Abstract
Background
The cell surface undergoes continuous change during cell movement. This is characterized by transient protrusion and partial or complete retraction of microspikes, filopodia, and lamellipodia. This requires a dynamic actin cytoskeleton, moesin, components of Rho-mediated signal pathways, rearrangement of membrane constituents and the formation of focal adhesion sites. While the immunofluorescence distribution of endogenous moesin is that of a membrane-bound molecule with marked enhancement in some but not all microextensions, the C-terminal fragment of moesin co-distributes with filamentous actin consistent with its actin-binding activity. By taking advantage of this property we studied the spontaneous protrusive activity of live NIH3T3 cells, expressing a fusion of GFP and the C-terminal domain of moesin.
Results
C-moesin-GFP localized to stress fibers and was enriched in actively protruding cellular regions such as filopodia or lamellipodia. This localization was reversibly affected by cytochalasin D. Multiple types of cytoskeletal rearrangements were observed that occurred independent of each other in adjacent regions of the cell surface. Assembly and disassembly of actin filaments occurred repeatedly within the same space and was correlated with either membrane protrusion and retraction, or no change in shape when microextensions were adherent.
Conclusions
Shape alone provided an inadequate criterion for distinguishing between retraction fibers and advancing, retracting or stable filopodia. Fluorescence imaging of C-moesin-GFP, however, paralleled the rapid and dynamic changes of the actin cytoskeleton in microextensions. Regional regulatory control is implicated because opposite changes occurred in close proximity and presumably independent of each other. This new and sensitive tool should be useful for investigating mechanisms of localized actin dynamics in the cell cortex.
Background
Lamellipodia, filopodia, retraction fibers and microspikes are dynamic and often transient membraneous structures on the surface of most cells. They can readily be observed in spreading, moving and dividing cultured cells, but also in migrating cells during development and inflammation, or in invading cancer cells in vivo. Recent evidence suggests that small GTPases of the rho family regulate this protrusive cell surface activity [1,2]. Using a permeabilized Swiss3T3 cell system, Mackay et al. [3] have shown recently that moesin, and possibly its relatives ezrin or radixin, are necessary for cellular responses induced by rho, namely the formation of lamellipodia, focal adhesion complexes and stress fibers in serum-starved fibroblasts. One or more members of this protein family is also required for the formation of filopodia in growth cones of neuronal cells [4], but how moesin interacts with the actin cytoskeleton during the dynamic restructuring of the cell cortex has not been entirely resolved.
One suggestion has been that moesin needs to be in an "activated" form [5,6], a form capable to interact with actin filaments and to link filaments to sites in the plasma membrane. Recently, Nakamura et al. [7,8] have deduced from in vitro co-sedimentation experiments with cellular forms of moesin that phosphatidylinositides and phosphorylation of a single threonine residue together are needed to convert moesin from an inactive protein to one that binds F-actin. Similarly, substitution of threonine 558 with aspartate simulated phosphorylation and activated F-actin binding of a recombinant form of moesin. An allosteric change has been proposed as the mechanism for this activation [8,9], since de-phosphorylation with a specific phosphatase obliterates binding [10]. When expressed in cells, epitope-tagged versions of the C-terminal domain of ezrin co-distribute with stress fibers in fixed cells [11]. This suggests that this peptide fused to GFP could serve as a probe for the imaging of dynamic changes of the actin cytoskeleton and, at the same time, of protrusive activity in live cells [12].
Results
Moesin is a major component of delicate microextensions of the cell surface of NIH3T3 cells where it is frequently co-localized by immunofluorescence with ezrin and radixin [12,13,14,15]. Filopodia-like microextensions can be defined by video microscopy as actively growing and freely motile filopodia, as attached structures that do not move and that remain stable in length, or as structures that are retracting and become shorter. Immunofluorescence analysis of fixed cells does not differentiate between these different forms of microextensions. Furthermore, this technique shows that moesin and F-actin are not uniformly distributed and not even detectable in some instances in these structures (our unpublished results). To substantiate these findings in live cells and to determine the reason for the variation in F-actin content, the present study was undertaken.
C-moesin-GFP Binds to Actin Filaments Without Disrupting Microfilament Organization or Cell Behavior
To study changes in the actin cytoskeleton in microextensions of living cells required a new approach. We utilized the C-terminal domain of moesin fused to GFP for imaging, since the structure contains a high affinity binding site for F-actin and since previous results had shown co-distribution of this domain with filamentous actin [11,13,16,17]. When expressed in NIH3T3 cells, C-moesin-GFP most prominently decorated stress fibers regardless of level of expression (Figure 1b, 1d) and regardless of whether GFP was positioned at the N- or C-terminus of the C-moesin fragment. As expected, double-staining of fixed, C-moesin-GFP transfected cells with TRITC-phalloidin yielded precisely overlapping images. The intracellular structures visualized by C-moesin-GFP also included bundles and networks of F-actin in lamellipodia. This pattern could be reproduced by staining with rhodamine-labeled phalloidin. Without any doubt, this distribution was quite distinct from the membrane distribution of full-length moesin-GFP or N-moesin-GFP fusion proteins and from the immunofluorescence localization of the endogenous protein in untransfected control cells [13].
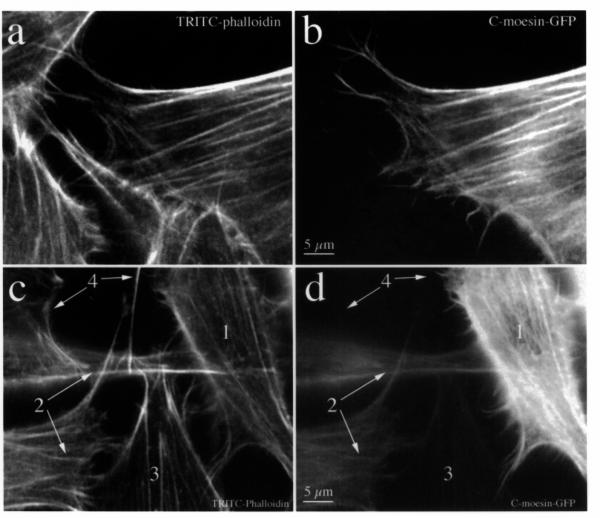
Figure 1.
C-moesin-GFP is co-localized with the microfilament cytoskeleton. Fields containing transfected and untransfected cells were imaged after staining with TRITC-phalloidin (a, c) and compared with images obtained by fluorescence of the same group of cells expressing C-moesin-GFP (b, d). The transfected cell in (b) is attached to several other untransfected cells. Its fluorescence pattern matches filopodia and stress fibers, and is identical in distribution to microfilaments stained with TRITC-phalloidin (a). Cells expressing different levels of C-moesin-GFP are seen within the same field (d, 1-4). The distribution of C-moesin-GFP is the same as that of TRITC-phalloidin (c), regardless of level of expression.
Side-by-side comparison of cells either expressing or not expressing C-moesin-GFP did not reveal obvious differences in overall shape, behaviour and surface activity (our unpublished data). Expression of the fusion protein also did not affect quality or intensity of TRITC-phalloidin staining of the cells when compared to adjacent untransfected cells. This implied that C-moesin-GFP does not compete with phalloidin for binding sites and that, at least during the observation period, microfilaments and stress fibers apparently assembled normally in the presence of C-moesin-GFP. A similar distribution to that of C-moesin was observed with corresponding fusion proteins of ezrin or radixin consistent with the structurally conserved F-actin binding sites of these fragments (Figure 2).
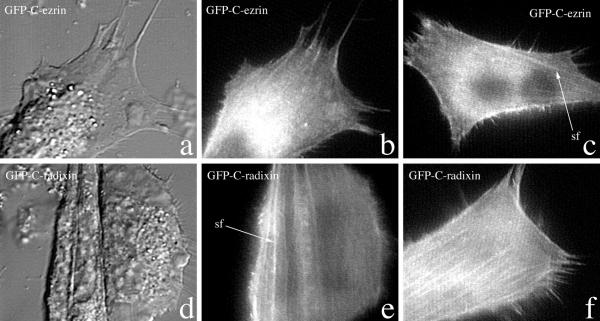
Figure 2.
Comparison of cells expressing the C-terminal domains of moesin, ezrin and radixin. NIH3T3 cells were transfected with C-terminal domain-GFP fusion proteins of ezrin and radixin As with moesin, no effect on cell behavior is seen 6 hours after transfection (a,b) Similar to C-moesin and consistent with identical amino acid sequences of the F-actin-binding region, both C-ezrin and C-radixin bind to actin filaments in stress fibers (sf) (b,c,e,f) and numerous microextensions. This localization is sensitive to cytochalasin D.
C-moesin-GFP as a Probe to Study Filopodial Microfilaments
By viewing cells for periods of up to several hours, and by comparing time-lapse video images obtained by DIC with fluorescence images we concluded that the intracellular distribution of C-moesin-GFP reflected the distribution of actin filaments that closely abut the plasma membrane. It is for this reason that the changes in fluorescence intensity and shape parallel the many discrete changes in cell surface architecture that were observed by DIC microscopy. F-actin-containing cellular structures could be monitored with a spatial resolution in the order of 200 nm, and a temporal resolution of 3-5 seconds even without the most sensitive camera (Figure 3).
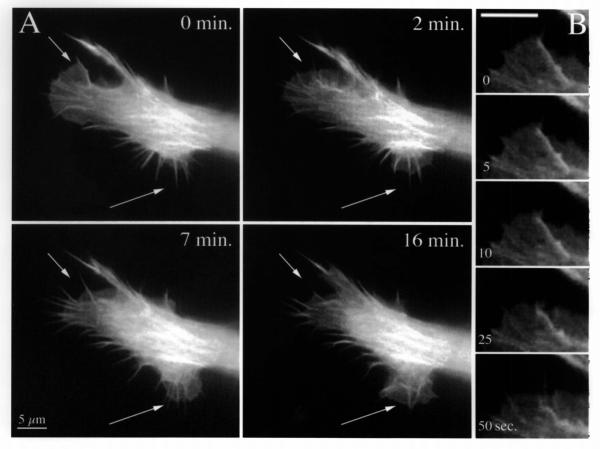
Figure 3.
The distribution of C-moesin-GFP parallels dynamic changes in the actin cytoskeleton. In (A), a live, fluorescent cell was imaged for 16 minutes. Note retraction of a lamellipodium (top arrow) and advance of another (lower arrow), as the cell changes direction and begins to migrate towards the bottom of the page. The retracting lamellipodium from (A) is shown at higher magnification and temporal resolution in (B). The fifty second sequence starts after the first panel of (A) and ends before the second panel. Note kinking, buckling and withdrawal of a rib-like microfilament bundle within the lamella. See also three related movies
An example of a moving pseudopodium, a large cell extension with advancing and retracting lamellae, is presented in Figure 3A, and an example of a smaller cortical region during retraction of a lamella is shown in Figure 3B. Multiple discrete changes occur in this relatively small area of the cell even during the short observation period. In the sequence shown in Figure 3A, the pseudopodium begins to alter its direction of migration. The first frame identifies a small lamella encompassing several ribbed filaments and a thinner pseudopodium terminating in a retraction fiber on the left, and a number of retraction fibers at the bottom edge. Multiple thick filament bundles can be seen within the body of the main pseudopodium. In the 16 minute sequence, the left hand lamella retracted, converting its ribs into retraction fibers. The lamella could be observed withdrawing into the body of the pseudopodium, while maintaining the same fluorescence intensity. The rate of lamellar withdrawal measured over different areas was 1.7 ± 0.5 μm/min.
By two minutes of observation, the cytoskeleton of the structure near the upper arrow in Figure 3A has collapsed and multiple kinks became apparent. A sequence of this process with higher temporal resolution is shown in Figure 3B. The sequence, showing bending and collapse of the microfilament bundle, began 30 seconds after the first panel of Figure 3A, and ended 50 seconds later. Seven minutes after this filopodium began to collapse, fluorescence accumulated in a new filopodium near the upper arrow and 9 minutes later this has disappeared again.
The main portion of the pseudopodium showed complex changes in its cytoskeletal fluorescence as some areas decreased in intensity and changed shape, while the tip maintained the same level of fluorescence. The thick bundles in the center of the main pseudopodium remained relatively constant during this time period. In contrast, the area at the lower edge of the cell began to protrude. Forward moving lamellar veils enveloped short filopodia by advancing at a rate of 0.85-1.5 μm/minute and the latter became ribs within the lamella. By 16 minutes, some of these were observed to bend upwards and to detach from the substrate, collapsing backwards into the cell.
A close-up look at another cell edge is shown in Figure 4A to illustrate that, even within a small area of the cell surface, occupying only a few microns, multiple, independent and simultaneous changes in cytoskeletal architecture could be visualized. The arrowheads in Figure 4A point to the same spot in all frames. The left-most arrowhead illustrates rapid lamellar advance followed by partial retraction and ruffling. The second and third arrowheads point to a filopodial microfilament core that advanced 4.6 μm within 10 minutes. Growth of this filopodial cytoskeleton (at 2nd arrowhead) was plotted over time and the graphic representation showed that elongation was not continuous but oscillatory (Figure 4B). The maximum rate of growth was 1.74 μm/min, but the overall rate of growth over 21 minutes was only 0.4 μm/min. Although there was considerable variation, similar growth rates and similar oscillatory behavior were observed for numerous other filopodia regardless of whether cells expressed C-moesin-GFP or not. The right most arrow head points to a second filopodial bundle that elongated and then retracted, illustrating that within this 8 μm stretch of the cellular edge at least three independent types of cytoskeletal rearrangements were taking place simultaneously.
Figure 4.
Multiple, simultaneous, and independent rearrangements of the microextension cytoskeleton are visualized by C-moesin-GFP. A 10 minute time-lapse sequence of the edge of a live, fluorescent cell is shown at high magnification in (A). The arrowheads point to the same spot in each time-frame. In (B), the growth of the middle filopodium from (A) was followed over time (17 minutes) and the changes in length/time are presented in this graph. See also three related movies
To investigate whether the shape of microextensions depended on microfilament content, we performed parallel analysis by comparing DIC images with fluorescence images of their cytoskeletal cores. As shown in Figure 5, when a microextension was attached to the substrate, its cytoskeletal core could be withdrawn and microfilaments could reenter the membranous sheath without necessarily inducing retraction or growth of the microextension. This obviously was not the case for all microextensions, since in growing and motile filopodia the fluorescence signal matched their shape very closely indicating that actin filaments filled a large amount of their cytoplasmic space.
Figure 5.
DIC and fluorescence time-lapse imaging of microextensions in live C-moesin-GFP transfected cells. C-moesin-GFP fluorescence is seen to withdraw from and to re-enter an attached microextension (arrows). Note that the fluorescence intensities of C-moesin-GFP in two closely spaced microextensions, and hence their actin filament contents, are independent of each other. This is illustrated on the right side of the figure by showing intensity distributions at three different points (indicated by lines B (for Base), M (for middle) and T (for Tip) in the four panel at the very bottom) along the two filopodia. See also three related movies
Cytochalasin D Changes the Distribution of C-moesin-GFP
The distribution of C-moesin-GFP appears to reflect rapid changes in microfilament organization in the moving cell edge. We, therefore, expected this distribution to be influenced by cytochalasin D. To investigate this, cells were treated with varying concentrations of cytochalasin D and imaged for different time periods before, during, and after exposure to the drug. Within seconds after addition of cytochalasin D, protrusive activity at cellular edges stopped as seen both by DIC and by fluorescence imaging. As time progressed, C-moesin-GFP fluorescence began to fade in membrane extensions that were attached to the substrate in a distal-to-proximal direction (Figure 6). This fading was incomplete and fluorescence remained associated with short segments and intracellular dots that varied in size and signal intensity that could be stained with phalloidin. Stress fibers also began to disappear, but this required more time than the disruption of microfilaments in microextensions. Bundles of the original stress fibers could still be recognized in a few cells 60 minutes after initiation of cytochalasin D treatment. The example in Figure 6A shows the cellular morphology by DIC and the distribution of C-moesin-GFP fluorescence in a cell treated with 20 μM cytochalasin D for 60 minutes. Other transfected cells showed qualitatively similar responses, but the rate at which dissolution of the cytoskeleton occurred varied.
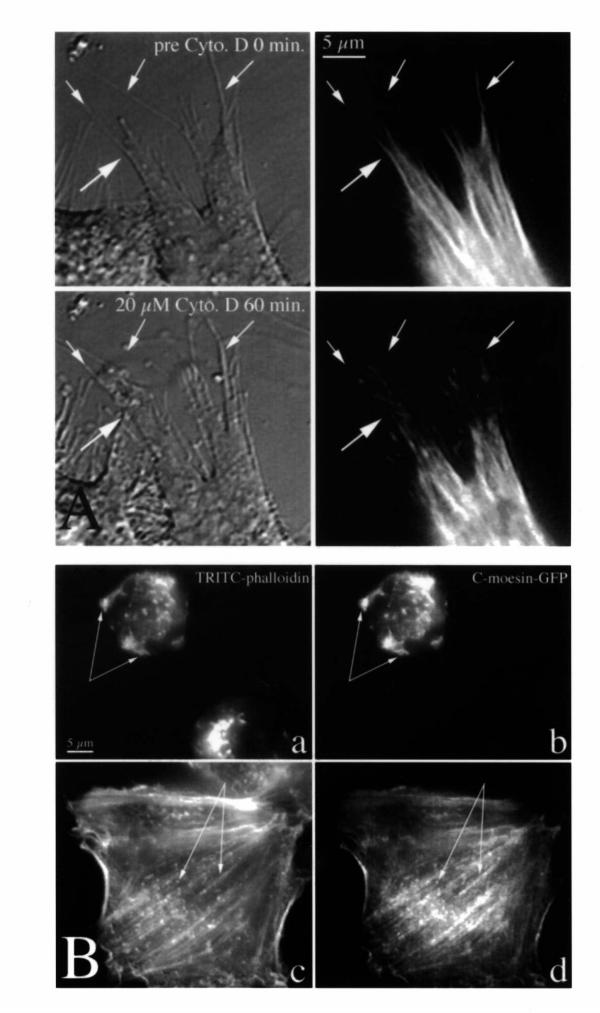
Figure 6.
C-moesin-GFP does not interfere with microfilament rearrangements during and after treatment with cytochalasin D. In (A), a transfected cell was imaged live by DIC (left) and C-moesin-GFP fluorescence (right) during treatment with cytochalasin D. The large arrow points at changes within one pseudopodium. The small arrows point to filopodia and retraction fibers. Note withdrawal and clumping of C-moesin-GFP fluorescence. In (B), cells were treated for 30 minutes with 20 μM cytochalasin D (a, b) and then fixed, or treated for 20 minutes and then allowed to recover for 1 hour after drug washout (c, d) before fixation. They were then stained with TRITC-phalloidin (a, c) and imaged for C-moesin-GFP fluorescence (b, d). Arrows point to identical spots in parallel images.
To test the effects of C-moesin-GFP on microfilament stability and assembly in vivo, we treated cells expressing C-moesin-GFP with cytochalasin D for different periods of time, stained the cells with TRITC-phalloidin, and scored transfected and untransfected cells for absence or presence of stress fibers. The example in Figure 6B (panels a & b) shows marked disruption of microfilament organization, but the distribution of TRITC-phalloidin and C-moesin-GFP was identical in both transfected and untransfected cells. The percentage of cells lacking stress fibers was the same for transfected (97%) and untransfected cells (95.5%) suggesting that C-moesin-GFP did not significantly protect microfilaments from the effects of cytochalasin D.
We also examined whether the presence of C-moesin-GFP interfered with recovery of microfilament structures after removal of cytochalasin D. One hour after drug washout, the cells expressing C-moesin-GFP showed a pattern of cytoskeletal organization that could not be distinguished from that of untransfected control cells (Figure 6B, panels c, d). The rate of recovery may have varied, however, since transfected cells tended to have thinner retraction fibers at that time.
Discussion
A General Method for Definition of Microextensions and for Analysis of Dynamic Changes in the Actin Cytoskeleton
The changes in cell surface topography and movement of cell surface microextensions in isolated cultured cells cannot easily be captured by images of fixed cells. To appreciate the many changes in shape and in the actin cytoskeleton that take place simultaneously, even within a relatively small area of the cell surface and within short time spans time lapse recording of live cells is necessary. This method clearly visualizes the dynamic nature of changes virtually over the entire cell surface the entire of the cell surface consisting of protrusion and retraction of microspikes, filopodial and lamellipodial structures. These changes occur continuously and do not appear to be synchronized [18]. At any given time, the cells appear irregularly shaped, with microspikes, broad lamellae, filopodia and retraction fibers extending from the main body towards the substrate, as well as others protruding from their dorsal cell surfaces. These protrusions are difficult to differentiate from one another functionally on the basis of shape alone, but time-lapse video microscopy at a focal plane near the surface of the culture dish allowed us to distinguish between growing and motile, or stable and retracting microextensions. We refer to advancing, motile and elongated microextensions as filopodia, and to fixed and static structures as retraction fibers. In the recent literature, such protrusions are summarily and often interchangeably described as filopodia, microvilli, retraction fibers, or pseudopodia without regard to potential structural or functional differences [19,20]. This deficiency is particularly evident, when comparing immunofluorescence results from different laboratories, because of the complex architecture of the cell surface and the multitude of shapes of cells of different types and from different organisms.
GFP was chosen as the fluorescent tag for imaging, because we found immunofluorescence of fixed cells to inadequately convey cytoskeletal dynamics of living cells and because of limited stability of the actin cytoskeleton during fixation and staining. The tagging of GFP has been successfully applied to a large number of cytoskeletal proteins [21]. Previous studies with ezrin-GFP showed its localization in ruffles and the leading edge of pseudopodia [22]. Relevant to our work were also experiments in D. melanogaster with the C-terminal fragment of a moesin-related protein fused to GFP [23]. The coding region was placed under the control of the hsp70 promoter for high level induction by heat shock of embryos. By Western blot flies with two copies of the transgene expressed detectable protein by 90 min, but 1-2 hours were required for fluorescence detection. Expression of the C-terminal domain had two early effects: 1) Upon induction, flies became paralyzed, but they soon recovered. This was thought to be caused by a temporary mechanical disruption of neuronal structures. 2) Long membrane processes appeared on specific cell surfaces within 2 hours after induction, but it could not be determined whether C-moesin stimulated growth of new structures, or whether it accumulated in normal structures that were difficult to discern by other means. Most importantly, however, the development of the embryo proceeded normally. This suggested that, at least in Drosophila, the C-moesin-GFP fusion protein did not interfere with cellular functions. In recent fibroblast experiments, however, Shaw et al. [16] noted that the C-domain was co-localized with stress fibers by immunofluorescence in unstimulated cells, but in response to the constitutively active form of Rho, RhoAV14, abnormally long and apical processes were formed. In insect cells, overexpression of the C-domain of ezrin enhanced cell adhesion and elicited membrane spreading that was accompanied by microspike and lamelliopodial extension and the formation of unusual, microtubule-containing thin processes, up to 200 μm in length [24]. Such unusual microextensions were not observed in our experiments.
The capability for direct analysis of living cells has significant and important advantages over immunofluorescence techniques. Results do not depend on exposed or available epitopes for antibody detection, and imaging of live cells is more reliable, since loss of fragile microextensions does not become an issue. We have observed such loss by direct microscopic observation of cells during "on stage" fixation and staining procedures for retrospective immunofluorescence analysis and have found that we could monitor, but not prevent such loss (our unpublished observations).
Cytoskeletal Dynamics in Transient Microextensions
The intracellular distribution of C-moesin-GFP and imaging of stress fibers and microextensions with this probe depended on filamentous actin as shown by the disruption of the normal pattern of F-actin in subcortex and stress fibers with the drug cytochalasin D. The changes faithfully reproduce what is typically seen by staining cells with phalloidin, namely withdrawal of actin from microextension and clumping within the cytoplasm. In our retrospective double-staining experiments we saw precise correspondence between the C-moesin-GFP fluorescence signal in the living cell and phalloidin in the same cell before, during, and after drug treatment. This strongly suggested that phalloidin and C-moesin do not compete for binding and probably occupy different binding sites on the filament.
Given the central importance of microfilament dynamics for cell movement, many attempts have been made to study actin rearrangements in vivo. Previous observations have mostly dealt with actin dynamics in lamellipodia using fluorescent analogues of actin and caged resorufin actin that were either microinjected or introduced into permeabilized cells [25,26]. These studies have shown that actin monomers are added at the distal edge of lamellipodia, probably contributing to the protrusive force at the cellular edge, and that actin filaments of the cortex are treadmilling or are in constant centripetal flux [27,28]. Filopodia and retraction fibers were rarely imaged, either because of choice of cell type, or because of difficulties in incorporating sufficient amount of labeled actin into sparse filaments of growing microextensions. More recent work with GFP-actin fusions, however, indicates that this probe is incorporated into filopodial tips [29].
Our images obtained with C-moesin-GFP indicate that cycles of microfilament protrusion and retraction occur in stationary retraction fibers, as well as in protruding and retracting microextensions. This may occur by actin polymerization or forward and backward movement of filaments, as has been observed in photoactivation experiments of fluorescent labeled actin in retraction fibers of spreading postmitotic PtK2 cells [30,31]. The rate of protrusion and retraction of lamellipodia and filopodia has been measured by many authors and it varied in different cell types [18,28,32,33,34]. The classical studies by Abercrombie et al. [32] reported measurements of fibroblast lamellar growth ranging from stationary to 1.8 μm/min. These authors also first documented that movement of the cell edge is oscillatory and is accomplished by advance and retraction of thin lamellae. The same phenomenon was observed in the movement of growth cones [35,36]. Our measurements of the rates of advance and retraction of a few microextensions determined from the C-moesin-GFP images are in accord with many of these earlier observations. This implies that the C-moesin-GFP distribution fairly accurately reflects the organization of the actin cytoskeleton and that it does not interfere with filament functions in filopodia. It also suggests that C-moesin-GFP may provide a sensitive new tool for studying spatial and temporal control mechanisms that regulate the actin cytoskeleton and its interactions with the plasma membrane in small segments of the cell cortex. The Rho family-Rho-GDI system (37,38), Ca(2+) (39,40) and phosphatidyl 4,5-biphosphate (41) are prime candidates for driving cellular processes by filamentous actin. Although as yet unknown, it is quite likely that filopodia play an important role in signaling and motility of fibroblasts similar to their function in neurite outgrowth.
Conclusions
Imaging of live NIH3T3 cells expressing the C-terminal F-actin binding domain of moesin fused to GFP before, during and after treatment with cytochalasin D, and retrospective analysis with fluorescent phalloidin are consistent with a pattern of actin microfilaments in different regions of the cells. The high sensitivity of this method allowed us to analyze dynamic and diverse changes that occur spontaneously in small areas of the cell surface and to distinguish microextensions according to their F-actin content, motility and life history. C-moesin-GFP may provide a sensitive new tool to study critical regulatory steps required to support the highly dynamic interactions between different cytoskeleton and membrane components, and to unravel spatial and temporal relationships.
Materials and Methods
Recombinant DNA Constructs
Rat C-moesin cDNA (amino acid residues 382-557) was prepared as a PCR product with flanking EcoR I sites and introduced into the EcoR I sites of the expression vector pCR™3-GFPmut2 [13,42]. The cloning resulted in the addition of 7 amino acid residues (RIRSRIP) at the junction between the moesin and GFP sequence. Human C-ezrin-GFP (amino acid residues 310-568) was constructed by ligating a GFP-encoding Hind III-Xma I fragment into pCR™3-ezrin that had been digested with Hind III and Xma I to delete the N-terminal portion of ezrin. Human C-radixin-GFP (amino acid residues 408-583) was made by ligating a GFP-encoding Kpn I-Pst I fragment into pCR™3-radixin that had been digested with Kpn I and Pst I to delete the N-terminal portion of radixin. The sequences of all the constructs were verified using the ABI PRISM™ DYE Terminator Cycle Sequencing ready reaction Kit (Perkin Elmer, Norwalk, CT). Plasmid DNA was prepared from Qiagen columns according to the manufacturer's instructions (Qiagen Inc., Chatsworth, CA).
Cell Culture and Electroporation
NIH3T3 cells were grown in Dulbecco's minimum essential medium (DMEM) supplemented with 10% fetal calf serum (growth medium), at 37°C, under 5% CO2 atmosphere. One day prior to transfection the cells were split and plated to reach 80-90% confluency on the following day. Cells were harvested and resuspended in growth medium at a concentration of 1.5-2 × 107 cells / ml. Two hundred microliters of cell suspension and 10 μg of plasmid DNA in 50 μl of phosphate buffered saline (PBS), pH 7.4, were mixed in a 0.4 cm electroporation cuvette (BioRad, Richmond, CA). Cells and DNA were incubated together for 10 minutes at room temperature prior to electroporation. Electroporation was carried out at 960 μF, 300 V and a resistance of 100 Ω (750 V/cm) using the BioRad Gene Pulser apparatus with capacitance extender. These conditions gave a time constant of 39-42. Immediately after electroporation cells were resuspended in growth medium and plated onto either glass coverslips (2.5 × 104 cells/ cm2) for microscopic analysis, or onto 3.5 cm Falcon plastic culture dishes (Beckton Dickinson, Lincoln, NJ) for biochemical analysis.
Antibodies and Reagents
Affinity purified polyclonal moesin specific rabbit antibodies (pAbMo) were isolated from crude pAsMoER serum by affinity chromatography as described [12]. pAbMo and polyclonal antibodies specific for ezrin (pAsE) and radixin (pAsR) were used for immunofluorescence and immunoblotting [12,14]. For double immunofluorescence experiments, chicken antibodies specific for moesin (ChG1, kindly provided by W. Lankes, Berlin, Germany) and FITC-conjugated goat anti-chicken antibodies were used. The GFP polyclonal antibody reagent was from Clontech Laboratories, Inc. (Palo Alto, CA). Horseradish peroxidase (HRPO)-conjugated goat anti rabbit antibodies were obtained from Boehringer Mannheim Biochemicals (Indianapolis, IN). TRITC (tetramethylrhodamine isothiocyanate)-labeled phalloidin and rhodamine-conjugated goat anti rabbit antibodies were from Jackson Immunochemicals (West Grove, PA).
Digitally Enhanced Video Differential Interference Contrast (DEV-DIC) microscopy of Live Cells
Prior to microscopic analysis, transfected cells were allowed to attach and spread on glass coverslips for 20 minutes to 6 hours in a CO2 tissue culture incubator. Most experiments were carried out between 4-7 hours after transfection. The coverslips were transferred to a perfusion chamber on the microscope stage that held two coverslips, separated by 200 μm spacers, and that allowed for exchange of medium and other reagents. The cells were viewed on a Zeiss Axiovert 35 inverted microscope with a Zeiss Plan-Apochromat 100× oil immersion lens and a short distance condenser. The microscope is also equipped with a heated stage, differential interference contrast (DIC) optics and epifluorescence. Filters and lightpaths were controlled with a filterwheel and shutters (Ludl Electronic Products Ltd., Hawthorne, NY). For GFP visualization, a single band excitation filter for FITC was used in combination with Pinkel#1 beamsplitter and emission filter (Chroma Technology Corp., Brattleboro, VT). Tissue culture medium without phenol red was kept warm and buffered in a CO2 incubator. The medium was replaced every 5 minutes through the perfusion chamber, and the stage temperature was kept at 37°C with an automatic thermostat. Images were collected using a B&W C2400 CCD camera with on-chip integration and an Argus 20 digital image processor (Hamamatsu Photonics, Japan).
Image Processing
For DEV-DIC images, the background from the optical system was subtracted, and the contrast enhanced with Argus 20 processor functions. On-chip-integration, background subtraction of white noise, and frame averaging was done for digital enhancement of fluorescence recordings. To assemble the figures, selected frames from the video tape were digitized on to a PowerTower™ 225 computer using a Miro video digitizing board at its highest resolution. The images were assembled into composites using Adobe Photoshop™4.0. Prints were made on a sublimation printer at 200 dpi resolution. Short digital movies corresponding to the figures were made using NIH Image™.
Supplementary Material
These movie files can be viewed with Quick Time, downloadable for free at http://www.apple.com/quicktime/download/
Acknowledgments
Acknowledgements
This work was supported by funds from the US Public Health Service 1RO1-41045, the Tobacco-Related Disease Research Program Grant 4TR-0316, Training Grants CA09302 and 2T32GM07365 (MRA), and partial fellowship support from the Janet M. Shamberger Fund to PL. We thank Laiqiang Huang for discussions and for providing some of the cDNA clones.
Pninit Litman and Manuel Ricardo Amieva contributed equally to this work.
Abbreviations
BSA, bovine serum albumin; C-terminal, carboxyterminal; DMEM, Dulbecco's minimal essential medium; DEV-DIC, digitally enhanced video-differential interference contrast; GFP, green fluorescent protein; N-terminal, aminoterminal; PBS, phosphate buffered saline; PCR, polymerase chain reaction; SDS PAGE, sodium dodecyl sulfate polyacrylamide electrophoresis; TRITC, tetramethyl rhodamine isothiocyanate.
Contributor Information
Pninit Litman, Email: pninit@peptor.co.il.
Manuel Ricardo Amieva, Email: amieva@leland.stanford.edu.
Heinz Furthmayr, Email: hfurthmayr@leland.stanford.edu.
References
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Mackay DJG, Esch F, Furthmayr H, Hall A. Rho- and rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ERM proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglini G, Kunda P, Quiroga S, Kosik K, Caceres A. Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J Cell Biol. 1998;143:443–455. doi: 10.1083/jcb.143.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita Sa, Tsukita Sh. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Amieva MR, Furthmayr H. Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem. 1995;270:31377–31385. doi: 10.1074/jbc.270.20.12319. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Huang L, Pestonjamasp K, Luna EJ, Furthmayr H. Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphoinositides. Mol Biol Cell. 1999;10:669–685. doi: 10.1091/mbc.10.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wong TYW, Lin RCC, Furthmayr H. Replacement of threonine 558, a critical site of phosphorylation of moesin in vivo, with aspartate activates F-actin binding of moesin. Regulation by conformational change. J Biol Chem. 1999;274:12803–12810. doi: 10.1074/jbc.274.18.12803. [DOI] [PubMed] [Google Scholar]
- Hishiya A, Ohnishi M, Tamura S, Nakamura F. Protein phosphatase 2C inactivates F-actin binding of platelet moesin. J Biol Chem. 1999;274:26705–26712. doi: 10.1074/jbc.274.38.26705. [DOI] [PubMed] [Google Scholar]
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva MR, Furthmayr H. Subcellular localization of moesin in dynamic filopodia, retraction fibers, and other structures involved in substrate exploration, attachment, and cell-cell contacts. Exp Cell Res. 1995;219:180–196. doi: 10.1006/excr.1995.1218. [DOI] [PubMed] [Google Scholar]
- Amieva MR, Litman P, Huang L, Ichimaru E, Furthmayr H. Disruption of dynamic cell surface architecture of NIH3T3 fibroblasts by the N-terminal domains of moesin and ezrin: in vivo imaging with GFP fusion proteins. J Cell Sci. 1999;112:111–125. doi: 10.1242/jcs.112.1.111. [DOI] [PubMed] [Google Scholar]
- Amieva MR, Wilgenbus KK, Furthmayr H. Radixin is a component of hepatocyte microvilli in situ. Exp Cell Res. 1994;210:140–144. doi: 10.1006/excr.1994.1021. [DOI] [PubMed] [Google Scholar]
- Henry MD, Gonzalez Agosti C, Solomon F. Molecular dissection of radixin: distinct and interdependent functions of the amino- and carboxy-terminal domains. J Cell Biol. 1995;129:1007–1022. doi: 10.1083/jcb.129.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Henry M, Solomon F, Jacks T. RhoA-dependent phosphorylation and relocalization of ERM proteins into apical membrane/actin protrusions in fibroblasts. Mol Biol Cell. 1998;9:403–419. doi: 10.1091/mbc.9.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestonjamasp K, Amieva MR, Strassel CP, Nauseef WM, Furthmayr H, Luna EJ. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol Biol Cell. 1995;6:247–259. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht-Buehler G, Lancaster RM. A quantitative description of the extension and retraction of surface protrusions in spreading 3T3 mouse fibroblasts. J Cell Biol. 1976;71:370–382. doi: 10.1083/jcb.71.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J. Are all pseudopods created equal ? Cell Motil Cytoskel. 1992;22:1–6. doi: 10.1002/cm.970220102. [DOI] [PubMed] [Google Scholar]
- Oshiro N, Fukata Y, Kaibuchi K. Phosphorylation of moesin by rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J Biol Chem. 1998;273:34663–34666. doi: 10.1074/jbc.273.52.34663. [DOI] [PubMed] [Google Scholar]
- Ludin B, Matus A. GFP illuminates the cytoskeleton. Trends Cell Biol. 1998;8:72–77. doi: 10.1016/S0962-8924(97)01169-0. [DOI] [PubMed] [Google Scholar]
- Lamb RF, Ozanne BW, Roy C, McGarry L, Stipp C, Mangeat P, Jay DG. Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr Biol. 1997;7:682–688. doi: 10.1016/S0960-9822(06)00295-8. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Martin M, Andreoli C, Sahuquet A, Montcourrier P, Algrain M, Mangeat P. Ezrin NH2-terminal domain inhibits the cell extension activity of the COOH-terminal domain. J Cell Biol. 1995;128:1081–1093. doi: 10.1083/jcb.128.6.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Hirokawa N. Actin dynamics in growth cones. J Neurosci. 1991;11:1918–1929. doi: 10.1523/JNEUROSCI.11-07-01918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons MH, Mitchison TJ. Control of actin polymerization in live and permeabilized fibroblasts. J Cell Biol. 1991;114:503–513. doi: 10.1083/jcb.114.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Comparison of actin and cell surface dynamics in motile fibroblasts. J Cell Biol. 1992;119:367–377. doi: 10.1083/jcb.119.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choida A, Jungbluth A, Sechi A, Murphy J, Ullrich A, Marriott G. The suitability and application of a GFP-actin fusion protein for long-term imaging of the organization and dynamics of the cytoskeleton in mammalian cells. Eur J Cell Biol. 1998;77:81–90. doi: 10.1016/S0171-9335(98)80075-7. [DOI] [PubMed] [Google Scholar]
- Cramer LP. Molecular mechanism of actin-dependent lamellipodia of motile cells. Front Biosci. 1997;2:D260–D270. doi: 10.2741/a189. [DOI] [PubMed] [Google Scholar]
- Cramer L, Mitchison TJ. Moving and stationary actin filaments are involved in spreading of postmitotic PtK2 cells. J Cell Biol. 1993;122:833–843. doi: 10.1083/jcb.122.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abercrombie M, Heaysman JE, Pegrum SM. The locomotion of fibroblasts in culture. I. Movements of the leading edge. Exp Cell Res. 1970;59:393–398. doi: 10.1016/0014-4827(70)90646-4. [DOI] [PubMed] [Google Scholar]
- Argiro V, Bunge MB, Johnson MI. A quantitative study of growth cone filopodial extension. J Neurosci Res. 1985;13:149–162. doi: 10.1002/jnr.490130111. [DOI] [PubMed] [Google Scholar]
- Felder S, Elson EL. Mechanics of fibroblast locomotion: quantitative analysis of forces and motions at the leading lamellas of fibroblasts. J Cell Biol. 1990;111:2513–2526. doi: 10.1083/jcb.111.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D, Chapman K. Analysis of microspike movements on the neuronal growth cone. J Neurosci. 1985;5:3204–3213. doi: 10.1523/JNEUROSCI.05-12-03204.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum-Finney B, Reichardt LF. Vinculin-deficient PC12 cell lines extend unstable lamellipodia and filopodia and have a reduced rate of neurite outgrowth. J Cell Biol. 1994;127:1071–1084. doi: 10.1083/jcb.127.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJG, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Takai Y. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem Biophys Res Commun. 1998;245:641–645. doi: 10.1006/bbrc.1998.8253. [DOI] [PubMed] [Google Scholar]
- Welnhofer EA, Zhao L, Cohan CS. Calcium influx alters actin bundle dynamics and retrograde flow in Helisoma growth cones. J Neurosci. 1999;19:7971–7982. doi: 10.1523/JNEUROSCI.19-18-07971.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Zucker RS, Bentley D. Induction of filopodia by direct local elevation of intracellular calcium ion concentration. J Cell Biol. 1999;145:1265–1275. doi: 10.1083/jcb.145.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidyl 4,5-biphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/S0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
These movie files can be viewed with Quick Time, downloadable for free at http://www.apple.com/quicktime/download/