007.047977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tao Q, Young LS, Woodman CB, Murray PG. Epstein-Barr virus (EBV) and its associated human cancers--genetics, epigenetics, pathobiology and novel therapeutics. Front Biosci. 2006;11:2672–2713. doi: 10.2741/2000. [DOI] [PubMed] [Google Scholar]
- 65.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 66.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. 2008;82:9094–9106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amon W, Farrell PJ. Reactivation of Epstein-Barr virus from latency. Rev Med Virol. 2005;15:149–156. doi: 10.1002/rmv.456. [DOI] [PubMed] [Google Scholar]
- 69.Rickinson AB, Lee SP, Steven NM. Cytotoxic T lymphocyte responses to Epstein-Barr virus. Curr Opin Immunol. 1996;8:492–497. doi: 10.1016/s0952-7915(96)80036-7. [DOI] [PubMed] [Google Scholar]
- 70.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81:9967–9975. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng H, Li LL, Hu DS, Deng XY, Cao Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell Mol Immunol. 2007;4:185–196. [PubMed] [Google Scholar]
- 75.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 76.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008 doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1–3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reeves M, Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol. 2008;325:297–313. doi: 10.1007/978-3-540-77349-8_17. [DOI] [PubMed] [Google Scholar]
- 79.Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 80.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;7:1684–1695. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 82.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. SUMO-1 modification of human cytomegalovirus IE1/IE72. J Virol. 2002;76:2990–2996. doi: 10.1128/JVI.76.6.2990-2996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grey F, Nelson J. Identification and function of human cytomegalovirus microRNAs. J Clin Virol. 2008;41:186–191. doi: 10.1016/j.jcv.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod’homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, et al. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Antman K, Chang Y. Kaposi’s sarcoma. N Engl J Med. 2000;342:1027–1038. doi: 10.1056/NEJM200004063421407. [DOI] [PubMed] [Google Scholar]
- 90.Aoki Y, Yarchoan R, Wyvill K, Okamoto S, Little RF, Tosato G. Detection of viral interleukin-6 in Kaposi sarcoma-associated herpesvirus-linked disorders. Blood. 2001;97:2173–2176. doi: 10.1182/blood.v97.7.2173. [DOI] [PubMed] [Google Scholar]
- 91.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Fraipont F, Nicholson AC, Feige JJ, Van Meir EG. Thrombospondins and tumor angiogenesis. Trends Mol Med. 2001;7:401–407. doi: 10.1016/s1471-4914(01)02102-5. [DOI] [PubMed] [Google Scholar]
- 95.Lawler J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J Cell Mol Med. 2002;6:1–12. doi: 10.1111/j.1582-4934.2002.tb00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Narizhneva NV, Razorenova OV, Podrez EA, Chen J, Chandrasekharan UM, DiCorleto PE, Plow EF, Topol EJ, Byzova TV. Thrombospondin-1 up-regulates expression of cell adhesion molecules and promotes monocyte binding to endothelium. FASEB J. 2005;19:1158–1160. doi: 10.1096/fj.04-3310fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ochiai S, Mizuno T, Deie M, Igarashi K, Hamada Y, Ochi M. Oxidative stress reaction in the meniscus of Bach 1 deficient mice: potential prevention of meniscal degeneration. J Orthop Res. 2008;26:894–898. doi: 10.1002/jor.20579. [DOI] [PubMed] [Google Scholar]
- 100.Randhawa P, Vats A, Shapiro R. The pathobiology of polyomavirus infection in man. Adv Exp Med Biol. 2006;577:148–159. doi: 10.1007/0-387-32957-9_10. [DOI] [PubMed] [Google Scholar]
- 101.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 102.Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, Pipas JM. Complete nucleotide sequence of polyomavirus SA12. J Virol. 2005;79:13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reich PR, Forget BG, Weissman SM. RNA of low molecular weight in KB cells infected with adenovirus type 2. J Mol Biol. 1966;17:428–439. doi: 10.1016/s0022-2836(66)80153-5. [DOI] [PubMed] [Google Scholar]
- 106.Maran A, Mathews MB. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology. 1988;164:106–113. doi: 10.1016/0042-6822(88)90625-3. [DOI] [PubMed] [Google Scholar]
- 107.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553–1564. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 109.Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. 2007;81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ghosh SS, Gopinath P, Ramesh A. Adenoviral vectors: a promising tool for gene therapy. Appl Biochem Biotechnol. 2006;133:9–29. doi: 10.1385/abab:133:1:9. [DOI] [PubMed] [Google Scholar]
- 112.Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86:751–755. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- 115.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 116.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weinberg MS, Morris KV. Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol. 2006;25:223–231. doi: 10.1089/dna.2006.25.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Purzycka KJ, Adamiak RW. The HIV-2 TAR RNA domain as a potential source of viral-encoded miRNA. A reconnaissance study. Nucleic Acids Symp Ser (Oxf) 2008:511–512. doi: 10.1093/nass/nrn259. [DOI] [PubMed] [Google Scholar]
- 119.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci U S A. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hakim ST, Alsayari M, McLean DC, Saleem S, Addanki KC, Aggarwal M, Mahalingam K, Bagasra O. A large number of the human microRNAs target lentiviruses, retroviruses, and endogenous retroviruses. Biochem Biophys Res Commun. 2008;369:357–362. doi: 10.1016/j.bbrc.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 122.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rosa MD, Gottlieb E, Lerner MR, Steitz JA. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wolin SL, Steitz JA. Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell. 1983;32:735–744. doi: 10.1016/0092-8674(83)90059-4. [DOI] [PubMed] [Google Scholar]
- 125.Bhat RA, Thimmappaya B. Two small RNAs encoded by Epstein-Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc Natl Acad Sci U S A. 1983;80:4789–4793. doi: 10.1073/pnas.80.15.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bhat RA, Thimmappaya B. Construction and analysis of additional adenovirus substitution mutants confirm the complementation of VAI RNA function by two small RNAs encoded by Epstein-Barr virus. J Virol. 1985;56:750–756. doi: 10.1128/jvi.56.3.750-756.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Spector DH. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 128.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 129.Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol. 2008;15:714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 131.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 132.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 133.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 134.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 136.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 137.Tam W, Dahlberg JE. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 138.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 139.Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eletto D, Russo G, Passiatore G, Del Valle L, Giordano A, Khalili K, Gualco E, Peruzzi F. Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J Cell Physiol. 2008;216:764–770. doi: 10.1002/jcp.21452. [DOI] [PMC free article] [PubMed] [Google Scholar]