Abstract
Despite the excellent image-contrast capability of MRI and the ability to synchronize MRI with the murine cardiac cycle, this technique is underused for assessing mouse models of cardiovascular disease because of its perceived cost and complexity. This perception stems, in part, from complications associated with the placement and adjustment of electrocardiographic leads that may interact with gradient pulses and the relatively long acquisition times required with traditional gating schemes. To improve the efficiency and reduce the cost and complexity of using cardiac MRI in mice, we combined wireless self-gating techniques (with which we derived cardiac synchronization signals from acquired data) with an imaging technique that acquires multislice cardiac cine images from four mice simultaneously. As a result, the wireless self-gated acquisitions minimized animal preparation time and improved image quality. The simultaneous acquisition of cardiac cine data from multiple animals greatly increased throughput and reduced costs associated with instrument access.
Keywords: cardiac MRI, multiple-mouse MRI, self-gating, imaging efficiency
INTRODUCTION
More people die annually from cardiovascular disease (CVD) than from any other cause. It accounts for one of every 3.4 deaths globally and one of every 2.8 deaths in the United States (1,2). Small animal models of CVD are often employed for the evaluation of novel diagnostic and therapeutic approaches to improving our understanding and treatment of disease. Noninvasive, imaging-based assessment of cardiac structure and function improves observational power by allowing disease or injury and response to therapy to be tracked longitudinally without requiring sacrifice at each observation. MRI, as one of such valuable diagnostic tools, can provide good image contrast between myocardium and blood.
The potential applicability of MRI-based investigations of small animal models of CVD is reduced because of cost and experimental complexity. Delays associated with the setup of electrocardiographic (ECG) equipment and the inefficient duty cycle of ECG-gated acquisitions limits the utility of traditional prospectively gated CMRI. Additionally, the use of metallic ECG leads introduces susceptibility artifacts and can pose problems with coupling to RF and gradient pulses that may distort the anatomy of interest, especially in small-animal investigations (3).
Self-gated (SG) imaging techniques allow cardiac phases to be identified from MR data, eliminating the need for ECG signals and systems. The first SG method, described by Spraggins (4), involved a rectilinear double-slice/double-echo pulse sequence, and gating data were obtained through a second non-phase-encoded readout navigator. Because of the increased acquisition time required by this technique, Larson et al. (5) developed a radial k-space sampling method that avoided the acquisition of extra gating data by extracting the motion synchronization signal directly from imaging data. A short second echo method by Crowe et al. (6) further optimized acquisition efficiency by using a conventional rectilinear sampling scheme that required only a slight increase in pulse repetition time (TR). Although these pulse sequences were developed to increase the acquisition efficiency in human imaging, they can also be adapted for small animal imaging (7) and substantially enhance CMRI in mice. Unfortunately, overall imaging times remain relatively long.
Improved animal throughput and reduced scan time can help reduce overall imaging time and cost for studies with a large number of animals or animal groups. In small-animal MRI investigations, scanning a large number of animals per time point is desirable to increase statistical confidence in the results. However, with single-animal imaging, throughput sets a limit on the number of animals that can be evaluated close to a particular point in time relative to injury or treatment. Furthermore, long imaging times can render the cost of such studies prohibitive to investigators. One way to increase throughput efficiency, thus reducing imaging time and cost, is to scan several animals at once (8). Multiple-animal imaging allows efficient utilization of normally unused MRI magnet bore space. Additionally, in multiple-animal imaging, each animal can be scanned using acquisition parameters that are identical to those required for single-animal studies (9,10). However, traditional prospective gating acquisition strategies are not possible with multiple animals because the cardiac cycle of each animal progresses at a unique frequency and phase, preventing synchronized triggering. Bishop et al. (11) employed multiple-animal imaging techniques to improve CMRI in mice in a hybrid SG technique by recording ECG signals from several animals and later using the stored data to aid in retrospective reconstruction. This approach, however, required ECG equipment to support several animals and a very long acquisition time (~1 hour). Thus, there is a need to further extend multiple-animal imaging with SG techniques to produce a more seamless and practical method for small-animal CMRI studies.
Our purpose was to investigate the feasibility of using a wireless SG multiple-mouse cine MRI technique to improve throughput and measurement efficiency in small-animal cardiac investigations. A linear array of volume coils developed to facilitate routine multi-animal imaging (10) was used to acquire cardiac cine data from four mice simultaneously with no sacrifice in image quality compared to single-animal imaging. A straightforward approach to retrospective reconstruction yielded good motion suppression and contrast, verifying that wireless retrospective gating can be combined with multiple-animal imaging techniques to substantially improve efficiency.
METHODS
Equipment
For imaging, we used a four-channel 4.7 T/40-cm Biospec scanner (Bruker BioSpin Corp., Billerica, MA) with a BGA26 actively shielded, water-cooled gradient coil system (950 mT m−1, 220-µs rise time). Four commercially available, shielded volume resonators (1P T7817 MRI; Bruker BioSpin) with a 3.5-cm inner diameter were strategically arranged in a linear configuration across the coronal plane and held in place with an acrylic former as described in (10) and illustrated in Figure 1. Coupling between coils was minimized by rotating adjacent elements to a point of maximal geometric isolation. The coils were spaced at twice the distance of the field of view prescribed along the phase-encoding direction (left/right), allowing signals from all coils to alias into the center of a single prescribed field of view (Figure 1). A custom set of four-animal imaging sleds was arranged to fit into the linear array of volume resonators via wheels guided along a rail system. An actively controlled transmit/receive switch system, working in conjunction with a power splitter, distributed excitation pulses to the four resonators.
FIGURE 1.
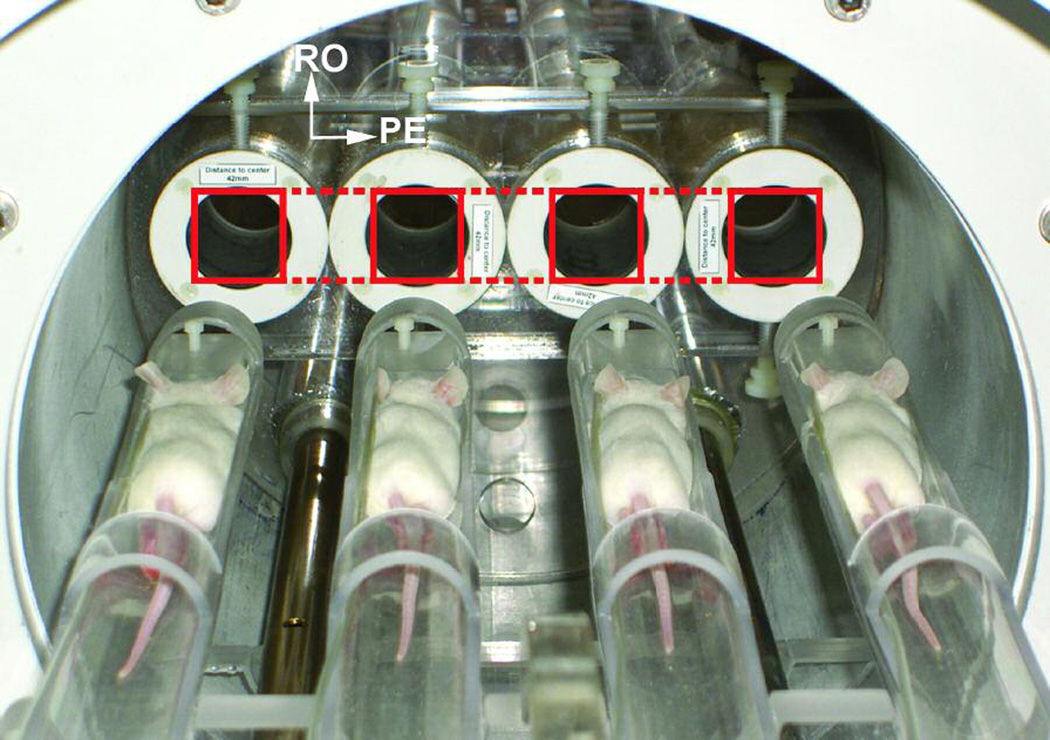
Geometry configuration for the imaging experiment. Coil resonators are held in the bore of the magnet. Imaging sleds holding the mice fit into the coils via wheels guided along a rail system. Dashed lines drawn over the coils illustrate the field aliasing that allows replication to phase encode all four mice simultaneously without the need for image cropping or rolling.
Animal Preparation
All imaging and animal procedures were approved by our Institutional Animal Care and Use Committee, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International.
Four anesthetized CD-1 mice were placed prone onto the sleds and coaligned along the field axis, remaining anesthetized during imaging with 1%–2% isoflurane in oxygen. Because of the consistent animal placement, the hearts of the mice were positioned at approximately the same location along the field axis. The whole array of sleds, loaded with mice, was brought into the proper imaging position using the rolling mechanism as described above.
Imaging
Anatomic positioning was determined by using a conventional fast low-angle shot (FLASH) sequence in one representative mouse. The imaging protocol called for simultaneous imaging of the four animals using a coronal scout sequence, which served as a reference for the prescription of slices in the multiple-animal SG CMRI acquisition. The SG CMRI acquisition used a fully refocused fast imaging with steady-state precession (TrueFISP) version of Spraggins’ double-slice method. Image data and gating (navigator) data were alternately acquired during each sequence TR, as illustrated in Figure 2. Uninterrupted image/navigator acquisitions ensured that a steady state was maintained during the scan. This continuity also allowed data to be acquired from the various cardiac phases throughout the whole cardiac cycle. The phase-encode value was incremented only after a given number of image/navigator repetitions were acquired. Imaging parameters were as follows: TR = 7.1 ms, echo time = 3.3 ms, flip angle = 30 degrees, acquisition matrix = 128 × 96 pixels, number of image/navigator repetitions = 120, field of view = 40 × 30 mm, and slice thickness = 1.5 mm.
FIGURE 2.
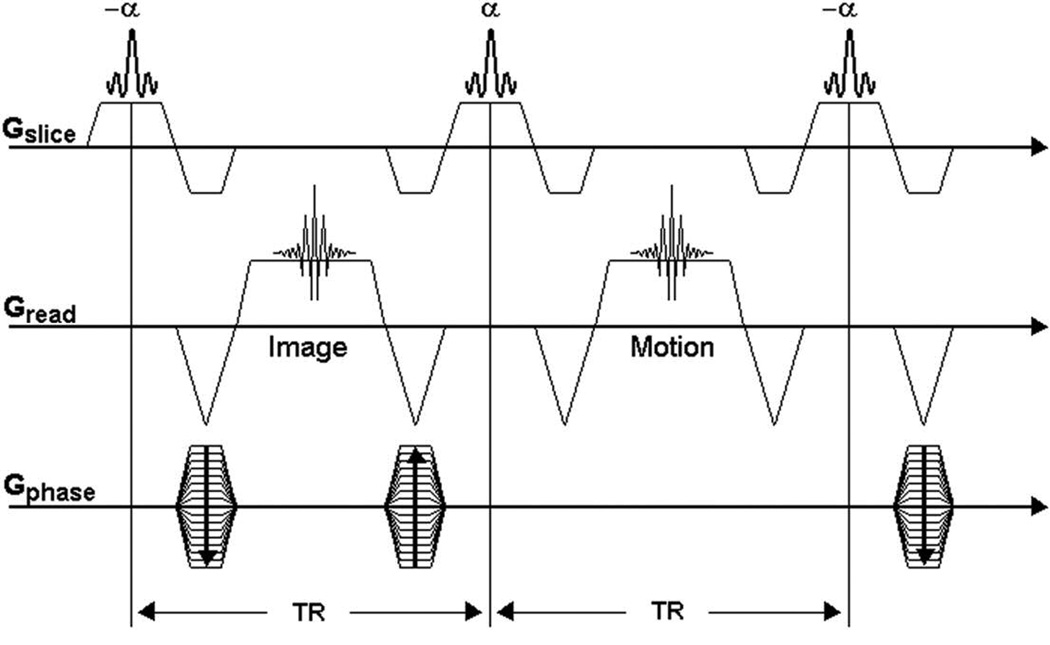
Self-gated true fast imaging with steady-state precession (TrueFISP) sequence based on the Spraggins’ double-slice sequence (4). The first echo is phase-encoded for image data; the second echo is non-phase-encoded (navigator) for gating. The maximum amplitude for the navigator signal oscillates synchronously with the cardiac motion. TR = repetition time.
Reconstruction
Offline reconstruction was performed using IDL software (ITT Industries, Inc., Boulder, CO). The echo peak magnitude of the navigator’s signal was plotted against the repetition number at each phase-encode value, giving rise to oscillatory graphs (Figure 3). Signal oscillations, previously shown to be synchronous with the cardiac cycle (4,12), were used to determine the cardiac phase associated with each image echo. The repetition numbers associated with peaks in the oscillatory graph were consecutively identified by performing an absolute maximum detection algorithm. These reference repetitions were mapped into the image data sets which were defined to correspond with the reference cardiac phase. Redundant data from these repetitions were used to perform signal averaging. Image data sets from nearby cardiac phases were similarly identified after mapping the repetition numbers correspondingly nearby to the reference set. The number of frames that completed a cardiac cycle for a given animal was determined by the number of repetitions between adjacent peaks on the oscillatory graph. Differences in this number between animals indicated different cardiac rates with uncertainties proportional to the TR. Respiratory motion was not accounted for.
FIGURE 3.
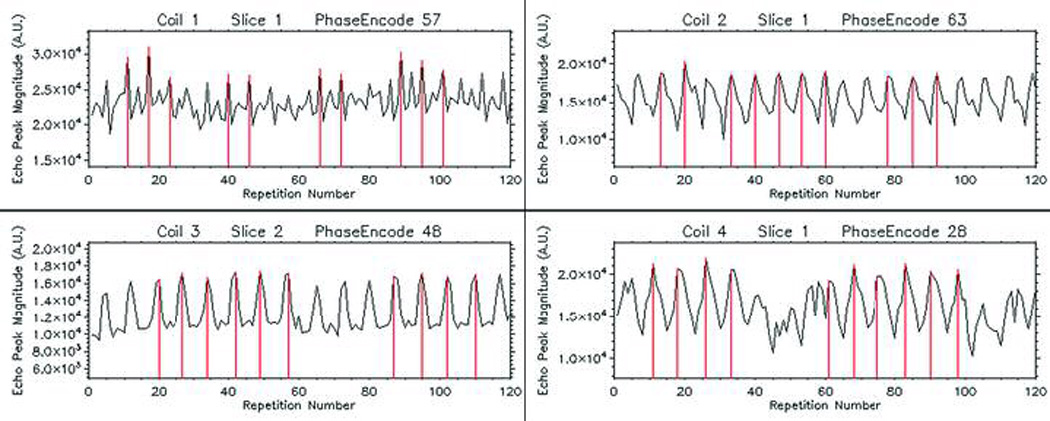
Representative self-gating signals from navigator echo peak magnitude. A maximum-detection algorithm established the reference phase of the cardiac cycle. Vertical lines indicate the repetition numbers used as references in identifying the same cardiac phases from among different cardiac cycles.
RESULTS
Representative echo peak magnitude SG signals from each coil are shown in Figure 3. The number of cycles per set of phase-encode repetitions ranged between 15 and 20 in this demonstration, with at least 10 cycles showing clear quasisymmetric peaks that identify the reference phase of the cardiac cycle.
Figure 4 shows representative cine frames (end-diastole) of each mouse cardiac cycle from the simultaneous four-animal data acquisition. Redundant data from ten cardiac cycles were averaged to improve the signal-to-noise ratio during an acquisition that required 2 min 44 sec per slice. 6–8 frames were reconstructed for each animal’s cardiac cycle, depending on individual heart rates. Cardiac muscle and blood can be clearly distinguished across the frames.
FIGURE 4.
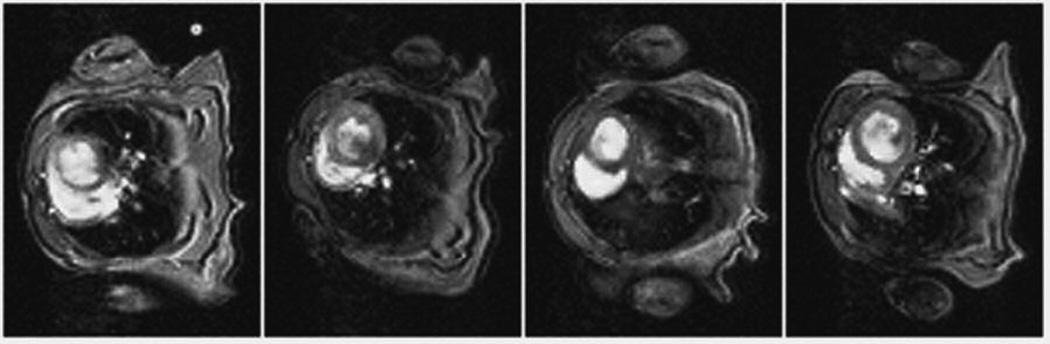
Representative self-gated cardiac cine frames acquired simultaneously with four coils. Images from left to right correspond to coils one through four, respectively. Each image was reconstructed from averaged oversampled data. Because interpolation was not used to derive additional frames, a fixed acquisition rate yielded a different number of cine frames for animals with different cardiac rates. The frames shown here correspond to end-diastole.
DISCUSSION AND CONCLUSION
We successfully integrated SG cine imaging strategies with multiple-animal imaging hardware to realize high-throughput CMRI in mice. Images from multiple-animal acquisitions have the same spatial resolution as do images from single-animal acquisitions. This demonstrates the convenience of the aliasing technique for avoiding enlarged fields of view, acquisition matrices, and imaging times to preserve spatial resolution in multiple-animal imaging. Images also show a high signal-to-noise ratio that is based on the average of oversampled data. Motion artifacts from heart pulsations are minimal, demonstrating the effectiveness of the SG procedure and data reordering. Minor movement artifacts are inherent of a relatively long TR during which variations of the heart frame occur. Image contrast between cardiac muscle and blood is sufficient for delineating the interior volume of ventricles for calculating the ejection fraction, stroke volume, and cardiac output used in assessing cardiac function. This demonstrates the feasibility of multiple-animal SG CMRI acquisitions to increase measurement efficiency without the need for ECG equipment.
The animal preparation time for imaging was about 5 min, and the simultaneous data acquisition time was 2 min 44 s per slice, demonstrating the feasibility of substantially increasing animal throughput efficiency. Total scan time for a complete study with eight slices, including localizers, animal preparation, and coil tuning, required about 40 min, leading to an equivalent scan time of 10 min per animal.
Further improvements to the method used here may be realized. Band-pass temporal filtering techniques may be implemented to improve the SG signal peak detection algorithm. However, our first-pass reconstruction method proved effective enough to successfully reorder the imaging data. This is attributed in part to the relatively strong gating signal of the TrueFISP sequence used in this demonstration. TR may be reduced by implementing the short second echo technique described by Crowe et al. (6) to increase the number of image/navigator pairs that are acquired per cardiac cycle. Note that flexibility in cine reconstruction is a virtue of retrospective gating strategies: a finer reordering scheme could be used to reconstruct a greater number of frames with less signal averaging, reducing error in selection of end-systolic and end-diastolic frames when quantitative measures such as ejection fraction are required. However, with the double-slice method, the relatively long TR still enables detection of the cardiac cycle while allowing the selection of a slice for gating to be placed away from the imaging slice. It should be noted that during this preliminary feasibility study, nontraditional images of the heart were obtained. This would have to be considered in any subsequent quantitative analysis of cardiac function, and no measures should be made that are based on assumptions of symmetry or motion. Possible solutions to this may be the use of three-dimensional imaging methods or alternate multi-animal imaging detector configurations to facilitate more traditional short- and long-axis views. For instance, instead of prescribing the slice axially, it could be skewed by some angle to alias along the hearts of staggered animals, yielding more traditional views of the heart.
The findings from this study suggest that MRI can be a time- and cost-effective imaging modality for evaluating CVD and cardiac injury in multiple-animal studies with small animal models. SG CMRI offers almost constant left ventricle wall signal intensity throughout the cardiac cycle and adequate signal-to-noise and contrast-to-noise ratios between blood and the myocardial wall (13). Furthermore, the breadth of applications in multiple-animal studies can be extensive, from near heart imaging to MR angiography, during which arterial pulsations degrade image quality because of motion artifacts (14,15).
In conclusion, we demonstrated the feasibility of wireless, truly SG cine CMRI of multiple animals to greatly increase throughput and measurement efficiency in CMRI of mice.
ACKNOWLEDGMENTS
We thank the Small Animal Imaging Facility at The University of Texas M. D. Anderson Cancer Center for technical support.
REFERENCES
- 1.American Heart Association. Heart Disease and Stroke Statistics. 2007:6. Update. [Google Scholar]
- 2.Minino AM, Heron MP, Smith BL. Deaths: Preliminary Data for 2004. Hyattsville, MD: National Center for Health Statistics; National Vital Statistics Reports. 2006;Vol. 54(No. 19):4. [PubMed]
- 3.Polson M, Barker A, Gardiner S. The effect of rapid rise-time magnetic fields on the ECG of the rat. Clin Phys Physiol Meas. 1982;3:231–234. doi: 10.1088/0143-0815/3/3/008. [DOI] [PubMed] [Google Scholar]
- 4.Spraggins T. Wireless retrospective gating: application to cine cardiac imaging. MRI. 1990;8:675–681. doi: 10.1016/0730-725x(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 5.Larson AC, White RD, Laub G, McVeigh ER, Li D, Simonetti OP. Self-gated cardiac cine MRI. Magn Reson Med. 2004;51:93–102. doi: 10.1002/mrm.10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowe ME, Larson AC, Zhang Q, Carr J, White RD, Li D, Simonetti OP. Automated rectilinear self-gated cardiac cine imaging. Magn Reson Med. 2004;52:782–788. doi: 10.1002/mrm.20212. [DOI] [PubMed] [Google Scholar]
- 7.Hiba B, Richard N, Janier M, Croisille P. Cardiac and respiratory double self-gated cine MRI in the mouse at 7 T. Magn Reson Med. 2006;55:506–513. doi: 10.1002/mrm.20815. [DOI] [PubMed] [Google Scholar]
- 8.Bock NA, Konyer NB, Henkelman RM. Multiple-mouse MRI. Magn Reson Med. 2003;49:158–167. doi: 10.1002/mrm.10326. [DOI] [PubMed] [Google Scholar]
- 9.Bock NA, Nieman BJ, Bishop JB, Henkelman RM. In vivo multiple-mouse MRI at 7 Tesla. Magn Reson Med. 2005;54:1311–1316. doi: 10.1002/mrm.20683. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez MS, Bankson JA. A practical method for 2D multiple-animal MRI. J Magn Reson Imaging. doi: 10.1002/jmri.21112. (in press) [DOI] [PubMed] [Google Scholar]
- 11.Bishop J, Feintuch A, Bock NA, Nieman B, Dazai J, Davidson L, Henkelman RM. Retrospective gating for mouse cardiac MRI. Magn Reson Med. 2006;55:472–477. doi: 10.1002/mrm.20794. [DOI] [PubMed] [Google Scholar]
- 12.Kim W, Min C, Kim D, Cho Z. Extraction of cardiac and respiratory motion cycles by use of projection data and its applications to NMR imaging. Magn Reson Med. 1990;13:25–37. doi: 10.1002/mrm.1910130105. [DOI] [PubMed] [Google Scholar]
- 13.Heijman E, Graaf W, Niessen P, Nauerth A, Eys G, Graaf L, Nicolay K, Strijkers GJ. Comparison between prospective and retrospective triggering for mouse cardiac MRI. NMR Biomed. 2007;20:439–447. doi: 10.1002/nbm.1110. [DOI] [PubMed] [Google Scholar]
- 14.Cassidy PJ, Schneider JE, Grieve SM, Lygate C, Neubauer S, Clarke K. Assessment of motion gating strategies for mouse magnetic resonance at high magnetic fields. J Magn Reson Imaging. 2004;19:229–237. doi: 10.1002/jmri.10454. [DOI] [PubMed] [Google Scholar]
- 15.Wiesmann F, Szimtenings M, Frydrychowics A, Illinger R, Hunecke A, Rommel E, Neubauer S, Haase A. High-resolution MRI with cardiac and respiratory gating allows for accurate in vivo artherosclerotic plaque visualization in the murine aortic arch. Magn Reson Med. 2003;50:69–74. doi: 10.1002/mrm.10500. [DOI] [PubMed] [Google Scholar]


