Summary
The mammalian target of rapamycin (mTOR) is a crucial effector in a complex signaling network commonly disrupted in cancer. mTOR exerts its multiple functions in the context of two different multiprotein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Loss of the tumor suppressor PTEN (phosphatase and tensin homolog deleted from chromosome 10) can hyperactivate mTOR through AKT and represents one of the most frequent events in human prostate cancer. We show here that conditional inactivation of mTor in the adult mouse prostate is seemingly inconsequential for this post-mitotic tissue. Conversely, inactivation of mTor leads to a marked suppression of Pten-loss-induced tumor initiation and progression in the prostate. This suppression is more pronounced than that elicited by the sole pharmacological abrogation of mTORC1. Acute inactivation of mTor in vitro also highlights the differential requirement of mTor function in proliferating and transformed cells. Collectively, our data constitute a strong rationale for developing specific mTOR kinase inhibitors targeting both mTORC1 and mTORC2 for the treatment of tumors triggered by PTEN deficiency and aberrant mTOR signaling.
Introduction
The mammalian target of rapamycin (mTOR) is a critical serine/threonine kinase that integrates diverse inputs, including signals from growth factors, nutrients, energy and stress, to regulate protein synthesis, cell growth, and proliferation (1).
mTOR was discovered in the early 1990’s as an inhibitory target of the bacterial macrolide rapamycin (2). Rapamycin has been a valuable tool to uncover many physiological functions of mTOR in health and disease. Over the years it has become clear that mTOR represents a critical node in a complex signaling network that is often deregulated in human cancer (3, 4).
In particular, an intricate relationship links mTOR to the AKT signaling cascade (5), a pathway that is reported to be hyperactive in many human cancers, including prostate cancer (CaP) (4). In human cancer the AKT pathway is commonly activated through the inactivation of at least one allele of the tumor suppressor PTEN, an event that occurs in 30% to 70% of primary CaP (6, 7).
In mammalian cells, mTOR affects AKT signaling at multiple levels as a component of two distinct protein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1, which is activated downstream of AKT, is sensitive to rapamycin-mediated inhibition. mTORC1 promotes cell growth largely through the phosphorylation and subsequent inactivation of eIF4E-binding proteins (4E-BPs) and by the phosphorylation and activation of ribosomal S6 kinase (S6K1) (1). mTORC2 is rapamycin-insensitive and has recently been demonstrated to phosphorylate AKT at Ser473 (8). This modification, in conjunction with AKT phosphorylation on Thr308 by the phosphoinositide-dependent kinase-1 (PDK1) triggers full activation of AKT in response to mitogenic stimuli (8).
Downstream of AKT, mTORC1 has been described as the most essential effector in driving cell proliferation and susceptibility to oncogenic transformation (9). This led to the targeting of mTORC1 as a therapeutic strategy in cancer, with rapamycin and its analogues (rapalogs) emerging as the first generation of mTOR inhibitors to enter cancer clinical trials. However, the rapalogs were only modestly successful, with different subsets of cancer showing a highly variable response to rapalogs treatment (3, 10). This unexpected outcome might be related to the inhibition of critical negative feedback loops arising from mTORC1 (11, 12). The recent finding that mTORC2 phosphorylates and activates AKT (8) suggests that the use of rapalogs may be limited by their preferential inhibition of mTORC1. This raises the question of whether inactivation of both mTORC1 and mTORC2, may represent a more effective therapeutic strategy.
Here we report that conditional genetic inactivation of mTor in the mouse prostate, which results in the abrogation of both mTORC1 and −2 functions, does not have major consequences on post-pubescent prostate gland physiology. Conversely, mTor inactivation represses prostate tumorigenesis driven by Pten loss, resulting in a reduction of intraepithelial neoplasia at early stages and the abrogation of invasive cancer at later time points. This reduction was markedly more efficient than that obtained by treatment with the mTORC1 preferential inhibitor RAD001. In line with this data, mTor deletion markedly affects proliferating cells in vitro. Hence, this study represents a proof of principle rationale for the therapeutic use of mTOR kinase inhibitors in the treatment of human cancers.
Results
mTor deletion has a negligible effect on mouse prostate morphology and function
mTOR has been reported to play a critical role in cell growth and proliferation in early mouse embryos (13, 14), consistent with results in S. cerevisiae (15) and D. Melanogaster (16, 17). Indeed, complete inactivation of mTor in mice results in embryonic lethality at E5.5, confirming the importance of mTOR in early development (13, 14). To analyze the consequences of mTor inactivation in a fully developed adult mouse tissue, we conditionally deleted mTor in the prostate, a tissue that is exquisitely sensitive to the Akt-mTOR signaling cascade (4).
We used the Cre/loxP approach (18) and specifically inactivated mTor in the prostates of mTorloxP/loxP mice by crossing them with Probasin-Cre4 (PB-Cre4) transgenic mice. PB-Cre4 transgenic mice specifically express Cre recombinase under the control of the ARR2 Probasin promoter in the prostate epithelium post-puberty (19). For simplicity, we refer to the resulting mTorloxP/loxP;PB-Cre4 offspring as mTorpc−/−. Polymerase chain reaction (PCR) analysis revealed the presence of the mTor recombined allele specifically in the three lobes of the prostate, namely anterior prostate (AP), ventral prostate (VP), and dorsolateral prostate (DLP), and a very low amount of recombination in the seminal vesicles (Fig. 1A), consistent with what has been previously reported for Probasin driven Cre expression (20). To determine the efficiency of mTor recombination in the prostates of these mice, first we compared mTor mRNA expression in the whole prostate of the mTorpc−/− mice to that of their wild-type (WT) littermates by quantitative reverse transcription polymerase chain reaction (RT-PCR) (Fig. 1B). Although we observed a significant reduction in the expression of mTor, the extent of recombination was underestimated because of non-epithelial WT contaminating tissue, preventing an accurate measure of mTor recombination in the prostatic epithelium, the compartment that expresses Probasin-driven Cre recombinase. Therefore, we used laser capture micro-dissection (LCM) to specifically extract prostatic epithelial cells from prostate cryosections of WT, mTorpc+/−, and mTorpc−/− mice (inset in Fig. 1C). DNA was extracted from these cells, and genomic quantitative real-time PCR analysis revealed that mTor is efficiently deleted in the prostates of the mTorpc−/− mice (Fig. 1C).
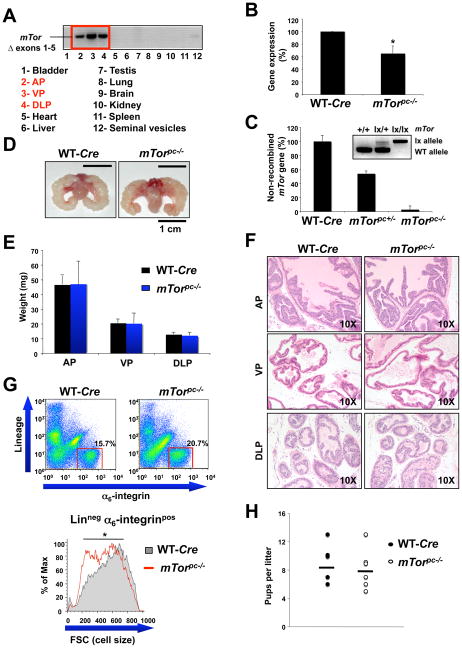
Figure 1. Effect of mTor conditional inactivation in the mouse prostate.
(A) PCR analysis to detect recombination of mTorΔ allele on DNA from several mouse tissues. AP, VP and DLP stand for anterior prostate, ventral prostate and dorsolateral prostate respectively. (B) mTor quantitative RT-PCR analysis of the prostate mRNA from WT and mTorpc−/− mice. Error bars show S.D. from three independent experiments. *, P < 0.05. (C) mTor genomic quantitative real-time PCR on DNA extracted from laser capture micro-dissected (LCM) epithelial cells of WT, mTorpc+/− and mTorpc−/− mice prostates. The inset shows the genotyping by PCR of the DNA extracted from the prostatic epithelial cells obtained by LCM and subsequently analyzed by genomic quantitative real-time PCR. (D) Representative images of prostates from mTorpc−/− and wild-type (WT) 3 month old mice. (E) Weights of prostate from mTorpc−/− and WT mice. Error bars show S.D. in the weight of 5 mice from each genotype. (F) Hematoxylin/Eosin (H&E) staining of AP, VP and DLP sections from the same mice showed in (D). (G) Upper panel: cell suspensions from mTorpc−/− and WT mouse prostates stained with lineage markers (CD31, CD45, Ter119) and for α6-integrin (CD49f). Gates indicate the Lineage negative, α6-integrin positive, basal epithelial cell population. Lower panel: representative overlay histogram of mTorpc−/− and WT basal epithelial cell Forward Scatter (FSC), an indicator of cell size. *, P < 0.05. (H) Size of the litters from WT and mTorpc−/− males at 3 months of age. There is not significant difference between the litters from WT and mTorpc−/− males.
Mice from mTorpc−/− and WT cohorts were euthanized at different time points (from 3 to 12 months of age) and their prostates extracted and characterized. We did not observe any macroscopic difference in the prostates of the mTorpc−/− mutants when compared to those of the WT age-matched controls at all time points (Fig. 1D). Additionally, no significant differences in the weights of the prostate lobes were observed (Fig. 1E). Moreover, histopathological analysis revealed that the prostates of the mTorpc−/− mice are very similar to those of age-matched WT controls and do not show any overt sign of involution or atrophy (Fig. 1F).
Functionally, the activity of the mTOR pathway is positively linked to cell size (21). We therefore sought to determine whether loss of mTor expression in the prostate of the mTorpc−/− mice resulted in a decrease of prostatic epithelial cell size. To this end, we attempted to define a discrete epithelial cell population using specific markers expressed on the cell surface. In this mouse model, the Cre recombinase needed for mTor excision is driven from the ARR2 Probasin promoter, which drives Cre expression in both the basal and luminal epithelial cell compartments of the prostate (22). The basal cell compartment of mouse prostate, which is thought to contain the epithelial progenitor cell population, expresses unique sets of both intracellular (cytokeratin5, p63) and cell surface (Sca1, α6-integrin) markers (22–24). We confirmed that the vast majority of basal cells of the AP did indeed co-express both cytokeratin5 (CK5) plus α6-integrin (Supplementary Fig. 1). Using flow cytometry to separate out this discrete population of cells we identified basal cells from single cell suspensions of prostate by staining for α6-integrin and non-epithelial, “Lineage” markers to label contaminating cells (CD31 for endothelial cells, CD45 and Ter119 for hematopoietic cells). Cell size for each population was determined from the Forward Scatter (FSC) parameter. When we compared the FSC profiles of the basal epithelial cell populations (α6-integrin positive, Lineage negative) from mTorpc−/− to that of WT controls we found that there was a minor but reproducible reduction in the cell size of the cells from mTorpc−/− mutants (average reduction in mean FSC-H of 7.5%) (Fig. 1G).
The reproductive capacity of mTorpc−/− was comparable to that of WT animals, as assessed by the size of the litters fathered by males of 3 months of age (Fig. 1H).
Overall, these data demonstrate that mTor deletion does not have a major impact on the morphology and function of the prostate, indicating the dispensable nature of mTor in a post-mitotic and non-proliferative context.
mTor deletion suppresses Pten-nul-driven prostate tumorigenesis more efficiently than does treatment with RAD001
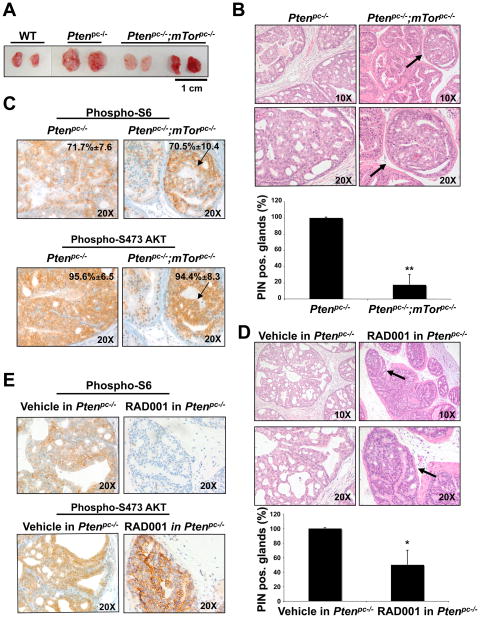
We next aimed to elucidate the effect of mTor inactivation in a mitotically active context. As noted, activation of mTOR is a central aspect of the pathway downstream of the PI3K/AKT signaling cascade, which is opposed by PTEN (25, 26). Conditional inactivation of Pten in the mouse prostate triggers prostatic intraepithelial neoplasia (PIN) that later evolves to a non-lethal invasive CaP with full penetrance at 6 months of age (27). We crossed the mTorpc−/− mice with PtenloxP/loxP;PB-Cre4 mice (Ptenpc−/−) and examined the incidence of prostate-specific lesions and invasive CaP in the resulting Ptenpc−/−;mTorpc−/− mice compared to that in age-matched Ptenpc−/− controls (n = 24 per cohort; 4 mice/time point). Three month old Ptenpc−/−;mTorpc−/− mice showed a reduction in prostate enlargement compared to Ptenpc−/− mice (Fig. 2A). Histopathological analysis of these mice revealed a marked deceleration in tumor initiation in Ptenpc−/−;mTorpc−/− mice as assessed by a decrease in the number of glands affected by PIN (14% of prostate glands analyzed were affected by PIN in Ptenpc−/−;mTorpc−/− mice versus 100% in the Ptenpc−/− mice, , 4 mice per genotype) (Fig. 2B, upper and lower panels).
Figure 2. Genetic inactivation of mTor suppresses Pten-null driven prostate tumorigenesis more efficiently than does treatment with RAD001.
(A) Representative images of prostates from 3 month old WT, Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice. (B) Upper panel: H&E staining of AP sections from 3 month old Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice. An example of residual PIN lesion in Ptenpc−/−;mTorpc−/− mouse prostate is indicated by the arrow and shown at higher magnification. Lower panel: percentage of glands affected by PIN in the prostate of 3 month old Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice. Error bars show S.D. in the number of glands affected by PIN in 4 mice from each genotype. **, P < 0.01. (C) IHC staining of PIN lesions present in the prostates of Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice showed in (B upper panel) with anti-phospho S6 and anti-phospho S473 Akt antibodies. Quantification (+/− SD) of phospho-S6 and phospho-Akt positive cells in the PIN lesions of Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice is indicated. (D) Upper panel: H&E staining of AP sections from 8 weeks old Ptenpc−/− mice treated with vehicle or RAD001 for 4 weeks. One of the residual PIN lesions in RAD001-treated Ptenpc−/− mouse prostate is indicated by the arrow and shown at higher magnification. Lower panel: percentage of glands affected by PIN in the prostate of 8 weeks old Ptenpc−/− mice treated with vehicle or RAD001 for 4 weeks. Error bars show S.D. in the number of glands affected by PIN in 4 vehicle-treated and 4 RAD001-treated Ptenpc−/− mice. *, P < 0.05. (E) IHC staining of PIN lesions present in the prostate of the Ptenpc−/− mice treated with vehicle or RAD001 shown in D upper panel) with antibodies directed against phospho-S6 and phospho S473 Akt.
We performed an immunohistochemical (IHC) analysis of mTORC1 and mTORC2 activity in PIN lesions in the prostates of Ptenpc−/−;mTorpc−/− mice, using the phosphorylated form of the ribosomal protein S6 (phospho-S6), as a read out of mTORC1 activation, and Akt phosphorylated on Ser473, as a read-out of mTORC2 activity. The percentage of phospho-AKT and phospho-S6 positive cells was indistinguishable between the PIN-affected glands of the Ptenpc−/− and the residual PIN lesions of the Ptenpc−/−;mTorpc−/− mice (Fig. 2C, upper and lower panels, respectively). Furthermore, IHC analysis for Pten showed that Pten recombination is similar in the residual PIN lesions of the Ptenpc−/−;mTorpc−/− mice compared with Ptenpc−/− mice (Supplementary Fig. 2A) whereas genomic quantitative real time-PCR for mTor on DNA extracted from LCM PIN lesions from the same mice revealed that mTor is not deleted in these lesions (Supplementary Fig. 2B). Taken together, these results suggest that the residual PIN-affected glands in the prostates of the Ptenpc−/−;mTorpc−/− mice arise from Pten-recombined cells that have escaped mTor recombination.
To compare the antitumor activity of preferential inhibition of mTORC1 relative to that of complete mTor inactivation, we treated a cohort of 4-week old Ptenpc−/− mice with RAD001 (cohorts of 4 mice per treatment). There was a much less pronounced reduction in the number of PIN positive glands in the prostates of Ptenpc−/− mice treated for 4 weeks as compared with Ptenpc−/−;mTorpc−/− mice (56% of prostate glands affected by PIN in the RAD001-treated Ptenpc−/− mice versus full penetrance in the vehicle-treated Ptenpc−/− mice, Fig. 2D). Thus, the preferential targeting of mTORC1 by RAD001 is less effective at reducing PIN lesions than is genetic inactivation of mTor, in which the function of both mTORC1 and mTORC2 is eliminated.
IHC analysis revealed a reduction in phospho-S6 staining of the residual PIN lesions of the RAD001-treated Ptenpc−/− mice compared with that of the PIN-affected glands of the vehicle- (Fig. 2E, upper panel), whereas the PIN-affected glands of the RAD001-treated Ptenpc−/− mice showed strong AKT phosphorylation on Ser473 (Fig. 2E, lower panel).
Overall, these results imply that mTORC1 inhibition is only partially effective in opposing cancer initiation driven by Pten-loss. We found that PIN lesions can develop in the absence of mTORC1 activity in the prostates of RAD001-treated Ptenpc−/− mice, as demonstrated by the lack of S6 phosphorylation in these lesions. Notably, phosphorylation of Akt on Ser473 is maintained in these lesions, indicating that mTORC2 remains active. However, in the prostates of the Ptenpc−/−;mTorpc−/− mice, PIN lesions were never found to display concomitant inactivation of mTORC1 and mTORC2, indicative of the essential nature of both mTOR multiprotein complexes for prostate tumorigenesis in the Ptenpc−/− mice.
mTor inactivation opposes Pten-loss-induced tumor progression from in situ to invasive cancer lesions
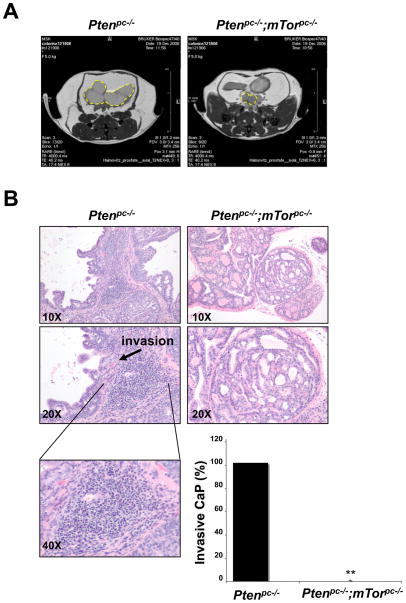
To further analyze the effect of mTor inactivation on Pten-loss driven CaP, we followed cohorts of Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice up to 12 months of age with monthly magnetic resonance imaging (MRI) analysis. MRI detected the presence of tumors masses in the prostates of 6 month old Ptenpc−/− mice [Fig. 3A, left panel and (27)]. These tumors were markedly smaller in the Ptenpc−/−;mTorpc−/− cohort than in the Ptenpc−/− mice (Fig. 3A, right panel). Moreover, histopathological analysis revealed that, as previously reported (27), at 6 months of age the Ptenpc−/− mice develop focal signs of invasive CaP at full penetrance. In contrast, prostates from mice with combined inactivation of Pten and mTor showed no signs of invasion (Fig. 3B, 4 mice per genotype). Additionally, invasive CaPs in Ptenpc−/− mutants exhibited a strong inflammatory response (Fig. 3B, lower left panel) with a marked reaction of the stroma surrounding the epithelial glands, features that were completely absent from the Ptenpc−/−;mTorpc−/− prostates (Fig. 3B). Furthermore, the Ptenpc−/−;mTorpc−/− mice only develop invasive CaP from 10 months of age and at low penetrance (20%) (4 mice per genotype were analyzed, p<0.01).
Figure 3. mTor inactivation opposes Pten-loss induced tumor progression from in situ to invasive cancer lesions.
(A) MRI analysis of prostatic tumors (dashed yellow circles) in Ptenpc−/− and Ptenpc−/−;mTorpc−/− 6 month old mice. (B) H&E staining of AP sections from 6 month old Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice. Focal invasion in Ptenpc−/− mouse prostate is indicated by an arrow. Inflammatory infiltration in Ptenpc−/− mouse prostate is shown at higher magnification in the lower left panel. Lower right panel: percentage of Ptenpc−/− and Ptenpc−/−;mTorpc−/− 6 month old mice with signs of invasion in the prostate. Four mice from each genotyping were analyzed. **, P < 0.01.
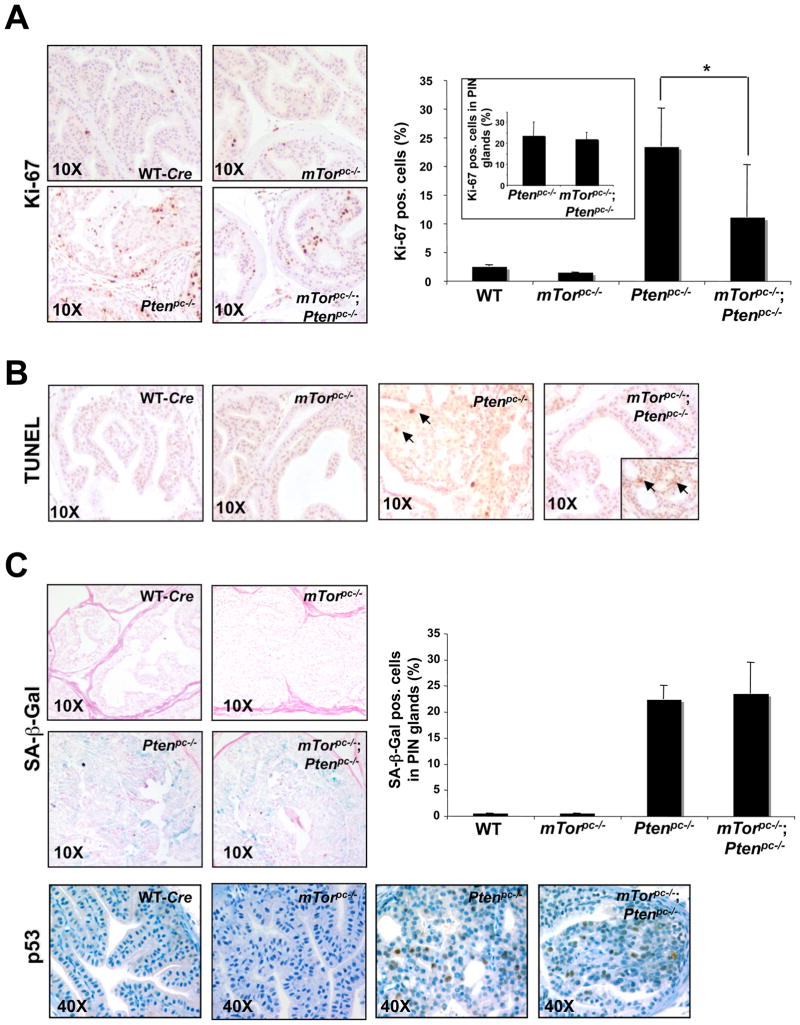
Figure 4. Biological outcome of mTor deletion in the WT and Ptenpc−/− prostate.
(A) Left panel: Ki-67 staining on prostate sections from 3 month old mice of the indicated genotypes. Right panel: quantification of Ki-67. Pos. stands for positive. Three different areas of one section were counted to determine an average and representative value for each slide. Slides from three independent mice were counted in this way to determine a standard deviation for the population. *, P < 0.05. The inset shows the Ki-67 quantification exclusively in the PIN lesions of the Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice prostates. (B) TUNEL assay for apoptosis on prostate sections from 3 month old mice of the indicated genotypes. The arrows show examples of TUNEL positive cells. The inset in the Ptenpc−/−;mTorpc−/− prostate section shows the TUNEL staining in a residual PIN lesion. (C) Upper left panel: senescence-associated β-Galactosidase staining (SA β-Gal) on prostate sections from 3 month old mice of the indicated genotypes. Upper right panel: quantification of the SA β-Gal staining in the glands affected by PIN of the prostate sections from 3 month old mice of the indicated genotypes. Quantifications were done on three representative sections from three mice. There is not significant difference between the PIN lesions of Ptenpc−/− and Ptenpc−/−;mTorpc−/− prostates. Pos. stands for positive. Lower panel: IHC for p53 on the same mice showed in the upper panel.
Together, these findings show that inactivation of both mTORC1 and mTORC2 opposes Pten-loss-induced tumor initiation and the subsequent progression from in situ to invasive cancer lesions.
mTor inactivation results in growth arrest in a mitotically-active context
Because the residual cancer lesions present in the Ptenpc−/−;mTorpc−/− prostates showed signs of having escaped mTor recombination, we hypothesized that Ptenpc−/−;mTorpc−/− double null cells lack the ability to support neoplastic transformation. We evaluated a number of different parameters in vivo to understand the fate of these cells. IHC analysis of the proliferation marker Ki-67 in prostates from Ptenpc−/−;mTorpc−/− mice showed a reduction in the overall proliferative rate compared to that in Ptenpc−/− mice (Fig. 4A). This proliferation was mainly restricted to the residual PIN lesions (note that the proliferation index for PIN lesions in prostates from both Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice is similar, as shown by the Ki-67 quantification in the inset of Fig. 4A), indicating that mTor deletion profoundly affects the ability of Pten−/− cells to proliferate and initiate tumorigenesis.
We next determined whether the Ptenpc−/−;mTorpc−/− cells that fail to proliferate engage an apoptotic response. Apoptosis was evaluated in cells in the prostates of WT, mTorpc−/−, Ptenpc−/− and Ptenpc−/−;mTorpc−/− mice using a standard TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) assay. No substantial apoptotic response was observed in the normal glands of the prostates of any of these genotypes (Fig. 4B), although a very slight incidence of apoptosis (~1%) was observed in PIN lesions, consistent with a higher cellular turn-over in the context of tumorigenesis (Fig. 4B).
Ptenpc−/− tumors display a robust p53-dependent senescence response, which opposes tumor progression (20). Abrogation of senescence, as demonstrated in the prostates of Ptenpc−/− mice that also have inactivation of Trp53, allows Pten-null tumors to achieve their full proliferative potential and drives the rapid progression of the PIN lesions to invasive CaP (20). Thus, we investigated whether cellular senescence could account for the slowed progression from in situ neoplastic lesions to invasive cancer in the prostates of the Ptenpc−/−;mTorpc−/− mice. To this end, we analyzed senescence-associated β-galactosidase (SA-β-Gal) activity (28), together with induction of p53 in the Ptenpc−/−;mTorpc−/− mice. At 3 months of age, SA-β–Gal staining in the PIN lesions of Ptenpc−/−;mTorpc−/− prostates was indistinguishable from that in the prostates of the Ptenpc−/− mice (Fig. 4C, upper left and right panels). This result is consistent with the notion that the PIN glands observed in the Ptenpc−/−;mTorpc−/− prostates retain the features of the Pten null PIN lesions since they have escaped mTor inactivation (Fig. 2C and Supplementary Fig. 2). The similar degree of senescence in both genotypes was confirmed by IHC analysis for p53 (Fig. 4C lower panel).
In summary, these data suggest that the majority of cells that experience combined Pten;mTor loss in the prostate epithelium of Ptenpc−/−;mTorpc−/− mice undergo a block in proliferation. This markedly decreases the rate of PIN initiation. The cells that lose Pten, while retaining mTor, have the propensity to proliferate more or to stochastically engage a cellular senescence response (20). This in turn opposes the progression of these PIN lesions to invasive CaP (20). The reduction in the overall number of PIN lesions in prostates from the Ptenpc−/−;mTorpc−/− mice explains why invasive lesions are virtually absent at six months of age. These PIN lesions arise from Pten null cells, which retain mTor function through escape from Cre-mediated mTor recombination. As a result, it is likely that these escaper PIN lesions maintain the potential to bypass senescence through loss of the p53 response. Indeed, consistent with this interpretation, a few invasive CaP lesions are also observed in Ptenpc−/−;mTorpc−/− mice at later time points (10 months). Because our data in vivo suggest that mTor deletion has a strong impact in a mitotically active context (Pten-loss driven neoplastic transformation), we sought to characterize the effects of acute mTor inactivation in greater detail in an in vitro model. We infected mTorloxP/loxP, PtenloxP/loxP and PtenloxP/loxP;mTorloxP/loxP mouse embryonic fibroblasts (MEF) with a Cre-PURO-IRES-GFP (Cre-Puromycin resistant-Internal Ribosome Entry Site-Green Fluorescent Protein)-encoding retrovirus (experimental timeline shown in Supplementary Fig. 3A). The efficiency of Cre-mediated loxP recombination, assessed by Western blot analysis (Fig. 5A) and genomic PCR (Supplementary Fig. 3B), showed elimination of both Pten and mTor. It should be noted that the abundance of both phospho-S6 and phospho-Akt is decreased upon mTor deletion in both the Pten-WT and Pten-recombined context, consistent with the functional loss of mTor (Fig. 5A). The remaining phospho-S6 and phospho-Akt may be due to residual mTOR protein remaining after recombination (Fig. 5A).
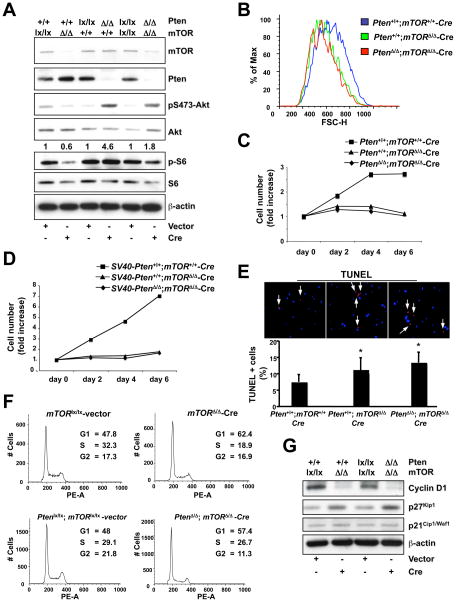
Figure 5. Biological outcome of mTor deletion in mouse embryonic fibroblasts (MEFs).
(A) Western blot analysis of lysates of mTorloxP/loxP, PtenloxP/loxP and PtenloxP/loxP;mTorloxP/loxP MEFs infected with PURO-IRES-GFP (vector) or Cre-PURO-IRES-GFP (Cre) (see experimental timeline shown in Supplementary Fig. 3A). Quantification of the phospho-Akt/total Akt ratio is shown. Quantifications were done by densitometry analysis performed with ImageJ software. For each genotyping the phopho-Akt/total Akt value of Cre-infected cells is normalized against the corresponding vector-infected control. (B) Flow cytometric analysis (Forward Scatter, FSC-H) of the mTor-null (Pten+/+;mTorΔ/Δ-Cre) and Pten;mTor-double null MEFs (PtenrΔ/Δ;mTorΔ/Δ-Cre) compared to the WT (Pten+/+;mTor+/+-Cre). (C) Cell proliferation curve analysis of the same MEFs analyzed in (B) followed over a 6-day period. (D) Cell proliferation curve analysis of mTorloxP/loxP and PtenloxP/loxP;mTorloxP/loxP primary MEFs firstly immortalized with SV40 large-T antigen, and subsequently infected with PURO-IRES-GFP (vector) or Cre-PURO-IRES-GFP (Cre). (E) TUNEL assay on the same MEFs analyzed in (B). *, P < 0.05. (F) Flow cytometric analysis of mTorloxP/loxP and PtenloxP/loxP;mTorloxP/loxP MEFs infected with PURO-IRES-GFP (vector) or Cre-PURO-IRES-GFP (Cre) (see experimental timeline shown in Supplementary Fig. 3A). In order to accurately evaluate cell cycle populations, data was gated to exclude the sub-G1 population. (G) Western blot analysis on the lysates of mTorloxP/loxP and PtenloxP/loxP;mTorloxP/loxP MEFs infected with PURO-IRES-GFP (vector) or Cre-PURO-IRES-GFP (Cre) (see experimental timeline shown in Supplementary Fig. 3A).
Flow cytometric analysis of the mTor-null (Pten+/+;mTorΔ/Δ-Cre) and mTor;Pten-double null MEFs (PtenrΔ/Δ;mTorΔ/Δ-Cre) revealed that both cell types display decreased cell size compared to WT (Pten+/+;mTor+/+-Cre) MEFs, as measured by the forward scatter parameter (Fig. 5B).
In addition, cell growth analysis revealed that mTor deletion affects the proliferative capability of MEFs both in WT (Pten+/+;mTorΔ/Δ-Cre) and Pten-null (PtenrΔ/Δ;mTorΔ/Δ-Cre) genetic backgrounds (Fig. 5C). These results corroborate our in vivo data, and highlight the fact that mTor deletion has a marked impact on mitotically active primary cells. We next immortalized primary MEFs using SV40 large-T antigen, and subsequently inactivated mTor alone or mTor and Pten together in these cells. Immortalization acts as a primary oncogenic event, which confers a strong proliferative advantage even in a scenario in which primary cells would normally undergo growth arrest. As shown in Fig. 5D, SV40-Pten+/+;mTorΔ/Δ-Cre-infected cells show complete inhibition of proliferation, indicating that mTor deletion opposes the strong proliferative potential bestowed on these cells by the SV40-Large T antigen. Moreover, even in the context of a second oncogenic hit, the inactivation of Pten, mTor deletion still acts to block proliferation (SV40-PtenΔ/Δ;mTorΔ/Δ-Cre in Fig. 5D).
We next sought analyze cell death in the Pten+/+;mTorΔ/Δ-Cre and PtenΔ/Δ;mTorΔ/Δ-Cre MEFs. TUNEL assays revealed a significant increase in the apoptotic index of Pten+/+;mTorΔ/Δ-Cre and PtenΔ/Δ;mTorΔ/Δ-Cre MEFs compared with that of WT MEFs (Fig. 5E), consistent with the idea that a prolonged growth arrest may result in an increase in cell death.
Cell cycle status was analyzed in the same cells by flow cytometric analysis (Fig. 5F) and by Western blot utilizing cell cycle-specific markers (Fig. 5G). Results from the flow cytometry revealed that mTor deletion in both the WT (Pten+/+;mTorΔ/Δ-Cre) and Pten-null (PtenΔ/Δ;mTorΔ/Δ-Cre) genetic backgrounds leads to G1-arrest (Fig. 5F). This G1-arrest is associated with a down-regulation of cyclin D1, a well-characterized cell cycle marker that is translationally regulated by mTOR (Fig. 5G) (29). An analysis of cyclin-dependent kinase inhibitors revealed that although there was no detectable change in p21Cip1/Waf1 (Fig. 5G), there was an increase in p27Kip1 abundance (new Fig. 5G). We also observed enhanced nuclear compartmentalization of p27Kip1 upon mTor deletion (Supplementary Fig. 3C) consistent with its function in regulating the cell-cycle (30).
Taken together, these results reinforce the concept that the functionality of mitotically active cells depends on the activity of mTor, whose deletion impairs tumor initiation and progression in a Pten-loss driven model of CaP. Our findings indicate that acute inactivation of mTor in vitro results in G1 growth arrest and a modest increase in apoptosis in the Pten+/+;mTorΔ/Δ-Cre and PtenΔ/Δ;mTorΔ/Δ-Cre MEFs.
Discussion
Overall, our results are consistent with a model in which the consequences of mTor deletion depend on the proliferative context of the cell. It is clear from previous knockout models that mTor plays a crucial role in the developing embryo (13, 14). Here, we show the consequences for mTor deletion in the adult prostate of the mouse. In contrast with what has been previously observed in the embryo, mTor had only minor effects on the adult mouse prostate, suggesting that mTor is dispensable in the post-mitotic setting.
However, our study of mTor inactivation in Pten-loss driven prostate tumorigenesis demonstrated the pivotal role of this protein in preserving the proliferative capability of mitotically active cells. Loss of mTor in this scenario markedly reduced the potential of Pten-null cells to initiate tumor formation. The essential requirement of mTor for tumorigenesis is also consistent with observations from mTor knock-out models, given the high mitotic index and proliferative demands of the developing embryo.
In vitro analysis in MEFs confirms the concept that inactivation of mTor impairs cell proliferation so profoundly that it cannot even be rescued by immortalization of these cells. Additionally, we show here that acute inactivation of mTor in these MEFs results in a combined G1 arrest and apoptosis.
The in vivo effect of inactivation of both mTORC1 and mTORC2 on prostate tumorigenesis driven by Pten-loss is much more pronounced than that observed upon treatment of Ptenpc−/− mice with the mTORC1 inhibitor, RAD001.
In a mouse model of PIN generated through overexpression of a constitutively active myr-AKT transgene in the prostate (31), RAD001 treatment completely reverts the PIN phenotype (32). In contrast, RAD001 treatment of our Ptenpc−/− mice, which show a more pronounced phenotype than that of the myr-AKT transgenic mice, is only partially effective. These data also emphasize that the type of murine preclinical model employed is critical for the extrapolation of the therapeutic potential of compounds studied. Furthermore, comparing results from mTORC1 inhibition by RAD001 and concomitant abrogation of mTORC1 and mTORC2 in Ptenpc−/− mice, we conclude that mTORC2 may play an essential role in prostate tumorigenesis driven by Pten-loss.
The critical role of mTORC2 in tumorigenesis is also highlighted by recent findings of Masri et al., showing that elevated mTORC2 activation, through overexpression of rictor, is observed in glioma cell lines and primary tumor cells (33).
Thus, taken these data strongly support the rationale for the development of inhibitors that target the catalytic subunit of mTOR. It is tempting to speculate that these inhibitors, through the targeting of a wider spectrum of mTOR downstream effectors, will hold added therapeutic potential over rapalogs, the first generation of mTOR inhibitors, which are currently being tested in several clinical trials. However, the differential outcome of mTOR abrogation in proliferating and non-proliferating tissues shown here warrants further studies to establish a dosage that will minimize potential toxicity of these compounds.
Materials and Methods
mTor and Pten mice
Previously generated mTorloxP/loxP and PtenloxP/loxP mice (13, 27) were crossed with the PB-Cre4 mice (19). All mouse work was performed in accordance with our IACUC approved protocol. For genotyping, tail DNA was subjected to PCR following the protocols previously described (13, 27). To detect the presence of the recombined mTorΔ allele, PCR analysis was carried out on DNA extracted from several tissues and the previously reported primers were used (13).
Quantitative mRNA RT-PCR
Total RNA was prepared from mice prostates using the Trizol method (Invitrogen). cDNA was obtained with Transcriptor (Roche). A taqman probe specific for the 5–6 exon boundary on mTor (note that exon 5 is deleted upon mTor recombination) was obtained from Applied Biosystems (Foster City, California). Amplifications were run in a 7900 Real-Time PCR System (Applied Biosystems). Each value was adjusted by using Glucuronidase B levels as reference.
Laser Capture Microdissection (LCM) and genomic quantitative real-time PCR
LCM was performed on 5 μm frozen sections using a VeritasTM Microdissection Instrument (Arcturus Bioscience Mt View, CA). Sections were cut from OCT-embedded samples on PEN Membrane Glass Slides (Arcturus Bioscience Mt View, CA), then fixed for 10 minutes in methanol, washed for 5 minutes in PBS, counterstained with Hematoxylin and dehydrated through washes in ethanol 95%, ethanol 100% and xylene. Sections were finally air-dried for 1–2 hours. LCM prostatic epithelial cells were collected on CapSure HS LCM Caps (Arcturus Bioscience Mt View, CA). DNA was extracted with QIAamp DNA Micro Kit (Qiagen) according to the manufacturer instructions. Quantitative real-time PCR was performed on a Roche LightCycler using the Quantitect SYBR Green PCR kit (Qiagen) and the following amplification protocol: 15 min at 95°C, 40 cycles (15 sec at 94°C - 20 sec at 57°C - 20 sec at 72°C) followed by determination/confirmation of amplicon melting temperature. Reactions were performed in triplicates, primer pairs were confirmed to yield a single amplicon band by 3% agarose gel electrophoresis. The following mouse primer set was used to amplify specifically a 185bp amplicon on exon 2 of mTor, which is deleted upon recombination: forward primer TCTGTGCACATCTTCCTTGC, reverse primer TGCTACCAGAGGCTGTCCTT. The following primers for β-actin were used as reference standards: forward primer GGCTGTATTCCCCTCCATCG, reverse primer CCAGTTGGTAACAATGCCATGT.
Flow cytometry
To generate a single cell suspension from mouse prostate, the lobes (AP VP, DLP) were dissected, minced, and cells extracted essentially as described (34). After passing sequentially through 70μm and 40μm cell strainers to remove debris and cell clumps, cells were resuspended in phosphate buffered saline (PBS) supplemented with 2% fetal bovine serum (FBS). Cells were then incubated with the following antibodies: biotin conjugated anti-CD31 (BD Bioscience), CD45, and Ter119 (eBioscience) plus Alexa647 conjugated anti-CD49f (α6 integrin, clone GoH3; Biolegend). Cells were washed and then incubated in Phycoerythrin conjugated streptavidin for the detection of biotin labeled antibodies. DAPI was used for live-dead discrimination and stained cells were analyzed on an LSRII (BD).
Western blot analysis and immunohistochemistry
Cell lysates were prepared with RIPA buffer [1 × PBS, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche)] and cleaned by centrifugation. The following antibodies were used for Western blot analysis: mouse anti-β-actin (AC-72; Sigma), anti- mTOR (Cell Signaling Technology), anti-Pten (Cell Signaling Technology), anti-cyclin D1 (BD Pharmingen), anti-cyclin B1 (Cell Signaling Technology), anti-p21 (C-19; Santa Cruz) and anti-p27 (Upstate).
For immunohistochemistry, prostate tissues were fixed in 10% neutral-buffered formalin (Sigma) overnight, subsequently washed once with PBS, transferred into 50% ethanol and stored in 70% ethanol. Prostate lobes were embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) in accordance with standard procedures. Sections were stained with the following antibodies: anti-phospho-S6 (S235/S236) (Cell Signaling Technology); anti-phospho-Akt (S473) (Cell Signaling Technology); anti-Pten (Ab-2; NeoMarkers), Ki-67 (Novacastra), (Nanotools), and anti-p53 (FL-393; Santa Cruz).
Treatment with RAD001
RAD001 (everolimus) was obtained from Novartis and was administered by oral gavage at 10mg/kg/day. The treatment started when the mice were 4 weeks old, prior to PIN onset, and maintained for 4 weeks.
MRI
Individual mice were subjected to MRI analysis to detect the presence of prostatic tumors. The analysis was carried out as previously described (20, 27).
TUNEL assay and senescence detection in vivo
For TUNEL assay, sections were pretreated with proteinase K (Sigma Aldrich). After that, the sections were incubated with Terminal transferase (TdT, Roche Diagnostics) and biotion 16-dUTP (Roche Diagnostics). Detection was performed with HRP-Streptavidin conjugated (1:100) and developed with DAB Peroxidase Substrate kit (SK-100, Vector lab). At the end, the slides were counterstained with Hematoxylin and mounted in permount. For detection of senescence in vivo, prostates were frozen in OTC and frozen sections 5 μm thick were stained for β-Galactosidase activity using the Senescence Detection Kit (Calbiochem). Quantifications were performed on three representative sections from three mice for each genotype.
MEFs generation, infection and analysis
Primary Mouse Embryonic Fibroblasts (MEFs) were prepared as described previously (35) and infected with a retrovirus expressing Cre-PURO-IRES-GFP or the corresponding empty control. Virus production and MEF infection were performed as previously described (20). Two days post-selection, cells were plated for growth curves, flow cytometric analysis, TUNEL and western blot as reported (20). TUNEL analysis was performed using the in situ cell death detection Kit (Roche). To obtain cell cycle profiles by flow cytometry, cells were trypsinized, harvested, and fixed and permeabilized in ice-cold 70% ethanol. After incubation on ice for 30 minutes or overnight at −20°C, fixed cells were washed twice with PBS and resuspended in Propidium iodide/RNase solution (BD Bioscience). Cells were kept at 4°C overnight prior to analysis on a BD LSRII flow cytometer.
Statistical analysis
Statistical evaluations were carried out using SigmaStat 2.03 (SPSS). For all analysis the level of statistical significance was set at p < 0.05 (indicated with one asterisk on the graphs) or p < 0.01 (indicated with two asterisks on the graphs). Unpaired student T-test was performed in order to determine the statistical significance.
Supplementary Material
Acknowledgments
We thank Rosa Bernardi, Min Sup Song and all the members of the Pandolfi laboratory for discussion and comments. We are grateful to the BIDMC pathology core for technical expertise in antibody characterization and immunohistochemistry. We are grateful for the MRI imaging work by MR small animal imaging core at MSKCC, which is partially supported by SAIRP (R24CA83084) and NIH Center Grant (P30 CA08748). We thank Novartis for RAD001. C.N. was supported in part by a fellowship from the American-Italian Cancer Foundation. A.C. is supported by an EMBO long-term fellowship. This work is supported by NIH/NCI grants (U01 CA-84292; R01 CA-82328) to P.P.P.
Footnotes
Authors’ contributions: C.N., A.C., A.A., R.M.H and P.P.P. designed and conceived the experiments. C.N., A.C., A.A., R.M.H. and Z.C. performed the experiments. C.N., J.G.C., A.C. and P.P.P. wrote the manuscript.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science (New York, NY. 1991;253:905–9. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 3.Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–46. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- 4.Majumder PK, Sellers WR. Akt-regulated pathways in prostate cancer. Oncogene. 2005;24:7465–74. doi: 10.1038/sj.onc.1209096. [DOI] [PubMed] [Google Scholar]
- 5.Bhaskar PT, Hay N. The two TORCs and Akt. Developmental cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Gray IC, Stewart LM, Phillips SM, Hamilton JA, Gray NE, Watson GJ, Spurr NK, Snary D. Mutation and expression analysis of the putative prostate tumour-suppressor gene PTEN. British journal of cancer. 1998;78:1296–300. doi: 10.1038/bjc.1998.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5246–50. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science (New York, NY. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 9.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–88. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 11.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and crosstalks. Oncogene. 2008;27:5527–41. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 13.Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Molecular and cellular biology. 2004;24:9508–16. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Molecular and cellular biology. 2004;24:6710–8. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Molecular biology of the cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science (New York, NY. 1999;285:2126–9. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes & development. 2000;14:2712–24. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nature reviews. 2002;2:251–65. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mechanisms of development. 2001;101:61–9. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–71. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Kasper S, Yuan J, Jin RJ, Zhang J, Ishii K, Wills ML, Hayward SW, Matusik RJ. Androgen-dependent prostate epithelial cell selection by targeting ARR(2)PBneo to the LPB-Tag model of prostate cancer. Laboratory investigation; a journal of technical methods and pathology. 2006;86:1074–88. doi: 10.1038/labinvest.3700463. [DOI] [PubMed] [Google Scholar]
- 23.Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, Wilson EL. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7180–5. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:181–6. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carracedo A, Salmena L, Pandolfi PP. SnapShot: PTEN signaling pathways. Cell. 2008;133:550 e1. doi: 10.1016/j.cell.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, Cordon-Cardo C, Pandolfi PP. Pten dose dictates cancer progression in the prostate. PLoS biology. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–22. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 30.Borriello A, Cucciolla V, Oliva A, Zappia V, Della Ragione F. p27Kip1 metabolism: a fascinating labyrinth. Cell Cycle. 2007;6:1053–61. doi: 10.4161/cc.6.9.4142. [DOI] [PubMed] [Google Scholar]
- 31.Majumder PK, Yeh JJ, George DJ, Febbo PG, Kum J, Xue Q, Bikoff R, Ma H, Kantoff PW, Golub TR, Loda M, Sellers WR. Prostate intraepithelial neoplasia induced by prostate restricted Akt activation: the MPAKT model. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7841–6. doi: 10.1073/pnas.1232229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, Loda M, Lane HA, Sellers WR. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 33.Masri J, Bernath A, Martin J, Jo OD, Vartanian R, Funk A, Gera J. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res. 2007;67:11712–20. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- 34.Barclay WW, Cramer SD. Culture of mouse prostatic epithelial cells from genetically engineered mice. The Prostate. 2005;63:291–8. doi: 10.1002/pros.20193. [DOI] [PubMed] [Google Scholar]
- 35.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.